Lactobacillus Sps in Reducing the Risk of Diabetes in High-Fat Diet-Induced Diabetic Mice by Modulating the Gut Microbiome and Inhibiting Key Digestive Enzymes Associated with Diabetes
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Culture of Bacterial Strains
2.2. Preparation of SEL
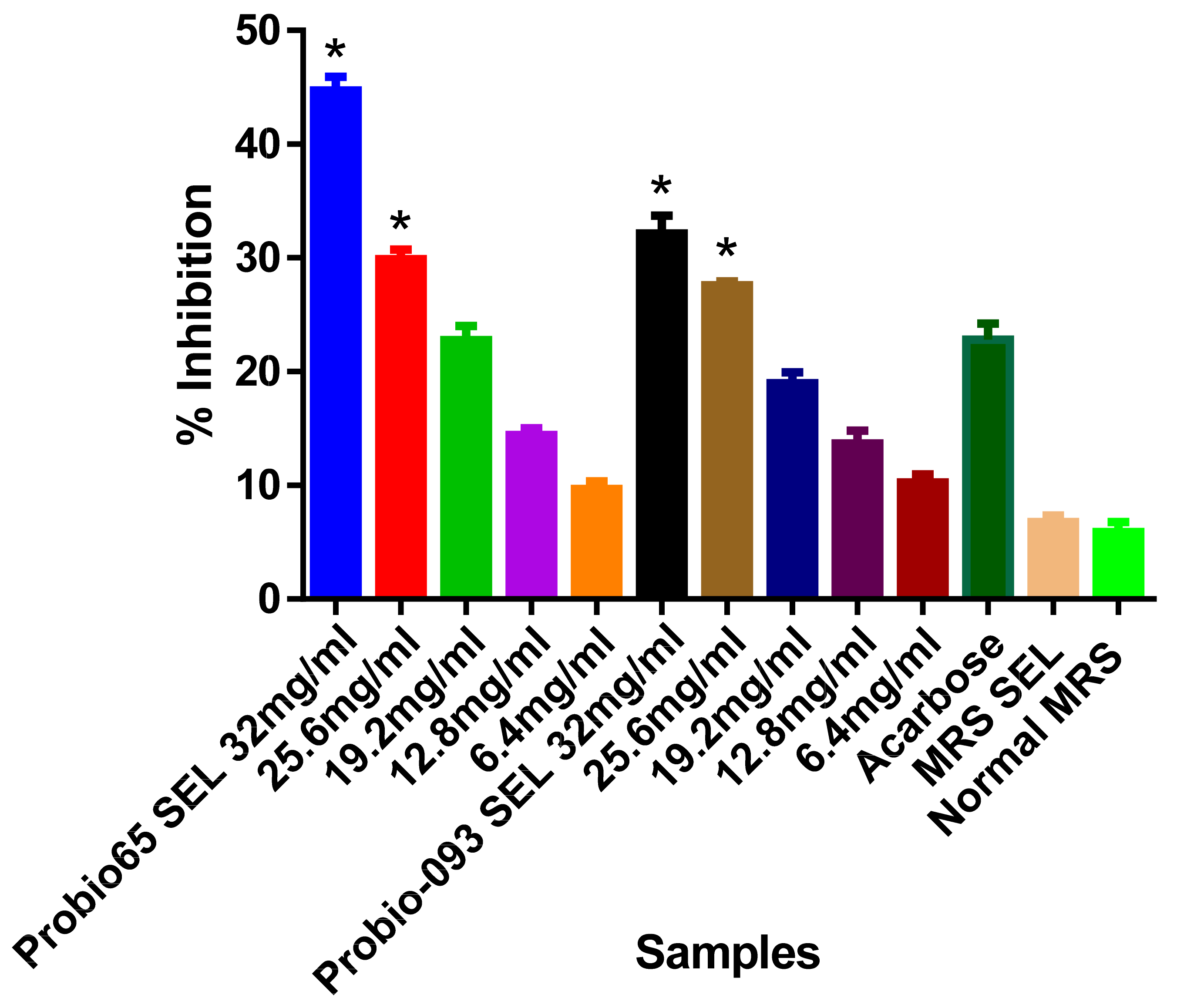
2.3. α-Amylase Inhibition
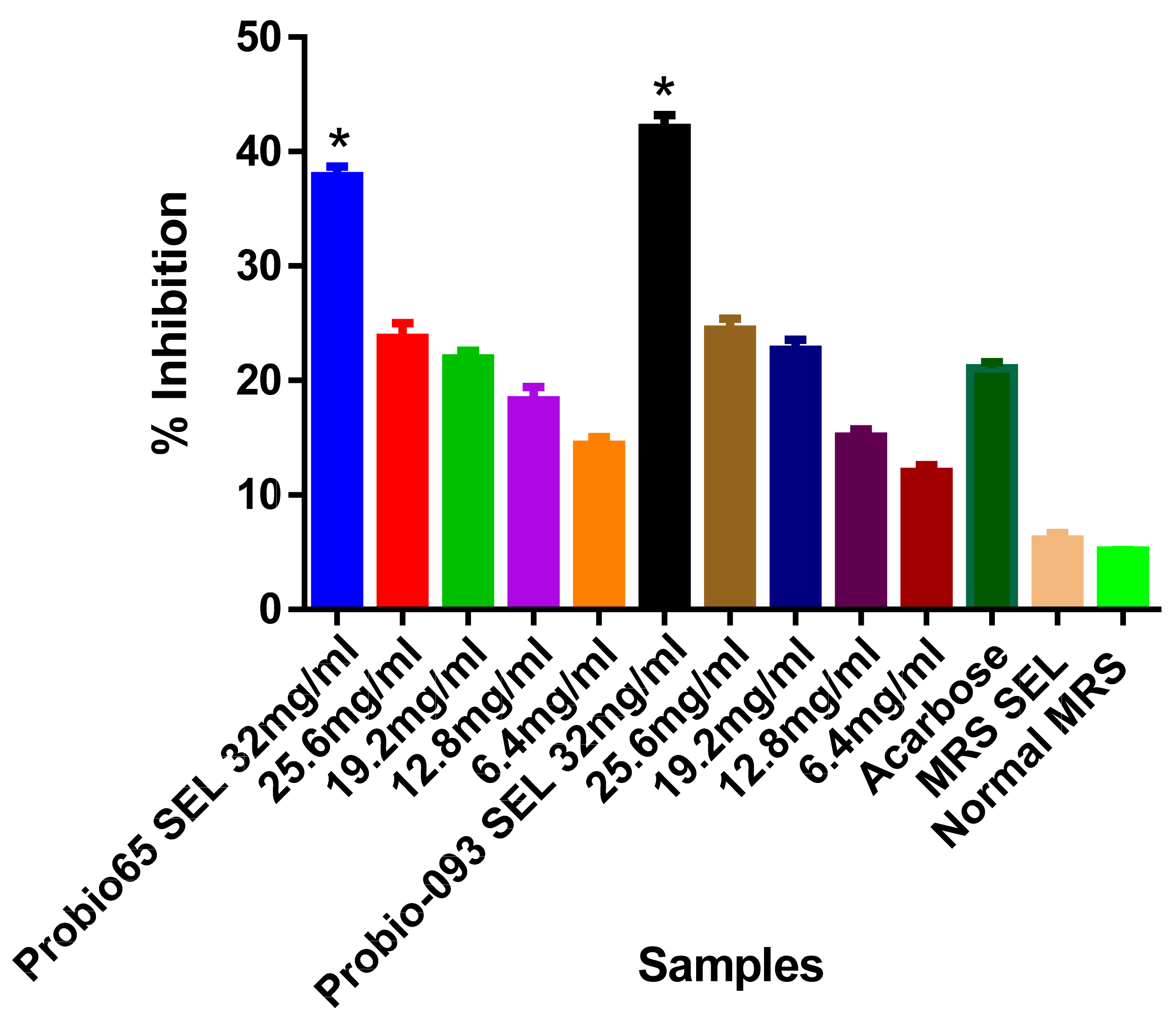
2.4. α-Glucosidase Inhibition
2.5. Animal Model and Diet
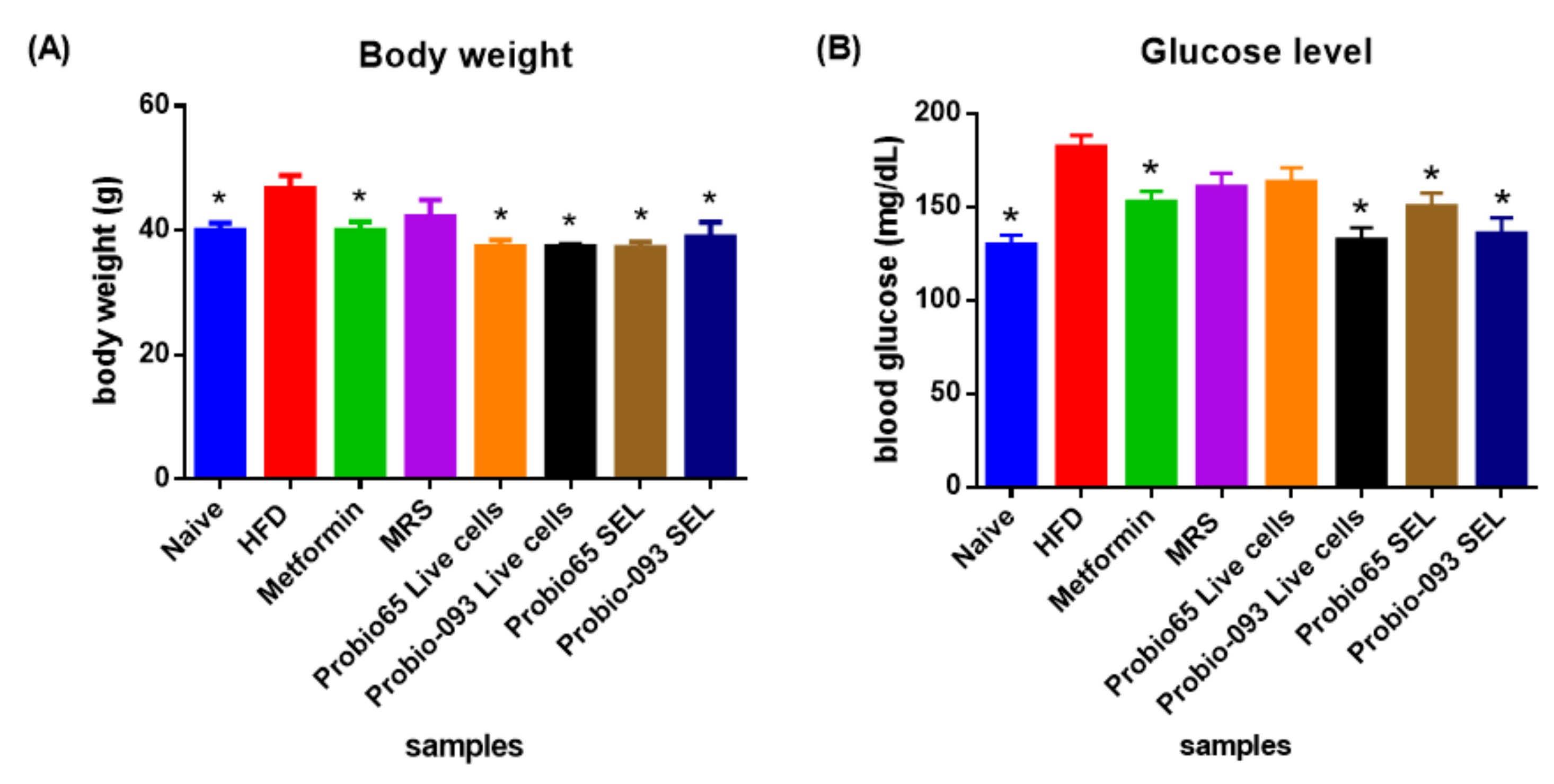
2.6. Bodyweight and Glucose Measurement
2.7. DNA Extraction from Fecal Pellets
2.8. Microbiome Analysis Using 16S rRNA Gene Target Primers with RT qPCR
2.9. Primers Efficiencies
2.10. Statistical Analysis
3. Results
3.1. Enzyme Inhibition Assays
3.2. Bodyweight and Glucose Level
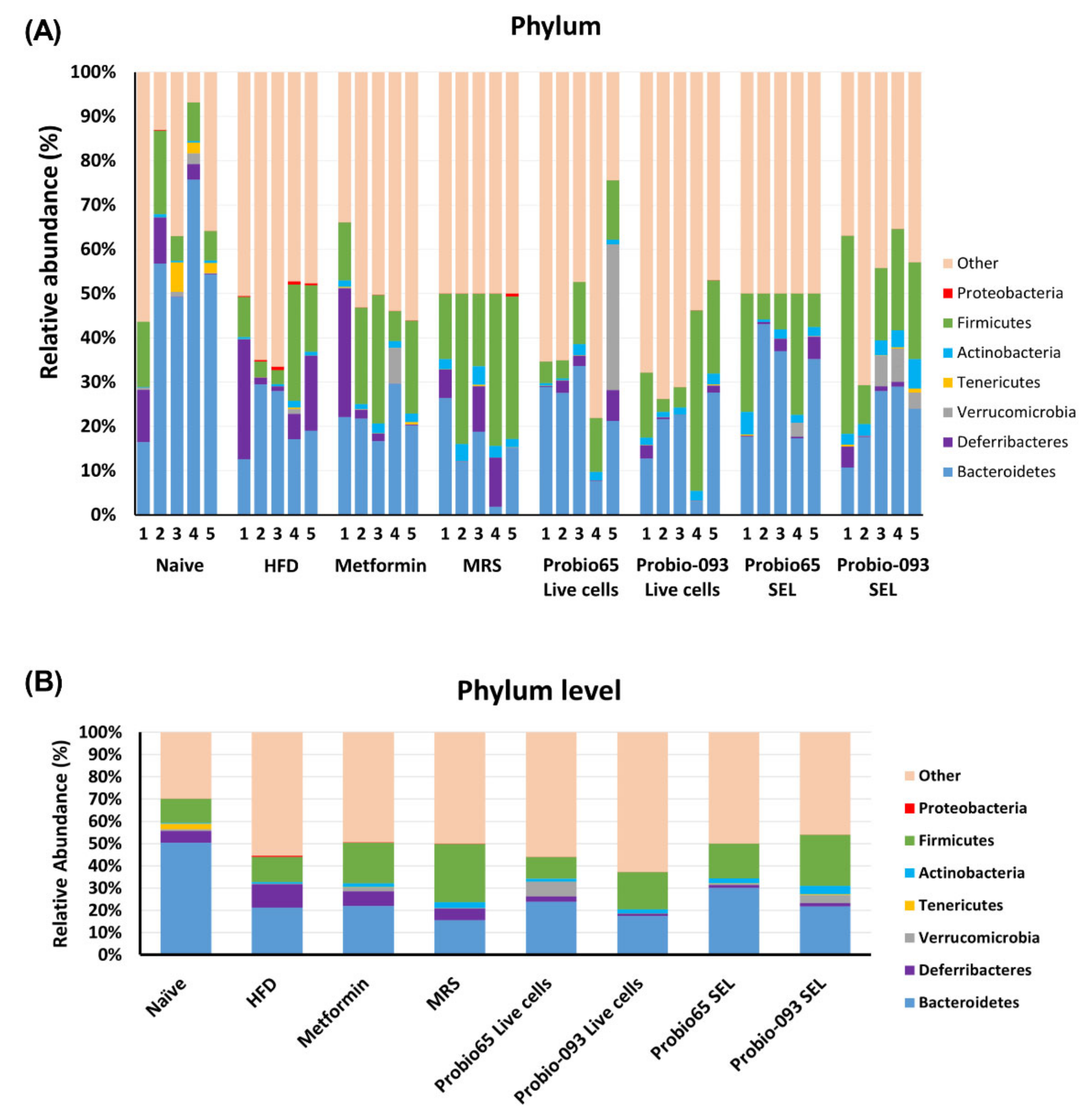
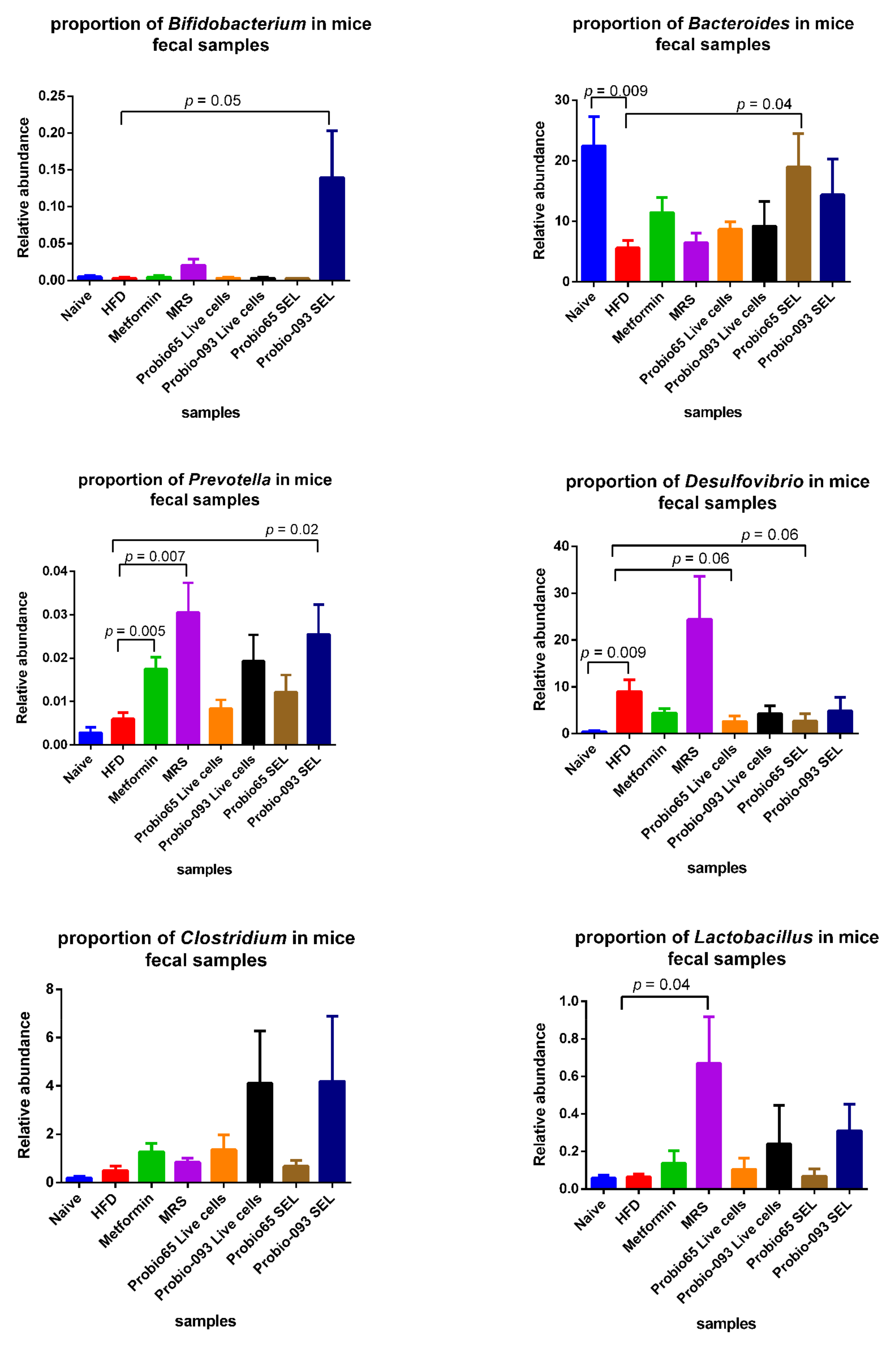
3.3. Gut Microbiota Composition at the Phylum Level
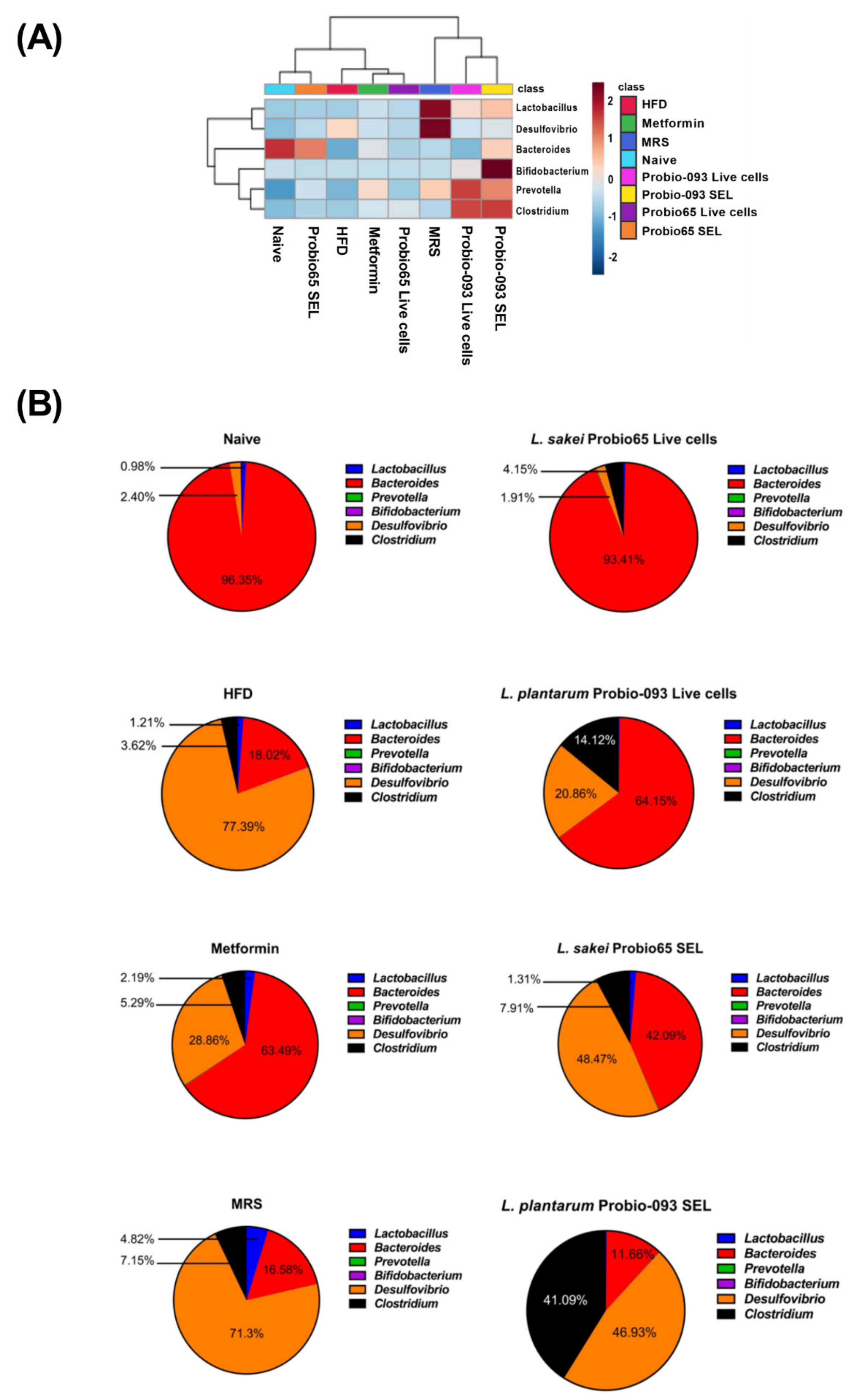
3.4. Microbiota Composition at the Genus Level
3.5. Microbiota Composition at the Genus Level into Graph Arrangements
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Surwit, R.S.; Kuhn, C.M.; Cochrane, C.; McCubbin, J.A.; Feinglos, M.N. Diet-lnduced Type II Diabetesin C57BL/6J Mice. J. Am. Diabetes Assoc. 1988, 37, 1163–1167. [Google Scholar] [CrossRef]
- Winzell, M.S.; Ahren, B. The high-fat diet-fed mouse: A model for studying mechanisms and treatment of impaired glucose tolerance and type 2 diabetes. Diabetes 2004, 53, S215–S219. [Google Scholar] [CrossRef]
- King, A.J.F. The use of animal models in diabetes research. Br. J. Pharmacol. 2012, 166, 877–894. [Google Scholar] [CrossRef] [PubMed]
- Sato, A.; Kawano, H.; Notsu, T.; Ohta, M.; Nakakuki, M.; Mizuguchi, K.; Itoh, M.; Suganami, T.; Ogawa, Y. Antiobesity effect of eicosapentaenoic acid in high-fat/high-sucrose diet-induced obesity: Importance of hepatic lipogenesis. Diabetes 2010, 59, 2495–2504. [Google Scholar] [CrossRef] [PubMed]
- Bailes, B.K. Diabetes mellitus and its chronic complications. AORN J. 2002, 76, 266–276, 278. [Google Scholar] [CrossRef] [PubMed]
- Standl, E.; Schnell, O. Alpha-glucosidase inhibitors 2012—Cardiovascular considerations and trial evaluation. Diab. Vasc. Dis. Res. 2012, 9, 163–169. [Google Scholar] [CrossRef]
- Balfour, J.A.; McTavish, D. Acarbose. Drugs 1993, 46, 1025–1054. [Google Scholar] [CrossRef] [PubMed]
- Ademiluyi, A.O.; Oboh, G. Aqueous extracts of Roselle (Hibiscus sabdariffa Linn.) varieties inhibit α-amylase and α-glucosidase activities in vitro. J. Med. Food 2013, 16, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Devaraj, S.; Yip, Y.M.; Panda, P.; Ong, L.L.; Wong, P.W.K.; Zhang, D.; Judeh, Z. Cinnamoyl Sucrose Esters as Alpha Glucosidase Inhibitors for the Treatment of Diabetes. Molecules 2021, 26, 469. [Google Scholar] [CrossRef] [PubMed]
