Systematic Characterization of TCP Gene Family in Four Cotton Species Revealed That GhTCP62 Regulates Branching in Arabidopsis
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sequence Retrieval and Information Statistics of TCP Proteins
2.2. Phylogenetic Analysis, Gene Structure and Conserved Motif
2.3. Analyses of Chromosomal Distribution and Collinearity
2.4. Plant Materials and Growth Conditions
2.5. Gene Expression Assays
2.6. Construction of Overexpression Vector and Subcellular Localization Vector
3. Results
3.1. Identification of TCP Gene Family in G. hirsutum
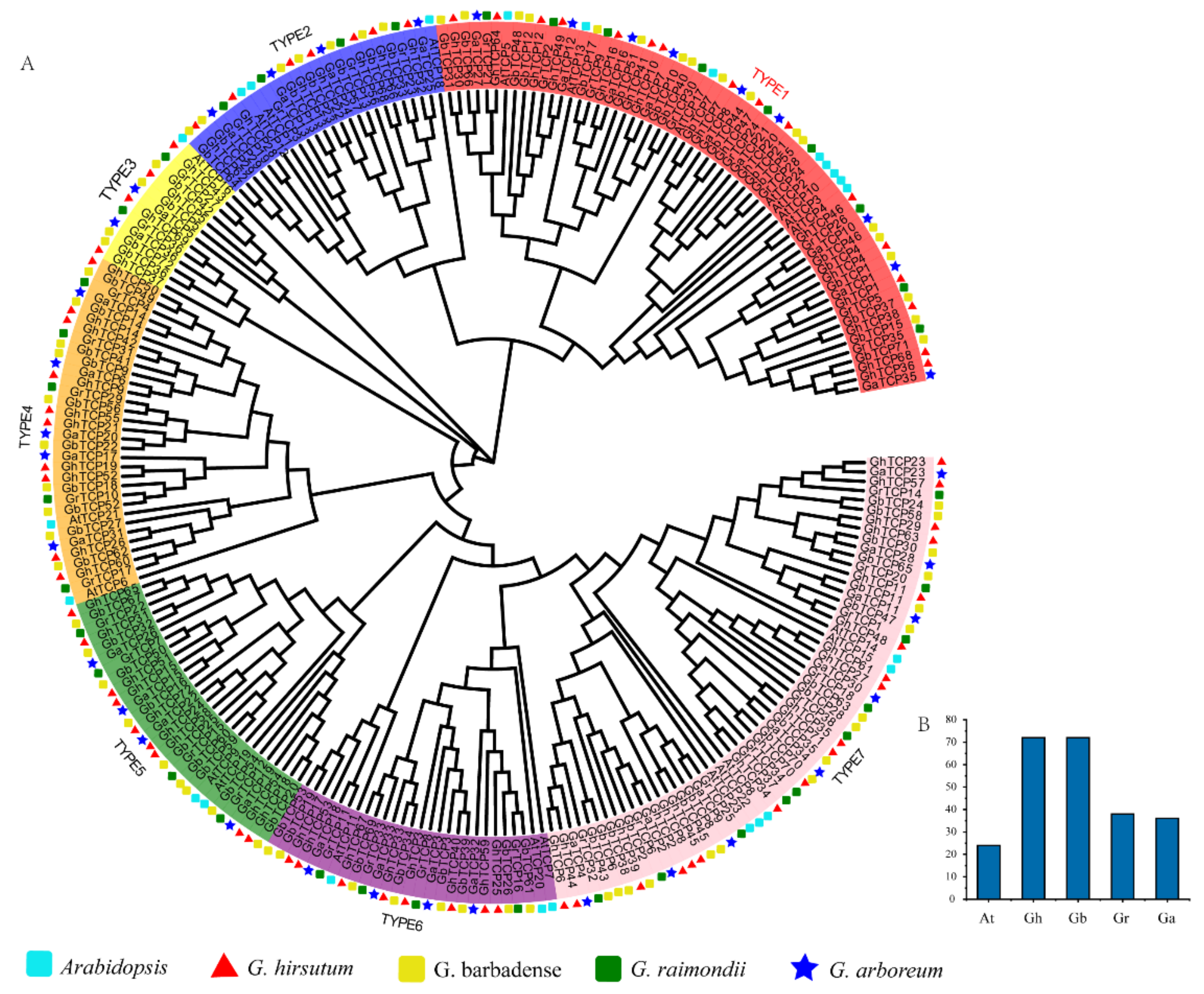
3.2. Phylogenetic Analysis of TCP Gene Family
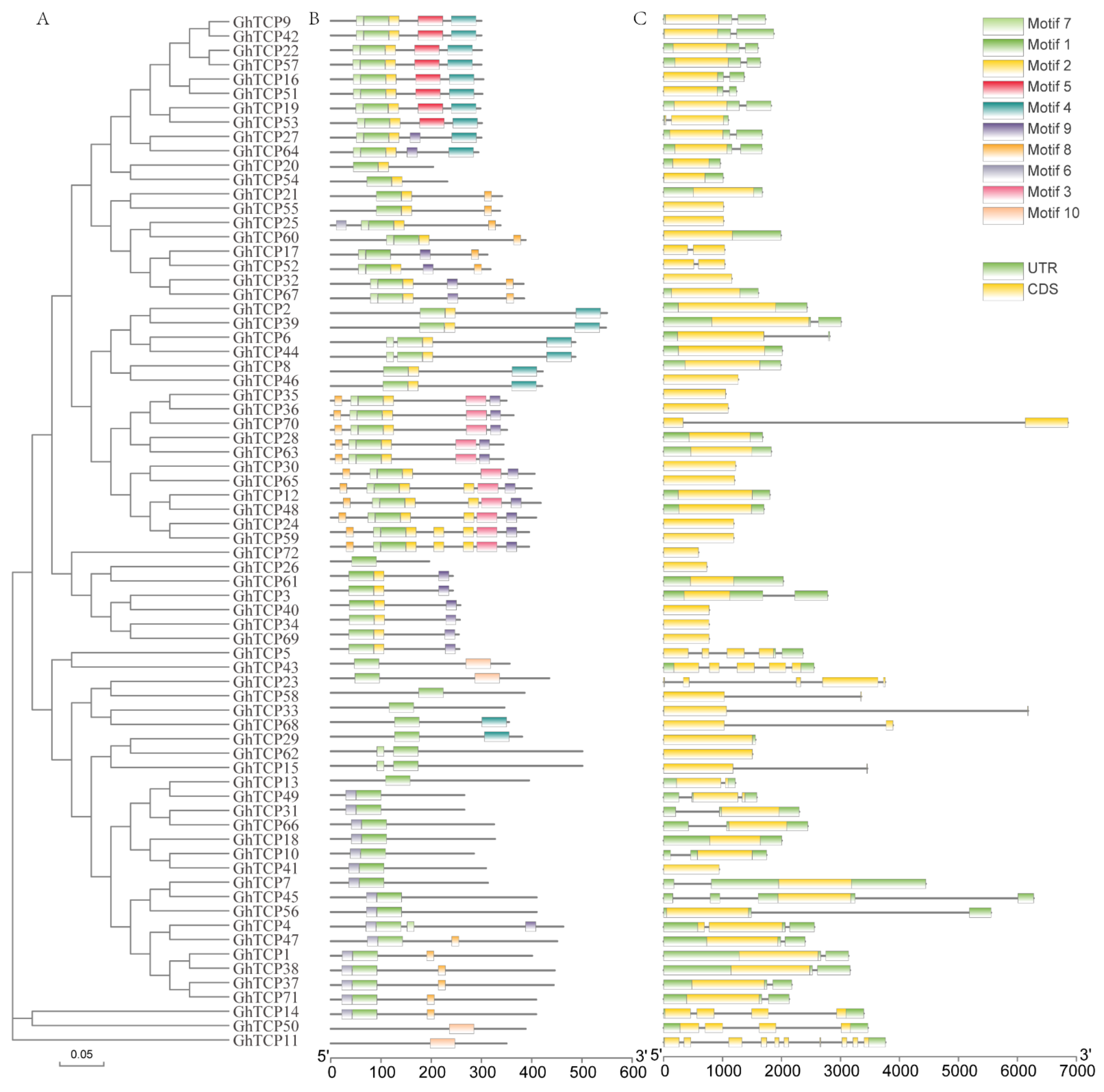
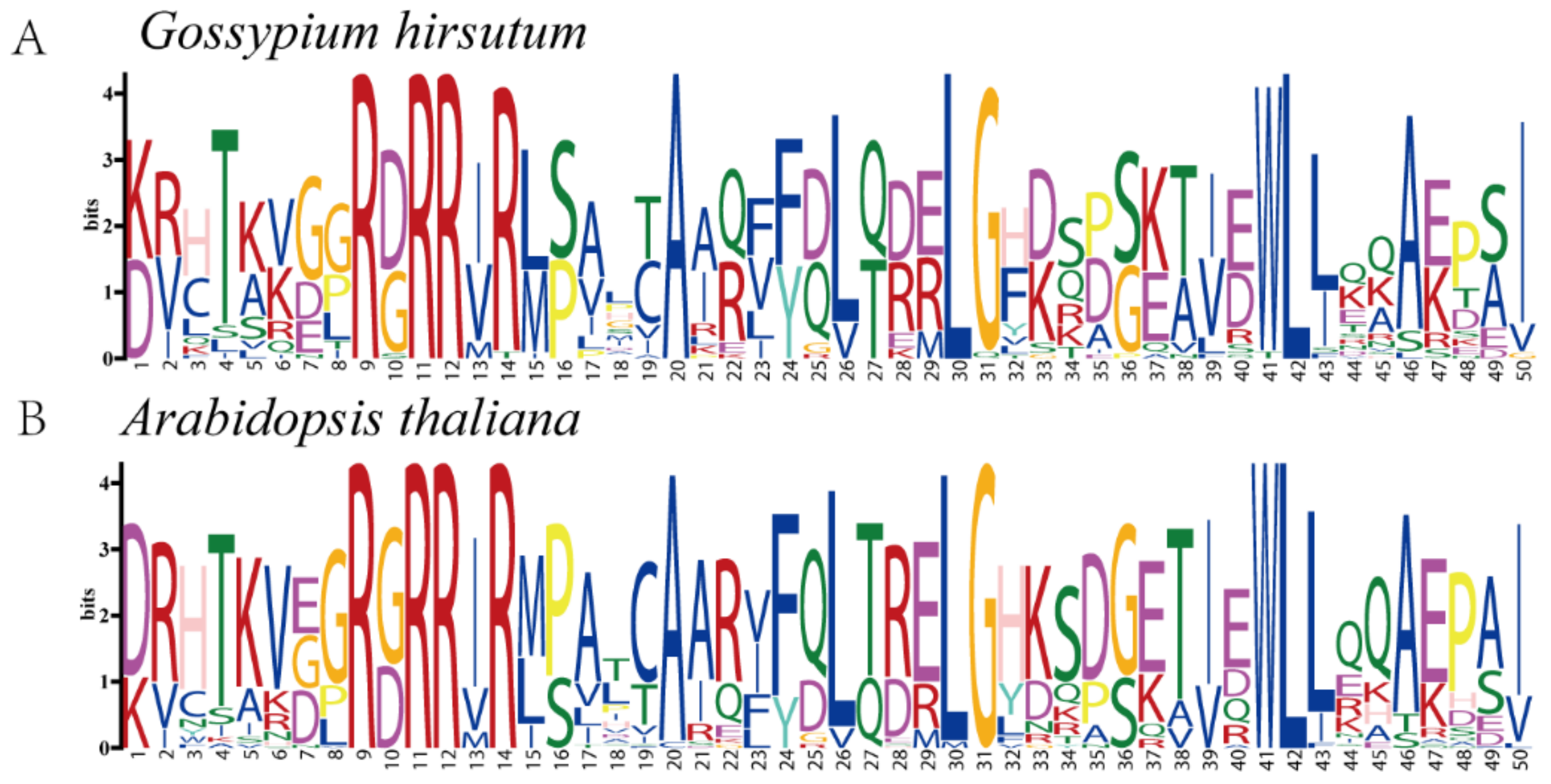
3.3. Gene Structure and Conserved Motifs
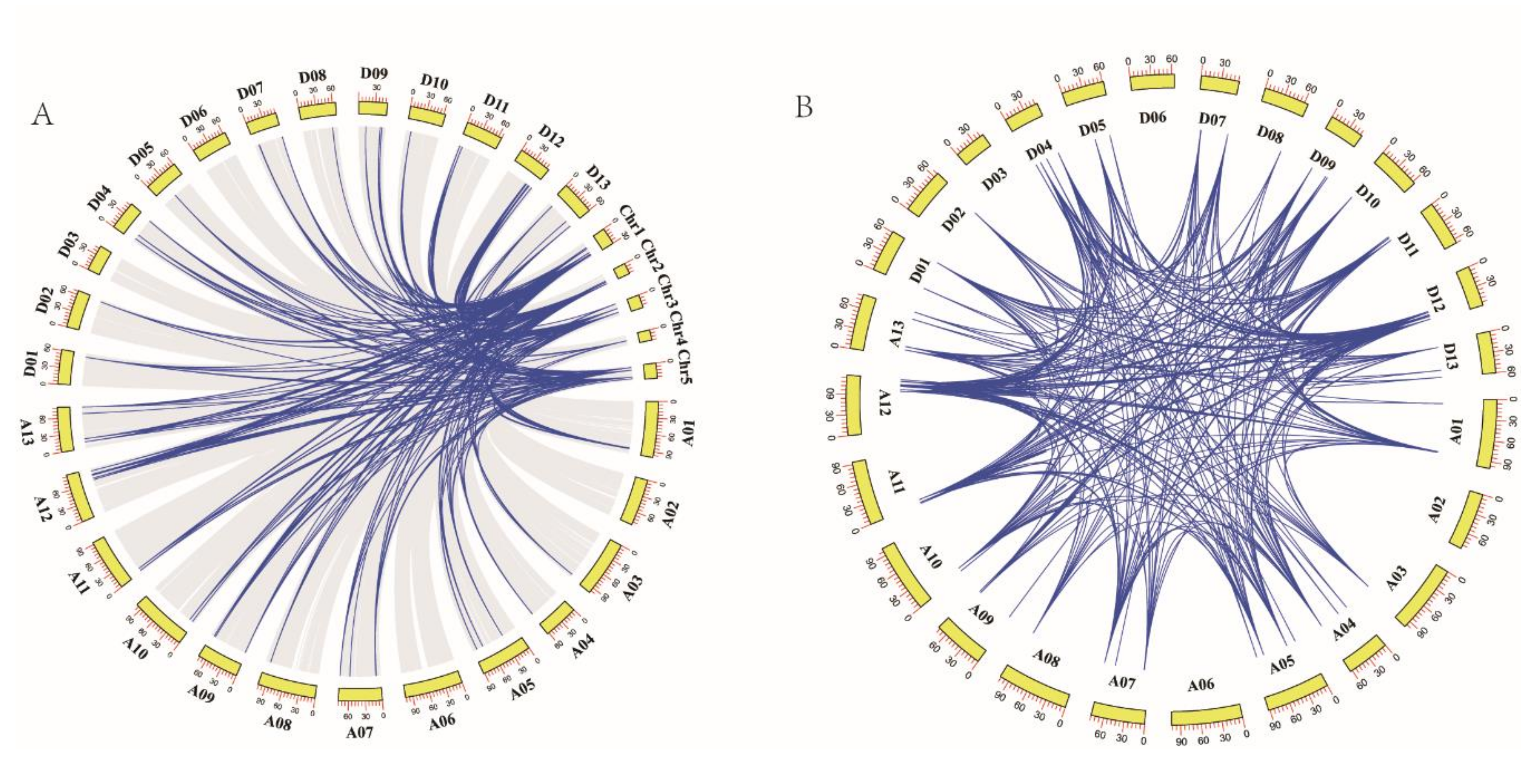
3.4. Chromosomal Distribution and Gene Collinearity Analysis
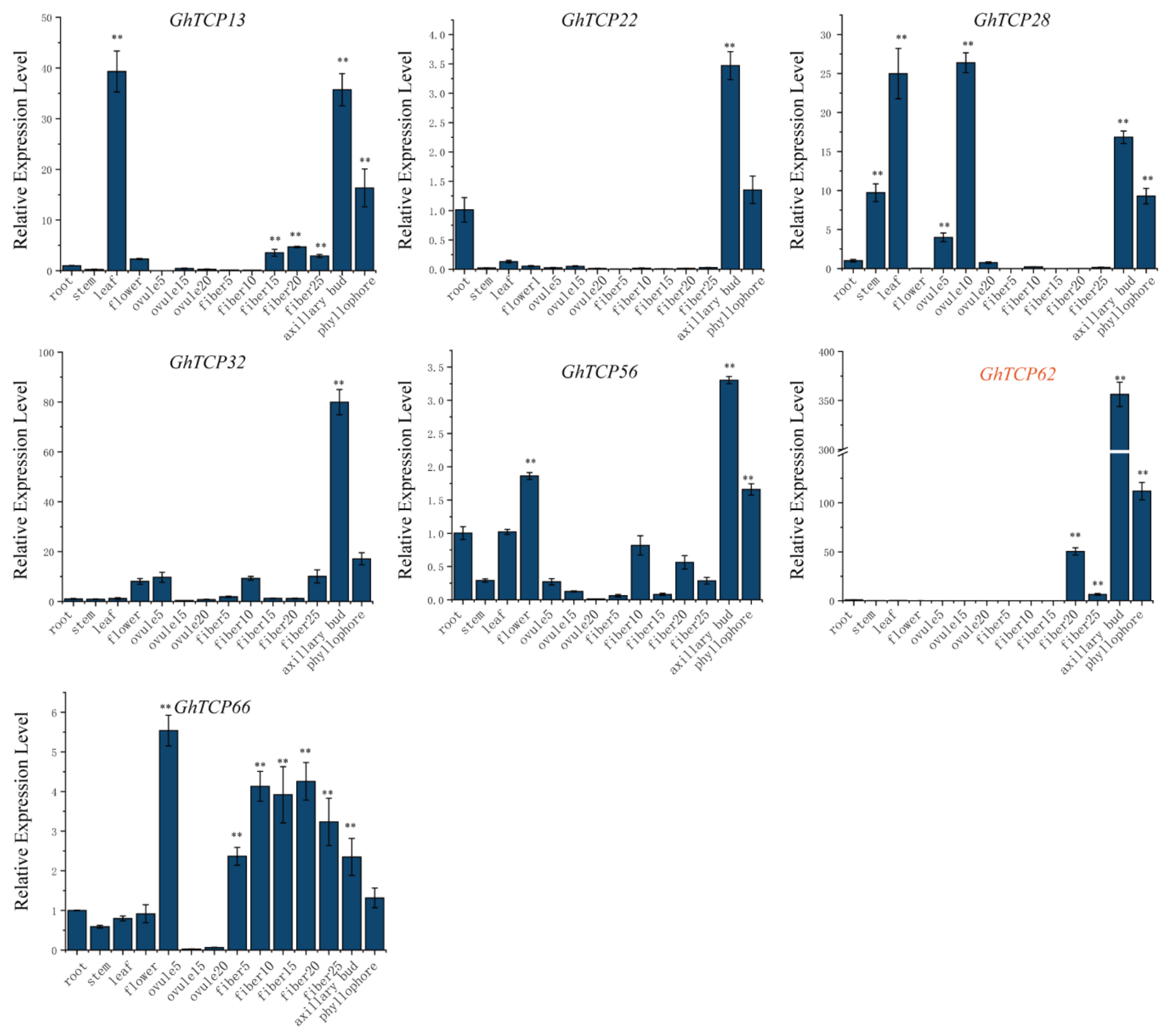
3.5. Expression Profiles of TCP Genes in G. hirsutum
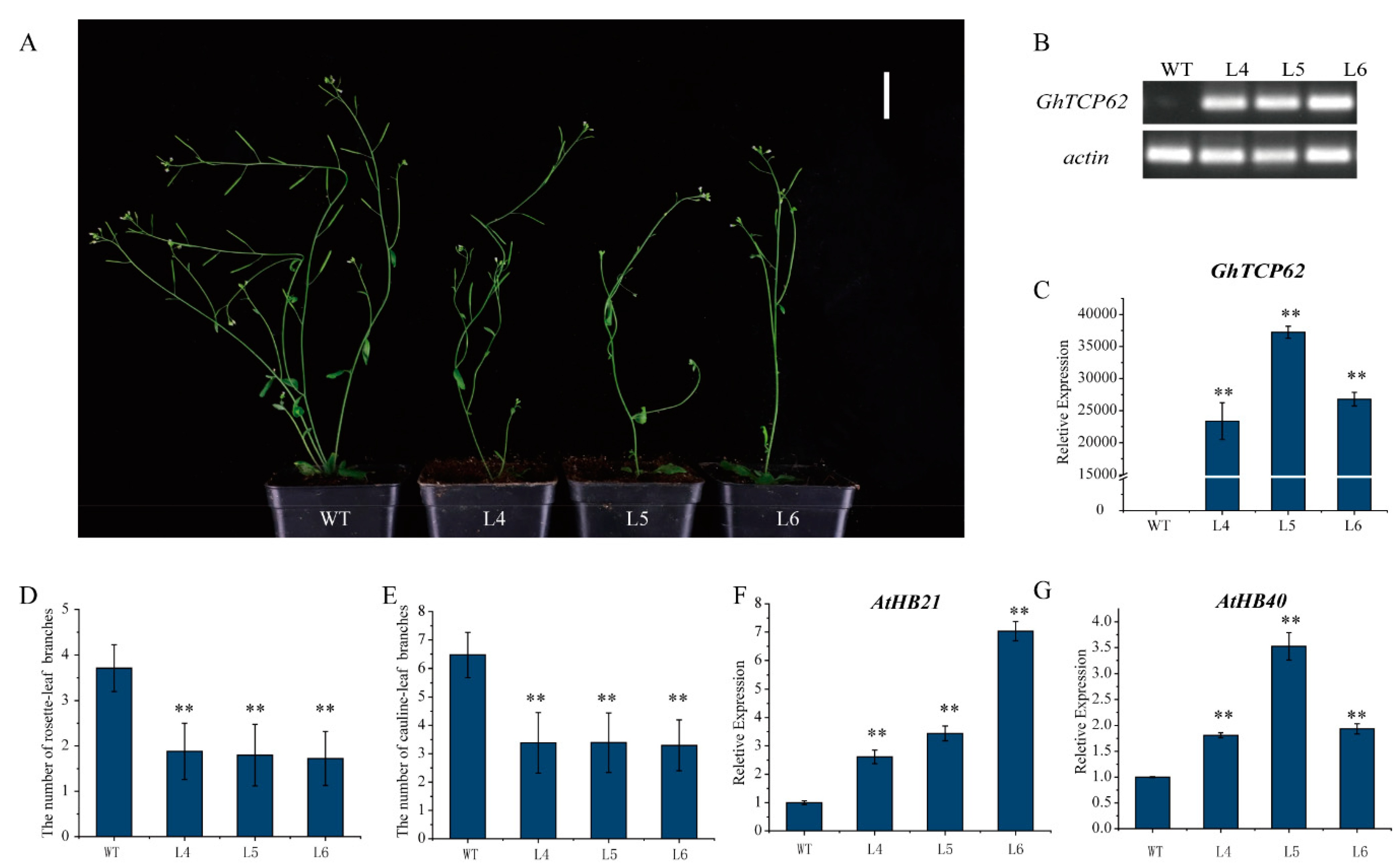
3.6. Overexpression of GhTCP62 in A. thaliana Inhibits Shoot Branching
4. Discussion
4.1. TCP Gene Plays an Important Role in Plants
4.2. Gene Replication Events Are the Main Reason for the Expansion of the TCP Gene Family
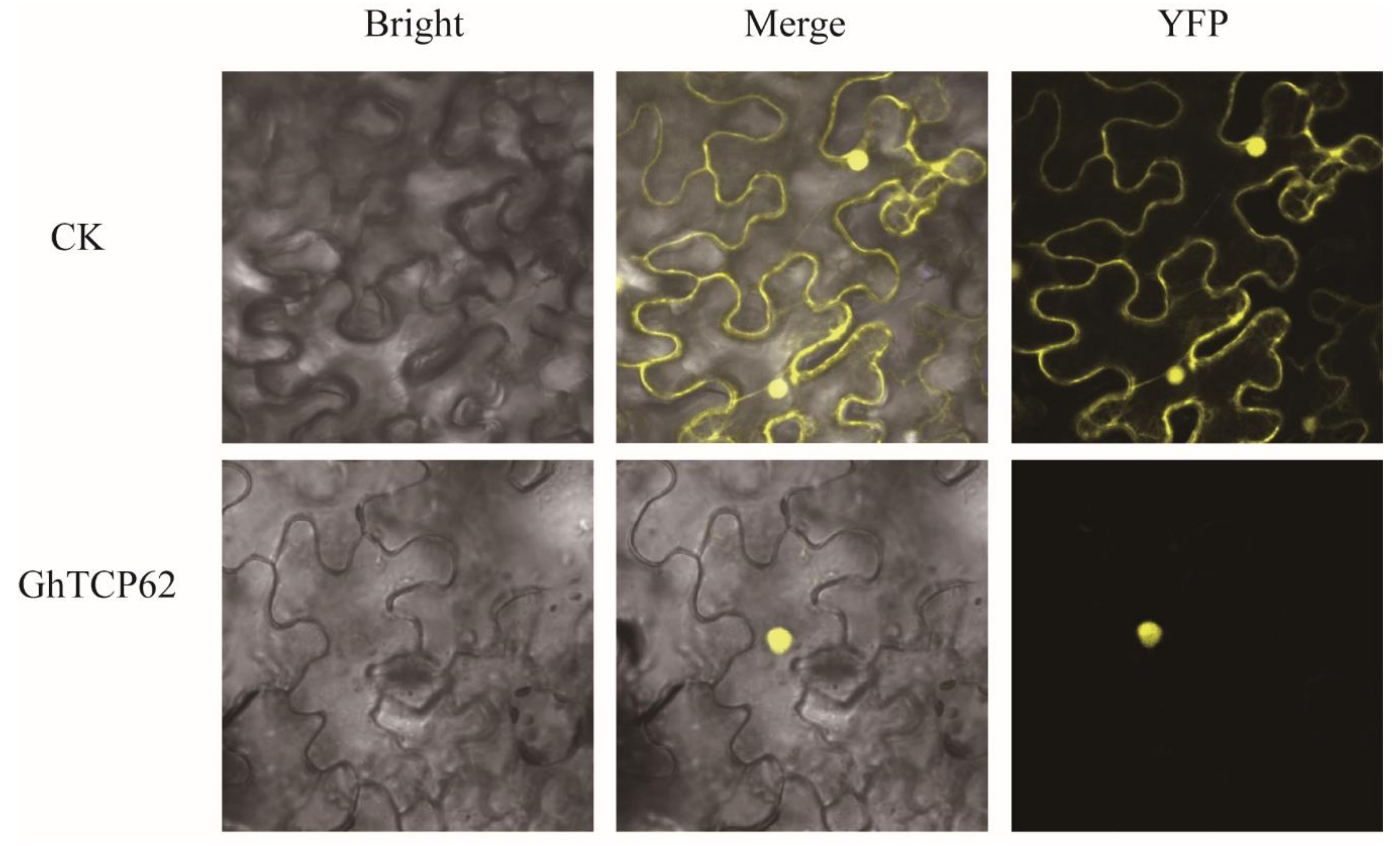
4.3. GhTCP62 Regulate Shoot Branching in Cotton
4.4. GhTCP62 Regulates Bud Activity and Branching Via HB21 and HB40 Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Barbier, F.F.; Dun, E.A.; Beveridge, C.A. Apical dominance. Curr. Biol. 2017, 27, R864–R865. [Google Scholar] [CrossRef] [Green Version]
- Kebrom, T.H.; Chandler, P.M.; Swain, S.M.; King, R.W.; Richards, R.A.; Spielmeyer, W. Inhibition of tiller bud outgrowth in the tin mutant of wheat is associated with precocious internode development. Plant Physiol. 2012, 160, 308–318. [Google Scholar] [CrossRef] [Green Version]
- Peng, J.; Richards, D.E.; Hartley, N.M.; Murphy, G.P.; Devos, K.M.; Flintham, J.E.; Beales, J.; Fish, L.J.; Worland, A.J.; Pelica, F. ‘Green revolution’genes encode mutant gibberellin response modulators. Nature 1999, 400, 256–261. [Google Scholar] [CrossRef] [PubMed]
- Agudamu; Yoshihira, T.; Shiraiwa, T. Branch development responses to planting density and yield stability in soybean cultivars. Plant Prod. Sci. 2016, 19, 331–339. [Google Scholar] [CrossRef] [Green Version]
- Green-Tracewicz, E.; Page, E.R.; Swanton, C.J. Shade avoidance in soybean reduces branching and increases plant-to-plant variability in biomass and yield per plant. Weed Sci. 2011, 59, 43–49. [Google Scholar] [CrossRef]
