Introducing Beneficial Alleles from Plant Genetic Resources into the Wheat Germplasm
Abstract
Simple Summary
Abstract
1. Introduction
2. Status of the Wheat Germplasm Conserved Ex Situ
3. Wheat Taxonomy, Domestication, and Genepool
3.1. Taxonomical Treatment of Triticum and Aegilops Taxa
| Ploidy Level | Common Name | Biological Status | Kernel Coverage | Genome Formula (Haploid) Considered in This Review | Taxon Name Considered in This Review | van Slageren [72] | Mac Key [70] | Dorofeev et al. [68] | Schiemann [71] |
|---|---|---|---|---|---|---|---|---|---|
| 2n = 2x = 14 | |||||||||
| Urartu wheat, wild Urartu einkorn | Wild | Hulled | Au | T. urartu Thumanjan ex Gandil. | T. urartu Thumanjan ex Gandil. | T. urartu Thumanjan ex Gandil. | T. urartu Thumanjan ex Gandil. | ||
| Wild einkorn | Wild | Hulled | Ab | T. boeoticum Boiss. | T. monococcum L. subsp. aegilopoides (Link) Thell. | T. monococcum L. subsp. boeoticum (Boiss.) Á. Löve et D. Löve | T. boeoticum Boiss. | T. boeoticum Boiss. em. Schiem. | |
| Einkorn, domesticated einkorn, small spelt | Domesticated | Hulled | Ab | T. monococcum L. | T. monococcum L. subsp. monococcum | T. monococcum L. subsp. monococcum | T. monococcum L. | T. monococcum L. | |
| Sinskaya’s wheat | Domesticated | Free-threshing | Ab | T. sinskajae A. Filat. et Kurkiev | T. sinskajae A. Filat. et Kurkiev | ||||
| 2n = 4x = 28 | |||||||||
| Wild emmer | Wild | Hulled | BA | T. dicoccoides (Körn. ex Asch. et Graebn.) Schweinf. | T. turgidum L. subsp. dicoccoides (Körn. ex Asch. et Graebn.) Thell. | T. turgidum subsp. dicoccoides (Körn. ex Asch. et Graebn.) Thell. | T. dicoccoides (Körn. ex Asch. et Graebn.) Schweinf. | T. dicoccoides Körn. | |
| Emmer | Domesticated | Hulled | BA | T. dicoccon Schrank * | T. turgidum L. subsp. dicoccum (Schrank ex Schübl.) Thell. | T. turgidum subsp. dicoccum (Schrank ex Schübl.) Thell. | T. dicoccum Schrank ex Schübl. | T. dicoccum Schübl. | |
| Persian wheat, dika | Domesticated | Free-threshing | BA | T. carthlicum Nevski | T. turgidum L. subsp. carthlicum (Nevski) Á. Löve et D. Löve | T. turgidum subsp. carthlicum (Nevski) Á. Löve et D. Löve | T. carthlicum Nevski | T. carthlicum Nevski | |
| Durum wheat, macaroni wheat | Domesticated | Free-threshing | BA | T. durum Desf. | T. turgidum L. subsp. durum (Desf.) Husn. | T. turgidum subsp. turgidum convar. durum (Desf.) Mac Key | T. durum Desf. | T. durum Desf. | |
| Polish wheat | Domesticated | Free-threshing | BA | T. polonicum L. | T. turgidum L. subsp. polonicum (L.) Thell. | T. turgidum subsp. turgidum convar. polonicum (L.) Mac Key | T. polonicum L. | T. polonicum L. | |
| Khorasan wheat, Turanian wheat | Domesticated | Free-threshing | BA | T. turanicum Jakubz. | T. turgidum L. subsp. turanicum (Jakubz.) Á. Löve et D. Löve | T. turgidum subsp. turgidum convar. turancium (Jakubz.) Mac Key | T. turanicum Jakubz. | T. orientale Perciv. | |
| Rivet, cone, English wheat, turgid wheat, poulard wheat | Domesticated | Free-threshing | BA | T. turgidum L. | T. turgidum L. subsp. turgidum | T. turgidum L. subsp. turgidum convar. turgidum | T. turgidum L. | T. turgidum L. | |
| Georgian wheat, Colchic emmer, Karamyschev’s wheat | Domesticated | Hulled | BA | T. karamyschevii Nevski | T. turgidum L. subsp. palaeocolchicum Á. Löve et D. Löve | T. turgidum subsp. georgicum (Dekapr. et Menabde) Mac Key | T. karamyschevii Nevski | ||
| Ethiopian wheat | Domesticated | Free-threshing | BA | T. aethiopicum Jakubz. | T. aethiopicum Jakubz. | ||||
| Espahanian wheat, Isfahanian emmer | Domesticated | Hulled | BA | T. ispahanicum Heslot | T. ispahanicum Heslot | T. ispahanicum Heslot | |||
| Jakubziner’s wheat | Domesticated | Free-threshing | BA | T. jakubzineri (Udachin et Schachm.) Udachin et Schachm. | T. jakubzineri (Udachin et Schachm.) Udachin et Schachm. | ||||