- Chiasson, J.L.; Josse, R.G.; Gomis, R.; Hanefeld, M.; Karasik, A.; Laakso, M.; STOP-NIDDM Trail Research Group. Acarbose for prevention of type 2 diabetes mellitus: The STOP-NIDDM randomised trial. Lancet 2002, 359, 2072–2077. [Google Scholar] [CrossRef] [PubMed]
- Barlow, G.M.; Yu, A.; Mathur, R. Role of the Gut Microbiome in Obesity and Diabetes Mellitus. Nutr. Clin. Pract. 2015, 30, 787–797. [Google Scholar] [CrossRef] [PubMed]
- Gravitz, L. Microbiome: The critters within. Nature 2012, 485, 12–13. [Google Scholar] [CrossRef] [PubMed]
- Tilg, H.; Kaser, A. Gut microbiome, obesity, and metabolic dysfunction. J. Clin. Investig. 2011, 121, 2126–2132. [Google Scholar] [CrossRef]
- Faith, J.J.; McNulty, N.P.; Rey, F.E.; Gordon, J.I. Predicting a human gut microbiota’s response to diet in gnotobiotic mice. Science 2011, 333, 101–104. [Google Scholar] [CrossRef]
- Jandhyala, S.M.; Talukdar, R.; Subramanyam, C.; Vuyyuru, H.; Sasikala, M.; Nageshwar Reddy, D. Role of the normal gut microbiota. World J. Gastroenterol. 2015, 21, 8787–8803. [Google Scholar] [CrossRef]
- Sommer, F.; Bäckhed, F. The gut microbiota—Masters of host development and physiology. Nat. Rev. Microbiol. 2013, 11, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, D.G.; Quigley, B.M. The y-intercept of the critical power function as a measure of anaerobic work capacity. Ergonomics 1991, 34, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Ostaff, M.J.; Stange, E.F.; Wehkamp, J. Antimicrobial peptides and gut microbiota in homeostasis and pathology. EMBO Mol. Med. 2013, 5, 1465–1483. [Google Scholar] [CrossRef]
- Ramakrishna, B.S. Role of the gut microbiota in human nutrition and metabolism. J. Gastroenterol. Hepatol. 2013, 28, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef]
- Arumugam, M.; Raes, J.; Pelletier, E.; Paslier, D.L.; Yamada, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.M.; et al. Enterotypes of the human gut microbiome. Nature 2011, 473, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Maskari, M.Y.A.; Waly, M.I.; Shuaibi, Y.S.; Ouhtit, A. Folate and vitamin B12 deficiency and hyperhomocysteinemia promote oxidative stress in adult type 2 diabetes. Nutrition 2012, 28, 23–26. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, H.B.; Bouzid, K.; Hassine, M.; Saadi, O.; Bahlous, A.; Abdelmoula, J.; Baccar, H.; Miled, F.B.M.B. Prevalence of non-conventional cardiovascular risk factors in Tunisian diabetics. Presse Med. 2014, 43. [Google Scholar] [CrossRef]
- Molina, M.; Gonzalez, R.; Folgado, J.; Real, J.T.; Martínez-Hervás, S.; Priego, A.; Lorente, R.; Chaves, F.J.; Ascaso, J.F. Correlation between plasma concentrations of homocysteine and diabetic polyneuropathy evaluated with the Semmes-Weinstein monofilament test in patients with type 2 diabetes mellitus. Med. Clin. 2013, 141, 382–386. [Google Scholar] [CrossRef] [PubMed]