- Jinge, T.; Chenglong, W.; Jinliang, X.; Lishuan, W.; Guanghui, X.; Weihao, W.; Dan, L.; Wenchao, Q.; Xu, H.; Qiuyue, C. Teosinte ligule allele narrows plant architecture and enhances high-density maize yields. Science 2019, 365, 658–664. [Google Scholar]
- McSteen, P.; Hake, S. barren inflorescence2 regulates axillary meristem development in the maize inflorescence. Development 2001, 128, 2881–2891. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Smith, S.M.; Li, J. Genetic regulation of shoot architecture. Annu. Rev. Plant Biol. 2018, 69, 437–468. [Google Scholar] [CrossRef]
- Knauer, S.; Javelle, M.; Li, L.; Li, X.; Ma, X.; Wimalanathan, K.; Kumari, S.; Johnston, R.; Leiboff, S.; Meeley, R. A high-resolution gene expression atlas links dedicated meristem genes to key architectural traits. Genome Res. 2019, 29, 1962–1973. [Google Scholar] [CrossRef] [Green Version]
- Guo, W.; Chen, L.; Herrera-Estrella, L.; Cao, D.; Tran, L.-S.P. Altering Plant Architecture to Improve Performance and Resistance. Trends Plant Sci. 2020, 25, 1154–1170. [Google Scholar] [CrossRef]
- Chen, L.; Yang, H.; Fang, Y.; Guo, W.; Chen, H.; Zhang, X.; Dai, W.; Chen, S.; Hao, Q.; Yuan, S. Overexpression of GmMYB14 improves high-density yield and drought tolerance of soybean through regulating plant architecture mediated by the brassinosteroid pathway. Plant Biotechnol. J. 2021, 19, 702–716. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 1998, 10, 1391–1406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, D.; Debnath, P.; Roohi; Sane, A.; Sane, V.A. Expression of the tomato WRKY gene, SlWRKY23, alters root sensitivity to ethylene, auxin and JA and affects aerial architecture in transgenic Arabidopsis. Physiol. Mol. Biol. Plants 2020, 26, 1187. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.; Zhang, J.; Wang, X.; Han, X.; Wei, B.; Wang, J.; Li, B.; Yu, H.; Huang, Q.; Gu, H. The WRKY transcription factor WRKY71/EXB1 controls shoot branching by transcriptionally regulating RAX genes in Arabidopsis. Plant Cell 2015, 27, 3112–3127. [Google Scholar] [CrossRef] [Green Version]
- Carrara, S.; Dornelas, M.C. TCP genes and the orchestration of plant architecture. Trop. Plant Biol. 2021, 14, 1–10. [Google Scholar] [CrossRef]
- Lan, J.; Qin, G. The regulation of CIN-like TCP transcription factors. Int. J. Mol. Sci. 2020, 21, 4498. [Google Scholar] [CrossRef]