| Araratian wild emmer, Armenian wild emmer | Wild | Hulled | GAt | T. araraticum Jakubz. | T. timopheevii subsp. armeniacum (Jakubz.) Mac Key ex van Slageren | T. timopheevii subsp. armeniacum (Jakubz.) Mac Key | T. araraticum Jakubz. | ||
| Militina’s wheat | Domesticated | Free-threshing | GAt | T. militinae Zhuk. et Migush. | T. militinae Zhuk. et Migush. | ||||
| Timofeev’s wheat | Domesticated | Hulled | GAt | T. timopheevii (Zhuk.) Zhuk. | T. timopheevii (Zhuk.) Zhuk. subsp. timopheevii | T. timopheevii (Zhuk.) Zhuk. subsp. timopheevii | T. timopheevii (Zhuk.) Zhuk. | T. timopheevii Zhuk. | |
| 2n = 6x = 42 | |||||||||
| Common wheat, bread wheat | Domesticated | Free-threshing | BAD | T. aestivum L. | T. aestivum L. subsp. aestivum | T. aestivum L. subsp. aestivum | T. aestivum L. | T. aestivum L. | |
| Club wheat | Domesticated | Free-threshing | BAD | T. compactum Host | T. aestivum subsp. compactum (Host) Mac Key | T. aestivum subsp. compactum (Host) Mac Key | T. compactum Host | T. compactum Host | |
| Macha wheat | Domesticated | Hulled | BAD | T. macha Dekapr. et Menabde | T. aestivum subsp. macha (Dekapr. et Menabde) Mac Key | T. aestivum subsp. macha (Dekapr. et Menabde) Mac Key | T. macha Dekapr. et Menabde | T. macha Dekapr. et Menabde | |
| Petropavlovsky’s wheat | Domesticated | Free-threshing | BAD | T. petropavlovskyi Udachin et Migush. | T. petropavlovskyi Udachin et Migush. | ||||
| Spelt wheat | Domesticated | Hulled | BAD | T. spelta L. | T. aestivum subsp. spelta (L.) Thell. | T. aestivum subsp. spelta (L.) Thell. | T. spelta L. | T. spelta L. | |
| Indian dwarf wheat, shot wheat | Domesticated | Free-threshing | BAD | T. sphaerococcum Perciv. | T. aestivum subsp. sphaerococcum (Perciv.) Mac Key | T. aestivum subsp. sphaerococcum (Perciv.) Mac Key | T. sphaerococcum Perciv. | T. sphaerococcum Perciv. | |
| Vavilov’s wheat | Domesticated | Hulled | BAD | T. vavilovii (Thumanjan) Jakubz. | T. vavilovii Jakubz. | T. vavilovii (Thumanjan) Jakubz. | |||
| Zhukovsky’s wheat | Domesticated | Hulled | GAtAb | T. zhukovskyi Menabde et Ericzjan | T. zhukovskyi Menabde et Ericzjan | T. zhukovskyi Menabde et Ericzjan | T. zhukovskyi Menabde et Ericzjan | ||
| Kilian et al. [60] and This Review | van Slageren [72] | Kimber et Sears [74] | Whitcombe [80] | Hammer [77,78] | Chennaveerayah [75] | Kihara [79] | Eig [76] | Zhukovsky [81] | |
|---|---|---|---|---|---|---|---|---|---|
| Subgenus Aegilops | |||||||||
| Section Aegilops | |||||||||
| 1 | Ae. biuncialis Vis. | Ae. biuncialis Vis. | T. macrochaetum (Shuttlew. et É. Huet ex Duval-Jouve) K. Richt. | Ae. lorentii Hochst. | Ae. lorentii Hochst. | Ae. biuncialis Vis. | Ae. biuncialis Vis. | Ae. biuncialis Vis. | Ae. biuncialis Vis. |
| 2 | Ae. columnaris Zhuk. | Ae. columnaris Zhuk. | T. columnare (Zhuk.) Ros. Morris et Sears | Ae. columnaris Zhuk. | Ae. columnaris Zhuk. | Ae. columnaris Zhuk. | Ae. columnaris Zhuk. | Ae. columnaris Zhuk. | Ae. columnaris Zhuk. |
| 3 | Ae. geniculata Roth | Ae. geniculata Roth | T. ovatum (L.) Raspail | Ae. ovata L. | Ae. geniculata Roth | Ae. ovata L. | Ae. ovata L. | Ae. ovata L. | Ae. ovata L. |
| subsp. geniculata | |||||||||
| subsp. gibberosa (Zhuk.) K. Hammer | |||||||||
| 4 | Ae. kotschyi Boiss. | Ae. kotschyi Boiss. | T. kotschyi (Boiss.) Bowden | Ae. kotschyi Boiss. | Ae. kotschyi Boiss. | Ae. kotschyi Boiss. | Ae. kotschyi Boiss. | Ae. kotschyi Boiss. | |
| 5 | Ae. neglecta Req. ex Bertol. | Ae. neglecta Req. ex Bertol. (4x and 6x) | T. triaristatum (Willd.) Godr. et Gren. (4x and 6x) | Ae. triaristata Willd. (4x and 6x) | Ae. neglecta Req. ex Bertol. | Ae. triaristata Willd. | Ae. triaristata Willd. (4x and 6x) | Ae. triaristata Willd. (4x and 6x) | Ae. triaristata Willd. (4x and 6x) |
| subsp. neglecta (4x) | subsp. neglecta (4x) | ||||||||
| subsp. recta (Zhuk.) K. Hammer (6x) | subsp. recta (Zhuk.) K. Hammer (6x) | ||||||||
| 6 | Ae. peregrina (Hack.) Maire et Weiller | Ae. peregrina (Hack.) Maire et Weiller | T. kotschyi (Boiss.) Bowden | Ae. peregrina (Hack.) Maire et Weiller | Ae. peregrina (Hack.) Maire et Weiller | Ae. variabilis Eig | Ae. variabilis Eig | Ae. variabilis Eig | |