- Ramos, R.V.; Laura, G.L.A.; Elina, M.C.B.; Donaji, B.A.A. Vitamins and type 2 diabetes mellitus. Endocr. Metab. Immune Disord. Drug Targets 2015, 15, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Jurado, N.I.N.; Salvado, J.S.; Gonzalez, M.A.M.; Bullo, M. Dietary phylloquinone intake and risk of type 2 diabetes in elderly subjects at high risk of cardiovascular disease. Am. J. Clin. Nutr. 2012, 96, 1113–1118. [Google Scholar] [CrossRef] [PubMed]
- Beulens, J.W.; Grobbee, D.E.; Sluijs, I.; Spijkerman, A.M.; van der Schouw, Y.T. Dietary phylloquinone and menaquinones intakes and risk of type 2 diabetes. Diabetes Care 2010, 33, 1699–1705. [Google Scholar] [CrossRef]
- Kallus, S.J.; Brandt, L.J. The intestinal microbiota and obesity. J. Clin. Gastroenterol. 2012, 46, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Van Treuren, W.; Dodd, D. Microbial Contribution to the Human Metabolome: Implications for Health and Disease. Annu. Rev. Pathol. 2020, 15, 345–369. [Google Scholar] [CrossRef]
- Guinane, C.M.; Cotter, P.D. Role of the gut microbiota in health and chronic gastrointestinal disease: Understanding a hidden metabolic organ. Ther. Adv. Gastroenterol. 2013, 6, 295–308. [Google Scholar] [CrossRef]
- Floch, M.H. Recommendations for probiotic use in humans-a 2014 update. Pharmaceuticals 2014, 7, 999–1007. [Google Scholar] [CrossRef] [PubMed]
- Pitocco, D.; Di Leo, M.; Tartaglione, L.; De Leva, F.; Petruzziello, C.; Saviano, A.; Pontecorvi, A.; Ojetti, V. The role of gut microbiota in mediating obesity and diabetes mellitus. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 1548–1562. [Google Scholar] [CrossRef]
- Thomas, C.M.; Versalovic, J. Probiotics-host communication: Modulation of signaling pathways in the intestine. Gut Microbes 2010, 1, 148–163. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Shi, B. Gut microbiota as a potential target of metabolic syndrome: The role of probiotics and prebiotics. Cell Biosci. 2017, 54. [Google Scholar] [CrossRef]
- Lee, B.J.; Bak, Y.-T. Irritable Bowel Syndrome, Gut Microbiota and Probiotics. J. Neurogastroenterol. Motil. 2011, 17, 252–266. [Google Scholar] [CrossRef]
- Rather, I.A.; Bajpai, V.K.; Huh, Y.S.; Han, Y.K.; Bhat, E.A.; Lim, J.; Peak, W.K.; Park, Y.H. Probiotic Lactobacillus sakei proBio-65 Extract Ameliorates the Severity of Imiquimod Induced Psoriasis-Like Skin Inflammation in a Mouse Model. Front. Microbiol. 2018, 9, 1021. [Google Scholar] [CrossRef]
- Rather, I.A.; Vivek, K.B.; Lew, L.C.; Majumder, R.; Nam, G.J.; Indugu, N.; Singh, P.; Kumar, S.; Hajra, N.H.; Sabir, J.S.M.; et al. Effect of a bioactive product SEL001 from Lactobacillus sakei probio65 on gut microbiota and its anti-colitis effects in a TNBS-induced colitis mouse model. Saudi J. Biol. Sci. 2020, 27, 261–270. [Google Scholar] [CrossRef]