- Doebley, J.; Stec, A. teosinte branched1 and the origin of maize: Evidence for epistasis and the evolution of dominance. Genetics 1995, 141, 333–346. [Google Scholar] [CrossRef]
- Dong, Z.; Wei, L.; Unger-Wallace, E.; Yang, J.; Chuck, G. Ideal crop plant architecture is mediated by tassels replace upper ears1, a BTB/POZ ankyrin repeat gene directly targeted by TEOSINTE BRANCHED1. Proc. Natl. Acad. Sci. USA 2017, 114, E8656. [Google Scholar] [CrossRef] [Green Version]
- Nakagawa, A.; Kitazawa, M.S.; Fujimoto, K. A design principle for floral organ number and arrangement in flowers with bilateral symmetry. Development 2020, 147, dev182907. [Google Scholar] [CrossRef]
- Kosugi, S.; Ohashi, Y. PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell 1997, 9, 1607–1619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selahattin, D. TCP Transcription Factors at the Interface between Environmental Challenges and the Plant’s Growth Responses. Front. Plant Sci. 2016, 7, 1930. [Google Scholar]
- Nicolas, M.; Cubas, P. TCP factors: New kids on the signaling block. Curr. Opin. Plant Biol. 2016, 33, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.M.; Wang, M.M.; Yang, J.; Wen, J.; Du, H. Evolutionary and Comparative Expression Analyses of TCP Transcription Factor Gene Family in Land Plants. Int. J. Mol. Sci. 2019, 20, 3591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crawford, B.C.; Nath, U.; Carpenter, R.; Coen, E.S. CINCINNATA Controls Both Cell Differentiation and Growth in Petal Lobes and Leaves of Antirrhinum. Plant Physiol. 2004, 135, 244–253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danisman, S.; Dijk, A.; Bimbo, A.; Wal, F.; Immink, R. Analysis of functional redundancies within the Arabidopsis TCP transcription factor family. J. Exp. Bot. 2013, 64, 5673–5685. [Google Scholar] [CrossRef] [Green Version]
- Yao, X.; Hong, M.; Jian, W.; Zhang, D. Genome-Wide Comparative Analysis and Expression Pattern of TCP Gene Families in Arabidopsis thaliana and Oryza sativa. J. Integr. Plant Biol. 2007, 49, 885–897. [Google Scholar] [CrossRef]
- Li, F.; He, X.; Zhang, Y.; Yi, Y. Identification and Bioinformatics Analysis of TCP Transcription Factor Family in Tomato. Mol. Plant Breed. 2018, 19, 847. [Google Scholar]
- Ding, S.; Cai, Z.; Du, H.; Wang, H. Genome-wide analysis of TCP family genes in Zea mays L. identified a role for ZmTCP42 in drought tolerance. Int. J. Mol. Sci. 2019, 20, 2762. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.J.; Wang, W.L.; Zhuang, J. TCP family genes control leaf development and its responses to hormonal stimuli in tea plant [Camellia sinensis (L.) O. Kuntze]. Plant Growth Regul. 2017, 83, 43–53. [Google Scholar] [CrossRef]