| subsp. peregrina | var. peregrina | subsp. peregrina | |||||||
| var. brachyathera (Boiss.) Maire et Weiller | subsp. cylindrostachys (Eig et Feinbrun) Maire et Weiller | ||||||||
| 7 | Ae. triuncialis L. | Ae. triuncialis L. | T. triunciale (L.) Raspail | Ae. triuncialis L. | Ae. triuncialis L. | Ae. triuncialis L. | Ae. triuncialis L. | Ae. triuncialis L. | Ae. triuncialis L. |
| subsp. triuncialis | var. triuncialis | subsp. triuncialis | |||||||
| subsp. persica (Boiss.) Zhuk. | var. persica (Boiss.) Eig | subsp. persica (Boiss.) Zhuk. | Ae. persica Boiss. | ||||||
| 8 | Ae. umbellulata Zhuk. | Ae. umbellulata Zhuk. | T. umbellulatum (Zhuk.) Bowden | Ae. umbellulata Zhuk. | Ae. umbellulata Zhuk. | Ae. umbellulata Zhuk. | Ae. umbellulata Zhuk. | Ae. umbellulata Zhuk. | Ae. umbellulata Zhuk. |
| subsp. umbellulata | |||||||||
| subsp. transcaucasica Dorof. et Migush. | subsp. transcaucasica Dorof. et Migush. | ||||||||
| Section Comopyrum | |||||||||
| 9 | Ae. comosa Sibth. et Sm. | Ae. comosa Sm. in Sibth. et Sm. | T. comosum (Sibth. et Sm.) K. Richt. | Ae. comosa Sibth. et Sm. | Ae. comosa Sibth. et Sm. | Ae. comosa Sibth. et Sm. | Ae. comosa Sibth. et Sm. | Ae. comosa Sibth. et Sm. | Ae. comosa Sibth. et Sm. |
| subsp. comosa | var. comosa | subsp. comosa | subsp. comosa | ||||||
| subsp. heldreichii (Boiss.) Eig (syn.: var. subventricosa Boiss.) | var. subventricosa Boiss. | subsp. heldreichii (Boiss.) Eig | subsp. heldreichii (Boiss.) Eig | subsp. heldreichii (Holzm.) Eig | |||||
| 10 | Ae. uniaristata Vis. | Ae. uniaristata Vis. | T. uniaristatum (Vis.) K. Richt. | Ae. uniaristata Vis. | Ae. uniaristata Vis. | Ae. uniaristata Vis. | Ae. uniaristata Vis. | Ae. uniaristata Vis. | Ae. uniaristata Vis. |
| Section Cylindropyrum | |||||||||
| 11 | Ae. cylindrica Host | Ae. cylindrica Host | T. cylindricum (Host) Ces., Pass. Et Gibelli | Ae. cylindrica Host | Ae. cylindrica Host | Ae. cylindrica Host | Ae. cylindrica Host | Ae. cylindrica Host | Ae. cylindrica Host |
| 12 | Ae. markgrafii (Greuter) K. Hammer | Ae. caudata L. | T. dichasians (Zhuk.) Bowden | Ae. caudata L. | Ae. markgrafii (Greuter) K. Hammer | Ae. caudata L. | Ae. caudata L. | Ae. caudata L. | Ae. caudata L. |
| Section Sitopsis | |||||||||
| 13 | Ae. bicornis (Forssk.) Jaub. et Spach | Ae. bicornis (Forssk.) Jaub. et Spach | T. bicorne Forssk. | Ae. bicornis (Forssk.) Jaub. et Spach | Ae. bicornis (Forssk.) Jaub. et Spach | T. bicorne Forssk. | Ae. bicornis (Forssk.) Jaub. et Spach | Ae. bicornis (Forssk.) Jaub. et Spach | Ae. bicornis (Forssk.) Jaub. et Spach |
| var. bicornis | |||||||||
| var. anathera Eig | |||||||||
| 14 | Ae. longissima Schweinf. et Muschl. | Ae. longissima Schweinf. et Muschl. | T. longissimum (Schweinf. et Muschl.) Bowden | Ae. longissima Schweinf. et Muschl. | Ae. longissima Schweinf. et Muschl. emend. Eig s.l. | T. longissimum (Schweinf. et Muschl.) Bowden subsp. longissimum | Ae. longissima Schweinf. et Muschl. | Ae. longissima Schweinf. et Muschl. | Ae. longissima Schweinf. et Muschl. emend. Eig |
| subsp. longissima | |||||||||
| 15 | Ae. sharonensis Eig | Ae. sharonensis Eig | Ae. sharonensis Eig | subsp. sharonensis (Eig) K. Hammer | subsp. sharonensis (Eig) Chennav. | Ae. sharonensis Eig | |||
| 16 | Ae. searsii Feldman et Kislev ex K. Hammer | Ae. searsii Feldman et Kislev ex K. Hammer | T. searsii (Feldman et Kislev) Feldman | Ae. searsii Feldman et Kislev | Ae. searsii Feldman et Kislev ex K. Hammer | ||||
| 17 | Ae. speltoides Tausch | Ae. speltoides Tausch | T. speltoides (Tausch) Gren. ex K. Richt. | Ae. speltoides Tausch | Ae. speltoides Tausch | T. speltoides Tausch | Ae. speltoides Tausch | Ae. speltoides Tausch | Ae. speltoides Tausch |
| subsp. speltoides | var. speltoides | subsp. speltoides | subsp. aucheri (Boiss.) Chennav. | ||||||