- Kusano, R.; Ogawa, S.; Matsuo, Y.; Tanaka, T.; Yazaki, Y.; Kouno, I. α-Amylase and lipase inhibitory activity and structural characterization of acacia bark proanthocyanidins. J. Nat. Prod. 2011, 74, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Hyun, T.K.; Kim, M.J. The inhibitory effects of ethanol extracts from sorghum, foxtail millet and proso millet on α-glucosidase and α-amylase activities. Food Chem. 2011, 124, 1647–1651. [Google Scholar] [CrossRef]
- Guo, X.; Xia, X.; Tang, R.; Zhou, J.; Zhao, H.; Wang, K. Development of a real-time PCR method for Firmicutes and Bacteroidetes in faeces and its application to quantify intestinal population of obese and lean pigs. Lett. Appl. Microbiol. 2008, 47, 367–373. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.W.; Chen, M.K.; Yang, B.Y.; Huang, X.J.; Zhang, X.R.; He, L.Q.; Zhang, J.; Hua, Z.C. Use of 16S rRNA Gene-Targeted Group-Specific Primers for Real-Time PCR Analysis of Predominant Bacteria in Mouse Feces. Appl. Env. Microbiol. 2015, 81, 6749–6756. [Google Scholar] [CrossRef] [PubMed]
- Hold, G.L.; Schwiertz, A.; Aminov, R.I.; Blaut, M.; Flint, H.J. Oligonucleotide probes that detect quantitatively significant groups of butyrate-producing bacteria in human feces. Appl. Environ. Microbiol. 2003, 69, 4320–4324. [Google Scholar] [CrossRef]
- Wise, M.G.; Siragusa, G.R. Quantitative analysis of the intestinal bacterial community in one- to three-week-old commercially reared broiler chickens fed conventional or antibiotic-free vegetable-based diets. J. Appl. Microbiol. 2007, 102, 1138–1149. [Google Scholar] [CrossRef] [PubMed]
- Layton, A.; McKay, L.; Williams, D.; Garrett, V.; Gentry, R.; Sayler, G. Development of Bacteroides 16S rRNA gene TaqMan-based real-time PCR assays for estimation of total, human, and bovine fecal pollution in water. Appl. Environ. Microbiol. 2006, 72, 4214–4224. [Google Scholar] [CrossRef]
- Larsen, N.; Vogensen, F.K.; van den Berg, F.W.J.; Nielsen, D.S.; Andreasen, A.S.; Pedersen, B.K. Gut Microbiota in Human Adults with Type 2 Diabetes Differs from Non-Diabetic Adults. PLoS ONE 2010, 5, e9085. [Google Scholar] [CrossRef] [PubMed]
- Yadav, H.; Jain, S.; Nagpal, R.; Marotta, F. Increased fecal viral content associated with obesity in mice. World J. Diabetes. 2016, 7, 316–320. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.P.; Li, L.L.; Han, X.; Li, Q.W.; Zhang, X.H.; Liu, J.J.; Wang, Y. Fecal microbiota transplantation improves metabolism and gut microbiome composition in db/db mice. Acta Pharmacol. Sin. 2020, 41, 678–685. [Google Scholar] [CrossRef]
- Hojo, M.; Asahara, T.; Nagahara, A.; Takeda, T.; Matsumoto, K.; Ueyama, H.; Matsumoto, K.; Asaoka, D.; Takahashi, T.; Nomoto, K.; et al. Gut Microbiota Composition Before and After Use of Proton Pump Inhibitors. Dig. Dis. Sci. 2018, 63, 2940–2949. [Google Scholar] [CrossRef] [PubMed]
- De Gregoris, T.B.; Aldred, N.; Clare, A.S.; Burgess, J.G. Improvement of phylum- and class-specific primers for real-time PCR quantification of bacterial taxa. J. Microbiol. Methods 2011, 86, 351–356. [Google Scholar] [CrossRef] [PubMed]