- Gao, G.; Kan, J.; Jiang, C.; Ahmar, S.; Zhang, J.; Yang, P. Genome-wide diversity analysis of TCP transcription factors revealed cases of selection from wild to cultivated barley. Funct. Integr. Genom. 2020, 21, 31–42. [Google Scholar] [CrossRef]
- Huo, Y.; Xiong, W.; Su, K.; Li, Y.; Sun, Z. Genome-Wide Analysis of the TCP Gene Family in Switchgrass (Panicum virgatum L.). Int. J. Genom. 2019, 2019, 8514928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, J.; Zhai, Z.; Li, Y.; Geng, S.; Song, G.; Guan, J.; Jia, M.; Wang, F.; Sun, G.; Feng, N. Genome-wide identification and expression profiling of the TCP family genes in spike and grain development of wheat (Triticum aestivum L.). Front. Plant Sci. 2018, 9, 1282. [Google Scholar] [CrossRef] [Green Version]
- Sarvepalli, K.; Nath, U. Hyper-activation of the TCP4 transcription factor in Arabidopsis thaliana accelerates multiple aspects of plant maturation. Plant J. Cell Mol. Biol. 2011, 67, 595–607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Zhang, H.; Mou, M.; Chen, Y.; Yu, D. Arabidopsis Class II TCP Transcription Factors Integrate with the FT–FD Module to Control Flowering. Plant Physiol. 2019, 181, 97–111. [Google Scholar] [CrossRef]
- Huang, G.; Huang, J.-Q.; Chen, X.-Y.; Zhu, Y.-X. Recent advances and future perspectives in cotton research. Annu. Rev. Plant Biol. 2021, 72, 437–462. [Google Scholar] [CrossRef] [PubMed]
- Zheng, K.; Ni, Z.; Qu, Y.; Cai, Y.; Yang, Z.; Sun, G.; Chen, Q. Genome-wide identification and expression analyses of TCP transcription factor genes in Gossypium barbadense. Sci. Rep. 2018, 8, 14526. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.; Li, Y.; Zhu, W.; Fu, X.; Han, X.; Wang, J.; Lin, H.; Ye, W. Identification, characterization, and expression patterns of tcp genes and microrna319 in cotton. Int. J. Mol. Sci. 2018, 19, 3655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hao, J.; Tu, L.; Hu, H.; Tan, J.; Deng, F.; Tang, W.; Nie, Y.; Zhang, X. GbTCP, a cotton TCP transcription factor, confers fibre elongation and root hair development by a complex regulating system. J. Exp. Bot. 2012, 63, 6267–6281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhan, J.; Chu, Y.; Wang, Y.; Diao, Y.; Zhao, Y.; Liu, L.; Wei, X.; Meng, Y.; Li, F.; Ge, X. The miR164-GhCUC2-GhBRC1 module regulates plant architecture through abscisic acid in cotton. Plant Biotechnol. J. 2021, 19, 1839–1851. [Google Scholar] [CrossRef]