| subsp. ligustica (Savign.) Zhuk. | var. ligustica (Savign.) Fiori | Ae. ligustica (Savign.) Coss. | subsp. ligustica (Savign.) Zhuk. | subsp. ligusticum (Savign.) Chennav. | Ae. ligustica (Savign.) Coss. | ||||
| Section Vertebrata | |||||||||
| 18 | Ae. crassa Boiss. (4x and 6x) | Ae. crassa Boiss. (4x and 6x) | T. crassum (Boiss.) Aitch. et Hemsl. (4x and 6x) | Ae. crassa Boiss. (4x and 6x) | Ae. crassa Boiss. (4x and 6x) | Ae. crassa Boiss. | Ae. crassa Boiss. (4x and 6x) | Ae. crassa Boiss. (4x and 6x) | Ae. crassa Boiss. (4x and 6x) |
| subsp. crassa | |||||||||
| 19 | Ae. vavilovii (Zhuk.) Chennav. (6x) | Ae. vavilovii (Zhuk.) Chennav. | T. syriacum Bowden | Ae. vavilovii (Zhuk.) Chennav. | subsp. vavilovii Zhuk. (6×) | Ae. vavilovii (Zhuk.) Chennav. | |||
| 20 | Ae. juvenalis (Thell.) Eig | Ae. juvenalis (Thell.) Eig | T. juvenale Thell. | Ae. juvenalis (Thell.) Eig | Ae. juvenalis (Thell.) Eig | Ae. juvenalis (Thell.) Eig | Ae. juvenalis (Thell.) Eig | Ae. juvenalis (Thell.) Eig | Ae. turcomanica Roshev. |
| Ae. turcomanica Roshev. | |||||||||
| 21 | Ae. tauschii Coss. | Ae. tauschii Coss. | T. tauschii (Coss.) Schmalh. | Ae. squarrosa L. | Ae. tauschii Coss. | Ae. squarrosa L. | Ae. squarrosa L. | Ae. squarrosa L. | Ae. squarrosa L. |
| subsp. tauschii | subsp. squarrosa | ||||||||
| subsp. strangulata (Eig) Tzvelev | |||||||||
| 22 | Ae. ventricosa Tausch | Ae. ventricosa Tausch. | T. ventricosum Ces., Pass. et Gibelli | Ae. ventricosa Tausch | Ae. ventricosa Tausch | Ae. ventricosa Tausch | Ae. ventricosa Tausch | Ae. ventricosa Tausch | Ae. ventricosa Tausch |
| Subgenus Amblyopyrum | |||||||||
| 23 | Ae. mutica Boiss. | Amblyopyrum muticum (Boiss.) Eig | T. tripsacoides (Jaub. et Spach) Bowden | Ae. mutica Boiss. | Ae. mutica Boiss. | Amblyopyrum muticum (Boiss.) Eig | Ae. mutica Boiss. | Ae. mutica Boiss. | Ae. mutica Boiss. |
| subsp. mutica | var. muticum | var. mutica | subsp. muticum | ||||||
| subsp. loliacea (Jaub et Spach) Zhuk. | var. loliaceum (Jaub. et Spach) Eig | var. loliacea (Jaub. et Spach) Eig | subsp. loliaceum (Jaub. et Spach) Á. Löve | ||||||
| Section/Subgenus | Diploid | Tetraploid | Hexaploid | |||
|---|---|---|---|---|---|---|
| Species | G | Species | G | Species | G | |
| Section Aegilops L. | Ae. umbellulata Zhuk. | U | Ae. biuncialis Vis. | UbMb | Ae. neglecta subsp. recta (Zhuk.) K. Hammer | UnXnNn |
| Ae. columnaris Zhuk. | UcXc | |||||
| Ae. geniculata Roth | UgMg | |||||
| Ae. kotschyi Boiss. | UkSk | |||||
| Ae. neglecta Req. ex Bertol. subsp. neglecta | UnXn | |||||
| Ae. peregrina (Hack.) Maire et Weiller | UpSp | |||||
| Ae. triuncialis L. | UtCt | |||||
| Section Comopyrum (Jaub. et Spach) Zhuk. | Ae. comosa Sibth. et Sm. | M | ||||
| Ae. uniaristata Vis. | N | |||||
| Section Cylindropyrum (Jaub. et Spach) Zhuk. | Ae. markgrafii (Greuter) K. Hammer | C | Ae. cylindrica Host | DcCc | ||
| Section Sitopsis (Jaub. et Spach) Zhuk. | Ae. bicornis (Forssk.) Jaub. et Spach | Sb | ||||
| Ae. longissima Schweinf. et Muschl. | Sl | |||||
| Ae. sharonensis Eig | Ssh | |||||
| Ae. searsii Feldman et Kislev ex K. Hammer | Ss | |||||
| Ae. speltoides Tausch | S | |||||
| Section Vertebrata Zhuk. emend. Kihara | Ae. tauschii Coss. | D | Ae. crassa Boiss. subsp. crassa (4x) | D1Xcr | Ae. crassa Boiss. subsp. crassa (6x) | D1D2Xcr |
| Ae. ventricosa Tausch | DvNv | Ae. vavilovii (Zhuk.) Chennav. | D1XcrSv | |||
| Ae. juvenalis (Thell.) Eig | D1XcrUj | |||||
| Subgenus Amblyopyrum | Ae. mutica Boiss. | T | ||||
3.2. Evolution and Domestication History
3.3. The Wheat Genepool Concept
4. Status of PGR Use for Wheat Improvement
5. PGR of Known Value to Be Incorporated in the Future: Breeders’ Needs
5.1. Diversification of Resistance Genes
5.2. Improved Tolerance to Drought, Heat, and Salinity
5.3. Organic Farming
5.4. Improved Nutritional Quality
6. Exploitation of PGR in Breeding: A Stepwise Approach
7. Approaches to Facilitate Introgressions from PGR
7.1. Introgression of Qualitative Traits from PGR