- Bessesen, D.H.; Van Gaal, L.F. Progress and challenges in anti-obesity pharmacotherapy. Lancet Diabetes Endocrinol. 2018, 6, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Luthra, T.; Agarwal, R.; Estari, M. A novel library of -arylketones as potential inhibitors of α-glucosidase: Their design, synthesis, in vitro and in vivo studies. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Serino, M.; Luche, E.; Gres, S.; Baylac, A.; Bergé, M.; Cenac, C.; Waget, A.; Klopp, P.; Iacovoni, J.; Klopp, C.; et al. Metabolic adaptation to a high-fat diet is associated with a change in the gut microbiota. Gut 2012, 61, 543–553. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Zhang, L.; Xue, J.; Yang, X.; Dong, X.; Sha, L.; Lei, H.; Zhang, X.; Zhu, L.; Wang, Z.; et al. Dietary inulin alleviates diverse stages of type 2 diabetes mellitus via anti-inflammation and modulating gut microbiota in db/db mice. Food Funct. 2019, 10, 1915–1927. [Google Scholar] [CrossRef] [PubMed]
- Sears, B.; Perry, M. The role of fatty acids in insulin resistance. Lipids Health Dis. 2015, 14, 121. [Google Scholar] [CrossRef]
- Heydemann, A. An Overview of Murine High Fat Diet as a Model for Type 2 Diabetes Mellitus. J. Diabetes Res. 2016, 2016, 14. [Google Scholar] [CrossRef] [PubMed]
- Unger, R.H. Minireview: Weapons of Lean Body Mass Destruction: The Role of Ectopic Lipids in the Metabolic Syndrome. Endocrinology 2003, 144, 5159–5165. [Google Scholar] [CrossRef]
- Takaoka, M.; Nagata, D.; Kihara, S.; Shimomura, I.; Kimura, Y.; Tabata, Y.; Saito, Y.; Nagai, R.; Sata, M. Periadventitial adipose tissue plays a critical role in vascular remodeling. Circ. Res. 2009, 105, 906–911. [Google Scholar] [CrossRef] [PubMed]
- Allen, J.M.; Berg Miller, M.E.; Pence, B.D.; Whitlock, K.; Nehra, V.; Gaskins, H.R.; White, B.A.; Fryer, J.D.; Woods, J.A. Voluntary and forced exercise differentially alters the gut microbiome in C57BL/6J mice. J. Appl. Physiol. 2015, 118, 1059–1066. [Google Scholar] [CrossRef] [PubMed]
- Spranger, J.; Kroke, A.; Möhlig, M.; Hoffmann, K.; Bergmann, M.M.; Ristow, M.; Boeing, H.; Pfeiffer, A.F. Inflammatory cytokines and the risk to develop type 2 diabetes: Results of the prospective population-based European Prospective Investigation into Cancer and Nutrition (EPIC)-Potsdam Study. Diabetes 2003, 52, 812–817. [Google Scholar] [CrossRef] [PubMed]
- Cani, P.D.; Amar, J.; Iglesias, M.A.; Poggi, M.; Knauf, C.; Bastelica, D.; Neyrinck, A.M.; Fava, F.; Tuohy, K.M.; Chabo, C.; et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 2007, 56, 1761–1772, . [Google Scholar] [CrossRef]
- Kawai, T.; Akira, S. TLR signaling. Cell Death Differ. 2006, 13, 816–825. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Kokoeva, M.V.; Inouye, K.; Tzameli, I.; Yin, H.; Flier, J.S. TLR4 links innate immunity and fatty acid-induced insulin resistance. J. Clin. Investig. 2006, 116, 3015–3025. [Google Scholar] [CrossRef]