- Wang, M.-Y.; Zhao, P.-M.; Cheng, H.-Q.; Han, L.-B.; Wu, X.-M.; Gao, P.; Wang, H.-Y.; Yang, C.-L.; Zhong, N.-Q.; Zuo, J.-R. The cotton transcription factor TCP14 functions in auxin-mediated epidermal cell differentiation and elongation. Plant Physiol. 2013, 162, 1669–1680. [Google Scholar] [CrossRef] [Green Version]
- Edgar R, C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, W369–W373. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [Green Version]
- Xiong, F.; Zhuo, F.; Reiter, R.J.; Wang, L.; Wei, Z.; Deng, K.; Song, Y.; Qanmber, G.; Feng, L.; Yang, Z. Hypocotyl elongation inhibition of melatonin is involved in repressing brassinosteroid biosynthesis in Arabidopsis. Front. Plant Sci. 2019, 10, 1082. [Google Scholar] [CrossRef]
- Liu, Z.; Qanmber, G.; Lu, L.; Qin, W.; Liu, J.; Li, J.; Ma, S.; Yang, Z.; Yang, Z. Genome-wide analysis of BES1 genes in Gossypium revealed their evolutionary conserved roles in brassinosteroid signaling. Sci. China Life Sci. 2018, 61, 1566–1582. [Google Scholar] [CrossRef]
- Li, J.; Yu, D.; Qanmber, G.; Lu, L.; Wang, L.; Zheng, L.; Liu, Z.; Wu, H.; Liu, X.; Chen, Q. GhKLCR1, a kinesin light chain-related gene, induces drought-stress sensitivity in Arabidopsis. Sci. China Life Sci. 2019, 62, 63–75. [Google Scholar] [CrossRef]
- Wang, R.; Liu, L.; Kong, Z.; Li, S.; Lu, L.; Chen, G.; Zhang, J.; Qanmber, G.; Liu, Z. Identification of GhLOG gene family revealed that GhLOG3 is involved in regulating salinity tolerance in cotton (Gossypium hirsutum L.). Plant Physiol. Biochem. 2021, 166, 328–340. [Google Scholar] [CrossRef]
- Qanmber, G.; Liu, J.; Yu, D.; Liu, Z.; Lu, L.; Mo, H.; Ma, S.; Wang, Z.; Yang, Z. Genome-wide identification and characterization of the PERK gene family in Gossypium hirsutum reveals gene duplication and functional divergence. Int. J. Mol. Sci. 2019, 20, 1750. [Google Scholar] [CrossRef] [Green Version]
- QANMBER, G.; Daoqian, Y.; Jie, L.; Lingling, W.; Shuya, M.; Lili, L.; Zuoren, Y.; Fuguang, L. Genome-wide identification and expression analysis of Gossypium RING-H2 finger E3 ligase genes revealed their roles in fiber development, and phytohormone and abiotic stress responses. J. Cotton Res. 2018, 1, 1. [Google Scholar] [CrossRef]