7.2. Introgression of Quantitative Traits from PGR
7.3. Introgressions from PGR without Known Characteristics
8. Global Initiatives for Promoting the Use of PGR for Crop Improvement
9. Summary and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- IDRC. Facts and Figures on Food and Biodiversity; Ottawa, Canada, 2010. Available online: https://www.idrc.ca/en/research-in-action/facts-figures-food-and-biodiversity (accessed on 21 September 2021).
- FAOSTAT. 2019. Available online: http://www.fao.org/faostat/en/#data/QC (accessed on 23 June 2021).
- European Union. Production, Yields and Productivity; European Union: Brussels, Belgium, 2018; pp. 1–11. [Google Scholar]
- Feldman, M. Origin of cultivated wheat. In The World Wheat Book. A History of Wheat Breeding; Bonjean, A.P., Angus, W.J., Eds.; Tec & Doc/Intersept Ltd.: London, UK; Paris, France; New York, NY, USA, 2001; pp. 3–56. [Google Scholar]
- Le Gouis, J.; Oury, F.-X.; Charmet, G. How changes in climate and agricultural practices influenced wheat production in Western Europe. J. Cereal Sci. 2020, 93, 102960. [Google Scholar] [CrossRef]
- Tidiane Sall, A.; Chiari, T.; Legesse, W.; Seid-Ahmed, K.; Ortiz, R.; van Ginkel, M.; Bassi, F.M. Durum wheat (Triticum durum Desf.): Origin, cultivation and potential expansion in Sub-Saharan Africa. Agronomy 2019, 9, 263. [Google Scholar] [CrossRef]
- Alsaleh, A.; Baloch, F.S.; Nachit, M.; Özkan, H. Phenotypic and genotypic intra-diversity among Anatolian durum wheat “Kunduru” landraces. Biochem. Syst. Ecol. 2016, 65, 9–16. [Google Scholar] [CrossRef]
- Khan, H. Genetic improvement for end-use quality in wheat. In Quality Breeding in Field Crops; Qureshi, A.M.I., Dar, Z.A., Wani, S.H., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 239–253. [Google Scholar]
- Mastrangelo, A.M.; Cattivelli, L. What makes bread and durum wheat different? Trends Plant Sci. 2021, 26, 677–684. [Google Scholar] [CrossRef]
- Mackay, T.F.C.; Falconer, D.S. Introduction to Quantitative Genetics, 4th ed.; Longman Group Ltd.: Harlow, UK, 1996; p. 480. [Google Scholar]
- Bassi, F.M.; Nachit, M.M. Genetic gain for yield and allelic diversity over 35 years of durum wheat breeding at ICARDA. Crop Breed. Genet. Genom. 2019, 1, e190004. [Google Scholar] [CrossRef]
- Tadesse, W.; Sanchez-Garcia, M.; Assefa, S.G.; Amri, A.; Bishaw, Z.; Ogbonnaya, F.C.; Baum, M. Genetic gains in wheat breeding and its role in feeding the world. Crop Breed. Genet. Genom. 2019, 1, e190005. [Google Scholar] [CrossRef]
- Austin, R.B.; Morgan, C.L.; Ford, M.A.; Blackwell, R.D. Contributions to grain yield from pre-anthesis assimilation in tall and dwarf barley phenotypes in two contrasting seasons. Ann. Bot. 1980, 45, 309–319. [Google Scholar] [CrossRef]
- Borlaug, N.E. Wheat breeding and its impact on world food supply. In Proceedings of the 3rd International Wheat Genetics Symposium, Canberra, Australia, 5–9 August 1968; pp. 1–36. [Google Scholar]
- Laidig, F.; Piepho, H.-P.; Drobek, T.; Meyer, U. Genetic and non-genetic long-term trends of 12 different crops in German official variety performance trials and on-farm yield trends. Theor. Appl. Genet. 2014, 127, 2599–2617. [Google Scholar] [CrossRef]
- Slafer, G.; Kernich, G. Have changes in yield (1900-1992) been accompanied by a decreased yield stability in Australian cereal production? Austral. J. Agric. Res. 1996, 47, 323–334. [Google Scholar] [CrossRef]
- Voss-Fels, K.P.; Stahl, A.; Wittkop, B.; Lichthardt, C.; Nagler, S.; Rose, T.; Chen, T.-W.; Zetzsche, H.; Seddig, S.; Majid Baig, M.; et al. Breeding improves wheat productivity under contrasting agrochemical input levels. Nat. Plants 2019, 5, 706–714. [Google Scholar] [CrossRef]
- Grassini, P.; Eskridge, K.M.; Cassman, K.G. Distinguishing between yield advances and yield plateaus in historical crop production trends. Nat. Commun. 2013, 4, 2918. [Google Scholar] [CrossRef]