- Imada, C. Enzyme inhibitors and other bioactive compounds from marine actinomycetes. Antonie Van Leeuwenhoek 2005, 87, 59–63. [Google Scholar] [CrossRef] [PubMed]
- Manivasagan, P.; Venkatesan, J.; Sivakumar, K.; Kim, S.K. Actinobacterial enzyme inhibitors—A review. Crit. Rev. Microbiol. 2015, 41, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Odamaki, T.; Kato, K.; Sugahara, H. Age-related changes in gut microbiota composition from newborn to centenarian: A cross-sectional study. BMC Microbiol. 2016, 16, 90. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, L.; Margolles, A.; Sanchez, B. Bile resistance mechanisms in Lactobacillus and Bifidobacterium. Front. Microbiol. 2013, 4, 396. [Google Scholar] [CrossRef] [PubMed]
- Imaoka, A.; Shima, T.; Kato, K.; Mizuno, S.; Uehara, T.; Matsumoto, S.; Setoyama, H.; Hara, T.; Umesaki, Y. Anti-inflammatory activity of probiotic Bifidobacterium: Enhancement of IL-10 production in peripheral blood mononuclear cells from µLcerative colitis patients and inhibition of IL-8 secretion in HT-29 cells. World J. Gastroenterol. 2008, 14, 2511–2516. [Google Scholar] [CrossRef] [PubMed]
- Almeida, C.C.; Lorena, S.L.; Pavan, C.R.; Akasaka, H.M.; Mesquita, M.A. Beneficial effects of long-term consumption of a probiotic combination of Lactobacillus casei Shirota and Bifidobacterium breve Yakult may persist after suspension of therapy in lactose-intolerant patients. Nutr. Clin. Pract. 2012, 27, 247–251. [Google Scholar] [CrossRef]
- Zanotti, I.; Turroni, F.; Piemontese, A.; Mancabelli, L.; Milani, C.; Viappiani, A.; Prevedini, G.; Sanchez, B.; Margolles, A.; Elviri, L.; et al. Evidence for cholesterol-lowering activity by Bifidobacterium bifidum PRL2010 through gut microbiota modulation. Appl. Microbiol. Biotechnol. 2015, 99, 6813–6829. [Google Scholar] [CrossRef] [PubMed]
- Kalliomäki, M.; Collado, M.C.; Salminen, S.; Isolauri, E. Early differences in fecal microbiota composition in children may predict overweight. Am. J. Clin. Nutr. 2008, 87, 534–538. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, C.L.; Onnerfält, J.; Xu, J.; Molin, G.; Ahrné, S.; Thorngren-Jerneck, K. The microbiota of the gut in preschool children with normal and excessive body weight. Obesity 2012, 20, 2257–2261. [Google Scholar] [CrossRef] [PubMed]
- Chambers, E.S.; Viardot, A.; Psichas, A.; Morrison, D.J.; Murphy, K.G.; Zac-Varghese, S.E.; MacDougall, K.; Preston, T.; Tedford, C.; Finlayson, G.S.; et al. Effects of targeted delivery of propionate to the human colon on appetite regulation, body weight maintenance and adiposity in overweight adults. Gut 2015, 64, 1744–1754. [Google Scholar] [CrossRef]
- Feng, Y.; Weng, H.; Ling, L.; Zeng, T.; Zhang, Y.; Chen, D.; Li, H. Modulating the gut microbiota and inflammation is involved in the effect of Bupleurum polysaccharides against diabetic nephropathy in mice. Int. J. Biol. Macromol. 2019, 132, 1001–1011. [Google Scholar] [CrossRef] [PubMed]