- Qanmber, G.; Ali, F.; Lu, L.; Mo, H.; Ma, S.; Wang, Z.; Yang, Z. Identification of histone H3 (HH3) genes in Gossypium hirsutum revealed diverse expression during ovule development and stress responses. Genes 2019, 10, 355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Qanmber, G.; Lu, L.; Wang, L.; Li, J.; Yang, Z.; Liu, Z.; Li, Y.; Chen, Q.; Mendu, V. Genome-wide analysis of cotton GH3 subfamily II reveals functional divergence in fiber development, hormone response and plant architecture. BMC Plant Biol. 2018, 18, 350. [Google Scholar] [CrossRef]
- Ali, F.; Qanmber, G.; Wei, Z.; Yu, D.; Li, Y.; Gan, L.; Li, F.; Wang, Z. Genome-wide characterization and expression analysis of geranyl geranyl diphosphate synthase genes of cotton (Gossypium spp.) in plant development and abiotic stresses. BMC Genom. 2020, 21, 561. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. Cell Mol. Biol. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cubas, P.; Lauter, N.; Doebley, J.; Coen, E. The TCP domain: A motif found in proteins regulating plant growth and development. Plant J. Cell Mol. Biol. 1999, 18, 215–222. [Google Scholar] [CrossRef] [Green Version]
- Li, S. The Arabidopsis thaliana TCP transcription factors: A broadening horizon beyond development. Plant Signal. Behav. 2015, 10, e1044192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koyama, T.; Furutani, M.; Tasaka, M.; Ohme-Takagi, M. TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell 2007, 19, 473–484. [Google Scholar] [CrossRef] [Green Version]
- Efroni, I.; Blum, E.; Goldshmidt, A.; Eshed, Y. A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell 2008, 20, 2293–2306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niwa, M.; Daimon, Y.; Kurotani, K.; Higo, A.; Pruneda-Paz, J.L.; Breton, G.; Mitsuda, N.; Kay, S.A.; Ohme-Takagi, M.; Endo, M.; et al. BRANCHED1 interacts with FLOWERING LOCUS T to repress the floral transition of the axillary meristems in Arabidopsis. Plant Cell 2013, 25, 1228–1242. [Google Scholar] [CrossRef] [Green Version]
- Roy, S.W.; Gilbert, W.J.N.R.G. The evolution of spliceosomal introns: Patterns, puzzles and progress. Nat. Rev. Genet. 2006, 7, 211–221. [Google Scholar]
- Dingwall, C.; Robbins, J.; Dilworth, S.M.; Roberts, B.; Richardson, W.D. The nucleoplasmin nuclear location sequence is larger and more complex than that of SV-40 large T antigen. J. Cell Biol. 1988, 107, 841–849. [Google Scholar] [CrossRef] [PubMed]
- Diao, Y.; Zhan, J.; Zhao, Y.; Liu, L.; Liu, P.; Wei, X.; Ding, Y.; Sajjad, M.; Hu, W.; Wang, P.; et al. GhTIE1 Regulates Branching Through Modulating the Transcriptional Activity of TCPs in Cotton and Arabidopsis. Front. Plant Sci. 2019, 10, 1348. [Google Scholar] [CrossRef]