- Juroszek, P.; von Tiedemann, A. Climate change and potential future risks through wheat diseases: A review. Eur. J. Plant Pathol 2013, 136, 21–33. [Google Scholar] [CrossRef]
- Ray, D.K.; Ramankutty, N.; Mueller, N.D.; West, P.C.; Foley, J.A. Recent patterns of crop yield growth and stagnation. Nat. Commun. 2012, 3, 1293. [Google Scholar] [CrossRef]
- Gerard, G.S.; Crespo-Herrera, L.A.; Crossa, J.; Mondal, S.; Velu, G.; Juliana, P.; Huerta-Espino, J.; Vargas, M.; Rhandawa, M.S.; Bhavani, S.; et al. Grain yield genetic gains and changes in physiological related traits for CIMMYT’s High Rainfall Wheat Screening Nursery tested across international environments. Field Crops Res. 2020, 249, 107742. [Google Scholar] [CrossRef]
- Khoury, C.K.; Brush, S.; Costich, D.E.; Curry, H.; de Haan, S.; Engels, J.M.M.; Guarino, L.; Hoban, S.; Mercer, K.L.; Miller, A.J.; et al. Crop genetic erosion: Understanding and responding to loss of crop diversity. New Phytol. 2021. [Google Scholar] [CrossRef]
- Rutkoski, J.E. Estimation of realized rates of genetic gain and indicators for breeding program assessment. Crop Sci. 2019, 59, 981–993. [Google Scholar] [CrossRef]
- Cobb, J.N.; Juma, R.U.; Biswas, P.S.; Arbelaez, J.D.; Rutkoski, J.; Atlin, G.; Hagen, T.; Quinn, M.; Ng, E.H. Enhancing the rate of genetic gain in public-sector plant breeding programs: Lessons from the breeder’s equation. Theor. Appl. Genet. 2019, 132, 627–645. [Google Scholar] [CrossRef]
- Ru, S.; Bernardo, R. Targeted recombination to increase genetic gain in self-pollinated species. Theor. Appl. Genet. 2019, 132, 289–300. [Google Scholar] [CrossRef]
- Voss-Fels, K.P.; Cooper, M.; Hayes, B.J. Accelerating crop genetic gains with genomic selection. Theor. Appl. Genet. 2019, 132, 669–686. [Google Scholar] [CrossRef]
- van Ginkel, M.; Ortiz, R. Cross the best with the best, and select the best: HELP in breeding selfing crops. Crop Sci. 2018, 58, 17–30. [Google Scholar] [CrossRef]
- Girma, E. Genetic erosion of wheat (Triticum spp.): Concept, research, results and challenges. J. Nat. Sci. Res. 2017, 7, 72–81. [Google Scholar]
- Asseng, S.; Ewert, F.; Martre, P.; Rötter, R.P.; Lobell, D.B.; Cammarano, D.; Kimball, B.A.; Ottman, M.J.; Wall, G.W.; White, J.W.; et al. Rising temperatures reduce global wheat production. Nat. Clim. Chang. 2015, 5, 143–147. [Google Scholar] [CrossRef]
- Daloz, A.S.; Rydsaa, J.H.; Hodnebrog, Ø.; Sillmann, J.; van Oort, B.; Mohr, C.W.; Agrawal, M.; Emberson, L.; Stordal, F.; Zhang, T. Direct and indirect impacts of climate change on wheat yield in the Indo-Gangetic plain in India. J. Agric. Food Res. 2021, 4, 100132. [Google Scholar] [CrossRef]
- Miedaner, T.; Juroszek, P. Climate change will influence disease resistance breeding in wheat in Northwestern Europe. Theor. Appl. Genet. 2021, 134, 1771–1785. [Google Scholar] [CrossRef]
- Dempewolf, H.; Baute, G.; Anderson, J.; Kilian, B.; Smith, C.; Guarino, L. Past and future use of wild relatives in crop breeding. Crop Sci. 2017, 57, 1070–1082. [Google Scholar] [CrossRef]
- Hasan, M.; Hasibuzzaman, A.S.M.; Abdullah, H.M.; Kallol, M.M.H. Genetic and genomic resources and their exploitation for unlocking genetic potential from the wild relatives. In Rediscovery of Genetic and Genomic Resources for Future Food Security; Salgotra, R.K., Zargar, S.M., Eds.; Springer: Singapore, 2020; pp. 193–210. [Google Scholar]
- Nair, K.P. Utilizing CWRs in major food crops to combat global warming. In Combating Global Warming: The Role of Crop Wild Relatives for Food Security; Springer International Publishing: Cham, Switzerland, 2019; pp. 61–78. [Google Scholar]
- Philipp, N.; Weichert, H.; Bohra, U.; Weschke, W.; Schulthess, A.W.; Weber, H. Grain number and grain yield distribution along the spike remain stable despite breeding for high yield in winter wheat. PLoS ONE 2018, 13, e0205452. [Google Scholar] [CrossRef] [PubMed]