- Bellahcene, M.; O’Dowd, J.F.; Wargent, E.T.; Zaibi, M.S.; Hislop, D.C.; Ngala, R.A.; Smith, D.M.; Cawthorne, M.A.; Stocker, C.J.; Arch, J.R. Male mice that lack the G-protein-coupled receptor GPR41 have low energy expenditure and increased body fat content. Br. J. Nutr. 2013, 109, 1755–1764. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Yu, H.; Xiao, X.; Hu, L.; Xin, F.; Yu, X. Inulin-type fructan improves diabetic phenotype and gut microbiota profiles in rats. PeerJ 2018, 6, e4446. [Google Scholar] [CrossRef]
- Loubinoux, J.; Mory, F.; Pereira, I.A.C.; Le Faou, A.E. Bacteremia Caused by a Strain of Desulfovibrio Related to the Provisionally Named Desulfovibrio fairfieldensis. J. Clin. Microbiol. 2000, 38, 1707. [Google Scholar] [CrossRef]
- Weglarz, L.; Dzierzewicz, Z.; Skop, B.; Orchel, A.; Parfiniewicz, B.; Wiśniowska, B.; Swiatkowska, L.; Wilczok, T. Desulfovibrio desulfuricans lipopolysaccharides induce endothelial cell IL-6 and IL-8 secretion and E-selectin and VCAM-1 expression. Cell Mol. Biol. Lett. 2003, 8, 991–1003. [Google Scholar]
- Beerens, H.; Romond, C. Sulfate-reducing anaerobic bacteria in human feces. Am. J. Clin. Nutr. 1977, 30, 1770–1776. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.J.; Lin, C.S.; Lu, C.C. Ganoderma lucidum reduces obesity in mice by modulating the composition of the gut microbiota. Nat. Commun. 2015, 6, 7489. [Google Scholar] [CrossRef]
- Cani, P.D.; Bibiloni, R.; Knauf, C.; Waget, A.; Neyrinck, A.M.; Delzenne, N.M.; Burcelin, R. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes 2008, 57, 1470–1481. [Google Scholar] [CrossRef] [PubMed]
- Spiljar, M.; Merkler, D.; Trajkovski, M. The Immune System Bridges the Gut Microbiota with Systemic Energy Homeostasis: Focus on TLRs, Mucosal Barrier, and SCFAs. Front. Immunol. 2017, 8, 1353. [Google Scholar] [CrossRef]


| Product Ingredients | g% | kcal% |
|---|---|---|
| Protein | 24 | 20 |
| Carbohydrate | 41 | 35 |
| Fat | 24 | 45 |
| Total | 89 | 100 |
| kcal/g | 4.73 | |
| Casein, 30 Mesh | 200 | 800 |
| L-Cystine | 3 | 12 |
| Corn Starch | 72.82 | 91 |
| Maltodextrin 10 | 100 | 400 |
| Sucrose | 172.8 | 691 |
| Cellulose, BW 200 | 50 | 0 |
| Soybean Oil | 25 | 225 |
| Lard | 177.5 | 1598 |
| Mineral Mix S10026 | 10 | 0 |
| DiCalcium Phosphate | 13 | 0 |
| Calcium Carbonate | 5.5 | 0 |
| Potassium Citrate, 1 H2O | 16.5 | 0 |
| Vitamin Mix V10001 | 10 | 40 |
| Choline Bitartrate | 2 | 0 |
| FD&C Red Dye #40 | 0.05 | 0 |
| FD&C Blue Dye #1 | 0 | 0 |
| FD&C Yellow Dye #5 | 0 | 0 |
| Total | 858.15 | 4057 |
| Target Primers | Sequence (5-3) | Tm (°C) | References |
|---|---|---|---|
| Bacteroidetes | (F) GGARCATGTGGTTTAATTCGATGAT | ||
| (R) AGCTGACGACAACCATGCAG | 58 | [40] | |
| Firmicutes | (F) GGAGYATGTGGTTTAATTCGAAGCA5 | ||
| (R) AGCTGACGACAACCATGCAC | 59.7 | [40] | |
| Actinobacteria | (F) TGTAGCGGTGGAATGCGC | ||
| (R) AATTAAGCCACATGCTCCGCT | 58.2 | [41] | |