- Yang, Y.; Nicolas, M.; Zhang, J.; Yu, H.; Guo, D.; Yuan, R.; Zhang, T.; Yang, J.; Cubas, P.; Qin, G. The TIE1 transcriptional repressor controls shoot branching by directly repressing BRANCHED1 in Arabidopsis. PLoS Genet. 2018, 14, e1007296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, C.; Finlayson, S.A. Abscisic Acid Is a General Negative Regulator of Arabidopsis Axillary Bud Growth. Plant Physiol. 2015, 169, 611–626. [Google Scholar] [CrossRef] [Green Version]
- Aguilar-Martínez, J.A.; Poza-Carrión, C.; Cubas, P. Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 2007, 19, 458–472. [Google Scholar] [CrossRef]
- Rubio-Somoza, I.; Zhou, C.M.; Confraria, A.; Martinho, C.; von Born, P.; Baena-Gonzalez, E.; Wang, J.W.; Weigel, D. Temporal control of leaf complexity by miRNA-regulated licensing of protein complexes. Curr. Biol. 2014, 24, 2714–2719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tatematsu, K.; Nakabayashi, K.; Kamiya, Y.; Nambara, E. Transcription factor AtTCP14 regulates embryonic growth potential during seed germination in Arabidopsis thaliana. Plant J. Cell Mol. Biol. 2008, 53, 42–52. [Google Scholar] [CrossRef] [PubMed]
- Pruneda-Paz, J.L.; Breton, G.; Para, A.; Kay, S.A. A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 2009, 323, 1481–1485. [Google Scholar] [CrossRef] [Green Version]
- Navaud, O.; Dabos, P.; Carnus, E.; Tremousaygue, D.; Hervé, C. TCP transcription factors predate the emergence of land plants. J. Mol. Evol. 2007, 65, 23–33. [Google Scholar] [CrossRef]
- Martín-Trillo, M.; Cubas, P. TCP genes: A family snapshot ten years later. Trends Plant Sci. 2010, 15, 31–39. [Google Scholar] [CrossRef]
- Lewis, J.M.; Mackintosh, C.A.; Shin, S.; Gilding, E.; Kravchenko, S.; Baldridge, G.; Zeyen, R.; Muehlbauer, G.J. Overexpression of the maize Teosinte Branched1 gene in wheat suppresses tiller development. Plant Cell Rep. 2008, 27, 1217–1225. [Google Scholar] [CrossRef] [PubMed]
- Qin, G.; Gu, H.; Zhao, Y.; Ma, Z.; Shi, G.; Yang, Y.; Pichersky, E.; Chen, H.; Liu, M.; Chen, Z.; et al. An indole-3-acetic acid carboxyl methyltransferase regulates Arabidopsis leaf development. Plant Cell 2005, 17, 2693–2704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schommer, C.; Palatnik, J.F.; Aggarwal, P.; Chételat, A.; Cubas, P.; Farmer, E.E.; Nath, U.; Weigel, D. Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 2008, 6, e230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aguilar-Martínez, J.A.; Sinha, N. Analysis of the role of Arabidopsis class I TCP genes AtTCP7, AtTCP8, AtTCP22, and AtTCP23 in leaf development. Front. Plant Sci. 2013, 4, 406. [Google Scholar] [CrossRef] [Green Version]