- Preece, C.; Peñuelas, J. A return to the wild: Root exudates and food security. Trends Plant Sci. 2020, 25, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Prohens, J.; Gramazio, P.; Plazas, M.; Dempewolf, H.; Kilian, B.; Díez, M.J.; Fita, A.; Herraiz, F.J.; Rodríguez-Burruezo, A.; Soler, S.; et al. Introgressiomics: A new approach for using crop wild relatives in breeding for adaptation to climate change. Euphytica 2017, 213, 158. [Google Scholar] [CrossRef]
- Sharma, M.; Punya; Gupta, B.B. Role of wild relatives for development of climate-resilient varieties. In Rediscovery of Genetic and Genomic Resources for Future Food Security; Salgotra, R.K., Zargar, S.M., Eds.; Springer: Singapore, 2020; pp. 303–314. [Google Scholar]
- Soriano, J.M.; Villegas, D.; Aranzana, M.J.; García del Moral, L.F.; Royo, C. Genetic structure of modern durum wheat cultivars and Mediterranean landraces matches with their agronomic performance. PLoS ONE 2016, 11, e0160983. [Google Scholar] [CrossRef] [PubMed]
- Zaïm, M.; El Hassouni, K.; Gamba, F.; Filali-Maltouf, A.; Belkadi, B.; Sourour, A.; Amri, A.; Nachit, M.; Taghouti, M.; Bassi, F.M. Wide crosses of durum wheat (Triticum durum Desf.) reveal good disease resistance, yield stability, and industrial quality across Mediterranean sites. Field Crops Res. 2017, 214, 219–227. [Google Scholar] [CrossRef]
- Schmolke, M.; Zimmermann, G.; Buerstmayr, H.; Schweizer, G.; Miedaner, T.; Korzun, V.; Ebmeyer, E.; Hartl, L. Molecular mapping of Fusarium head blight resistance in the winter wheat population Dream/Lynx. Theor. Appl. Genet. 2005, 111, 747–756. [Google Scholar] [CrossRef] [PubMed]
- Schmolke, M.; Zimmermann, G.; Schweizer, G.; Miedaner, T.; Korzun, V.; Ebmeyer, E.; Hartl, L. Molecular mapping of quantitative trait loci for field resistance to Fusarium head blight in a European winter wheat population. Plant Breed. 2008, 127, 459–464. [Google Scholar] [CrossRef]
- Zhu, Z.; Hao, Y.; Mergoum, M.; Bai, G.; Humphreys, G.; Cloutier, S.; Xia, X.; He, Z. Breeding wheat for resistance to Fusarium head blight in the Global North: China, USA, and Canada. Crop J. 2019, 7, 730–738. [Google Scholar] [CrossRef]
- Kilian, B.; Dempewolf, H.; Guarino, L.; Werner, P.; Coyne, C.; Warburton, M.L. Crop Science special issue: Adapting agriculture to climate change: A walk on the wild side. Crop Sci. 2021, 61, 32–36. [Google Scholar] [CrossRef]
- Singh, S.; Vikram, P.; Sehgal, D.; Burgueño, J.; Sharma, A.; Singh, S.K.; Sansaloni, C.P.; Joynson, R.; Brabbs, T.; Ortiz, C.; et al. Harnessing genetic potential of wheat germplasm banks through impact-oriented-prebreeding for future food and nutritional security. Sci. Rep. 2018, 8, 12527. [Google Scholar] [CrossRef] [PubMed]
- Leišová-Svobodová, L.; Michel, S.; Tamm, I.; Chourová, M.; Janovska, D.; Grausgruber, H. Diversity and pre-breeding prospects for local adaptation in oat genetic resources. Sustainability 2019, 11, 6950. [Google Scholar] [CrossRef]
- Rubio Teso, M.L.; Iriondo, J.M. In situ conservation assessment of forage and fodder CWR in Spain using phytosociological associations. Sustainability 2019, 11, 5882. [Google Scholar] [CrossRef]
- El Haddad, N.; Kabbaj, H.; Zaïm, M.; El Hassouni, K.; Tidiane Sall, A.; Azouz, M.; Ortiz, R.; Baum, M.; Amri, A.; Gamba, F.; et al. Crop wild relatives in durum wheat breeding: Drift or thrift? Crop Sci. 2021, 61, 37–54. [Google Scholar] [CrossRef]
- Sharma, S.; Paul, P.J.; Kumar, C.V.S.; Rao, P.J.; Prashanti, L.; Muniswamy, S.; Sharma, M. Evaluation and identification of promising introgression lines derived from wild Cajanus species for broadening the genetic base of cultivated pigeonpea [Cajanus cajan (L.) Millsp.]. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef]
- Aberkane, H.; Kishii, M.; Amri, A.; Payne, T.S.; Smale, M.; Jamora, N. Reaching into the Past to Tackle New Challenges: Improving Wheat by Conserving Wild ‘Goat Grass’; CGIAR Genebank Platform; Crop Trust|ICARDA|CIMMYT: Bonn, Germany, 2019; p. 4. [Google Scholar]