| Verrucomicrobia | (F) TCAKGTCAGTATGGCCCTTAT | ||
| (R) CAGTTTTYAGGATTTCCTCCGCC | 55.9 | [41] | |
| Tenericutes | (F) ATGTGTAGCGGTAAAATGCGTAA | ||
| (R) CMTACTTGCGTACGTACTACT | 57 | [41] | |
| Deferribacteres | (F) CTATTTCCAGTTGCTAACGG | ||
| (R) GAGHTGCTTCCCTCTGATTATG | 55.2 | [41] | |
| Proteobacteria | (F) CATTGACGTTACCCGCAGAAGAAGC | ||
| (R) CTCTACGAGACTCAAGCTTGC | 59.8 | [42] | |
| Lactobacillus | (F) AGCAGTAGGGAATCTTCCA | ||
| (R) CACCGCTACACATGGAG | 54.5 | [43] | |
| Bacteroides | (F) CGATGGATAGGGGTTCTGAGAGGA | ||
| (R) GCTGGCACGGAGTTAGCCGA | 54.5 | [44] | |
| Prevotella | (F) CACCAAGGCGACGATCA | ||
| (R) GGATAACGCCYGGACCT | 55.1 | [45] | |
| Bifidobacteria | (F) GCGTGCTTAACACATGCAAGTC | ||
| (R) CACCCGTTTCCAGGAGCTATT | 60.2 | [46] | |
| Desulfovibrio | (F) CCGTAGATATCTGGAGGAACATCAG | ||
| (R) ACATCTAGCATCCATCGTTTACAGC | 62.9 | [47] | |
| Clostridium | (F) AAATGACGGTACCTGACTAA | ||
| (R) CTTTGAGTTTCATTCTTGCGAA | 53.2 | [48] | |
| Universal | (F) AAACTCAAAKGAATTGACGG | ||
| (R) CTCACRRCACGAGCTGAC | 51.1 | [49] |
| Target Primers | Efficiency (%) |
|---|---|
| Bacteroidetes | 101.7 |
| Firmicutes | 100.5 |
| Actinobacteria | 101.3 |
| Verrucomicrobia | 95.2 |
| Tenericute | 105.3 |
| Deferribacteres | 97 |
| Proteobacteria | 101.7 |
| Lactobacillus | 93.4 |
| Bacteroides | 104.4 |
| Prevotella | 101.7 |
| Bifidobacteria | 101.7 |
| Desulfovibrio | 93.4 |
| Clostridium | 94.9 |
| Universal | 91.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gulnaz, A.; Nadeem, J.; Han, J.-H.; Lew, L.-C.; Son, J.-D.; Park, Y.-H.; Rather, I.A.; Hor, Y.-Y. Lactobacillus Sps in Reducing the Risk of Diabetes in High-Fat Diet-Induced Diabetic Mice by Modulating the Gut Microbiome and Inhibiting Key Digestive Enzymes Associated with Diabetes. Biology 2021, 10, 348. https://doi.org/10.3390/biology10040348
Gulnaz A, Nadeem J, Han J-H, Lew L-C, Son J-D, Park Y-H, Rather IA, Hor Y-Y. Lactobacillus Sps in Reducing the Risk of Diabetes in High-Fat Diet-Induced Diabetic Mice by Modulating the Gut Microbiome and Inhibiting Key Digestive Enzymes Associated with Diabetes. Biology. 2021; 10(4):348. https://doi.org/10.3390/biology10040348
Chicago/Turabian StyleGulnaz, Aneela, Jawad Nadeem, Jong-Hun Han, Lee-Ching Lew, Jae-Dong Son, Yong-Ha Park, Irfan A. Rather, and Yan-Yan Hor. 2021. "Lactobacillus Sps in Reducing the Risk of Diabetes in High-Fat Diet-Induced Diabetic Mice by Modulating the Gut Microbiome and Inhibiting Key Digestive Enzymes Associated with Diabetes" Biology 10, no. 4: 348. https://doi.org/10.3390/biology10040348
APA StyleGulnaz, A., Nadeem, J., Han, J.-H., Lew, L.-C., Son, J.-D., Park, Y.-H., Rather, I. A., & Hor, Y.-Y. (2021). Lactobacillus Sps in Reducing the Risk of Diabetes in High-Fat Diet-Induced Diabetic Mice by Modulating the Gut Microbiome and Inhibiting Key Digestive Enzymes Associated with Diabetes. Biology, 10(4), 348. https://doi.org/10.3390/biology10040348