- Finlayson, S.A.; Krishnareddy, S.R.; Kebrom, T.H.; Casal, J.J. Phytochrome regulation of branching in Arabidopsis. Plant Physiol. 2010, 152, 1914–1927. [Google Scholar] [CrossRef] [Green Version]
- Takeda, T.; Suwa, Y.; Suzuki, M.; Kitano, H.; Ueguchi-Tanaka, M.; Ashikari, M.; Matsuoka, M.; Ueguchi, C. The OsTB1 gene negatively regulates lateral branching in rice. Plant J. Cell Mol. Biol. 2003, 33, 513–520. [Google Scholar] [CrossRef] [PubMed]
- Finlayson, S.A. Arabidopsis Teosinte Branched1-like 1 regulates axillary bud outgrowth and is homologous to monocot Teosinte Branched1. Plant Cell Physiol. 2007, 48, 667–677. [Google Scholar] [CrossRef] [Green Version]
- Muhr, M.; Paulat, M.; Awwanah, M.; Brinkkötter, M.; Teichmann, T. CRISPR/Cas9-mediated knockout of Populus BRANCHED1 and BRANCHED2 orthologs reveals a major function in bud outgrowth control. Tree Physiol. 2018, 38, 1588–1597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teichmann, T.; Muhr, M. Shaping plant architecture. Front Plant Sci 2015, 6, 233. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; Yang, J.; Li, S.; Liu, L.; Qanmber, G.; Chen, G.; Duan, Z.; Zhao, N.; Wang, G. Systematic Characterization of TCP Gene Family in Four Cotton Species Revealed That GhTCP62 Regulates Branching in Arabidopsis. Biology 2021, 10, 1104. https://doi.org/10.3390/biology10111104
Liu Z, Yang J, Li S, Liu L, Qanmber G, Chen G, Duan Z, Zhao N, Wang G. Systematic Characterization of TCP Gene Family in Four Cotton Species Revealed That GhTCP62 Regulates Branching in Arabidopsis. Biology. 2021; 10(11):1104. https://doi.org/10.3390/biology10111104
Chicago/Turabian StyleLiu, Zhao, Jingyu Yang, Shengdong Li, Le Liu, Ghulam Qanmber, Guoquan Chen, Zhenyu Duan, Na Zhao, and Gang Wang. 2021. "Systematic Characterization of TCP Gene Family in Four Cotton Species Revealed That GhTCP62 Regulates Branching in Arabidopsis" Biology 10, no. 11: 1104. https://doi.org/10.3390/biology10111104
APA StyleLiu, Z., Yang, J., Li, S., Liu, L., Qanmber, G., Chen, G., Duan, Z., Zhao, N., & Wang, G. (2021). Systematic Characterization of TCP Gene Family in Four Cotton Species Revealed That GhTCP62 Regulates Branching in Arabidopsis. Biology, 10(11), 1104. https://doi.org/10.3390/biology10111104