- Hoisington, D.; Khairallah, M.; Reeves, T.; Ribaut, J.V.; Skovmand, B.; Taba, S.; Warburton, M. Plant genetic resources: What can they contribute toward increased crop productivity? Proc. Natl. Acad. Sci. USA 1999, 96, 5937–5943. [Google Scholar] [CrossRef]
- Sharma, S. Prebreeding using wild species for genetic enhancement of grain legumes at ICRISAT. Crop Sci. 2017, 57, 1132–1144. [Google Scholar] [CrossRef]
- Sharma, S.; Sharma, R.; Govindaraj, M.; Mahala, R.S.; Satyavathi, C.T.; Srivastava, R.K.; Gumma, M.K.; Kilian, B. Harnessing wild relatives of pearl millet for germplasm enhancement: Challenges and opportunities. Crop Sci. 2021, 61, 177–200. [Google Scholar] [CrossRef]
- Sharma, S.; Sharma, R.; Pujar, M.; Yadav, D.; Yadav, Y.; Rathore, A.; Mahala, R.S.; Singh, I.; Verma, Y.; Deora, V.S.; et al. Use of wild Pennisetum species for improving biotic and abiotic stress tolerance in pearl millet. Crop Sci. 2021, 61, 289–304. [Google Scholar] [CrossRef]
- Vanavermaete, D.; Fostier, J.; Maenhout, S.; De Baets, B. Deep scoping: A breeding strategy to preserve, reintroduce and exploit genetic variation. Theor. Appl. Genet. 2021. [Google Scholar] [CrossRef] [PubMed]
- FAO. Available online: www.fao.org/wiews-archive/germplasm_report.jsp (accessed on 14 June 2021).
- Knüpffer, H. Triticeae genetic resources in ex situ genebank collections. In Genetics and Genomics of the Triticeae, 1st ed.; Muehlbauer, G.J., Feuillet, C., Eds.; Plant Genetics and Genomics: Crops and Models; Springer: New York, NY, USA, 2009; Volume 7, part 1 (Genetics of the Triticeae); pp. 31–79. [Google Scholar]
- Kimber, G.; Feldman, M. Wild Wheat, an Introduction; Special Report; College of Agriculture, University of Missouri-Columbia: Columbia, MO, USA, 1987; Volume 353, p. 142. [Google Scholar]
- Kishii, M. An update of recent use of Aegilops species in wheat breeding. Front. Plant Sci. 2019, 10, 585. [Google Scholar] [CrossRef] [PubMed]
- Kilian, B.; Mammen, K.; Millet, E.; Sharma, R.; Graner, A.; Salamini, F.; Hammer, K.; Özkan, H. Aegilops. In Wild Crop Relatives: Genomics and Breeding Resources. Cereals; Kole, C., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 1–76. [Google Scholar]
- Castaneda-Alvarez, N.P.; Khoury, C.K.; Achicanoy, H.A.; Bernau, V.; Dempewolf, H.; Eastwood, R.J.; Guarino, L.; Harker, R.H.; Jarvis, A.; Maxted, N.; et al. Global conservation priorities for crop wild relatives. Nat. Plants 2016, 2, 16022. [Google Scholar] [CrossRef] [PubMed]
- Shaw, P.D.; Weise, S.; Obreza, M.; Raubach, S.; Mccouch, S.; Kilian, B.; Werner, P. Database solutions for genebanks and germplasm collections. In Plant Genetic Resources in the 21th Century. The OMICS Era; Ghamkhar, K., Williams, W., Brown, A.H.D., Eds.; Apple Academic Press, Inc.: Palm Bay, FL, USA, 2021; in press. [Google Scholar]
- Weise, S.; Oppermann, M.; Maggioni, L.; van Hintum, T.; Knüpffer, H. EURISCO: The European search catalogue for plant genetic resources. Nucleic Acids Res. 2017, 45, D1003–D1008. [Google Scholar] [CrossRef] [PubMed]
- Raubach, S.; Kilian, B.; Dreher, K.; Amri, A.; Bassi, F.M.; Boukar, O.; Cook, D.; Cruickshank, A.; Fatokun, C.; El Haddad, N.; et al. From bits to bites: Advancement of the Germinate platform to support prebreeding informatics for crop wild relatives. Crop Sci. 2021, 61, 1538–1566. [Google Scholar] [CrossRef]
- Shaw, P.D.; Raubach, S.; Hearne, S.J.; Dreher, K.; Bryan, G.; McKenzie, G.; Milne, I.; Stephen, G.; Marshall, D.F. Germinate 3: Development of a common platform to support the distribution of experimental data on crop wild relatives. Crop Sci. 2017, 57, 1259–1273. [Google Scholar] [CrossRef]
- Dash, S.; Campbell, J.D.; Cannon, E.K.S.; Cleary, A.M.; Huang, W.; Kalberer, S.R.; Karingula, V.; Rice, A.G.; Singh, J.; Umale, P.E.; et al. Legume information system (LegumeInfo.org): A key component of a set of federated data resources for the legume family. Nucleic Acids Res. 2016, 44, D1181–D1188. [Google Scholar] [CrossRef]
- König, P.; Beier, S.; Basterrechea, M.; Schüler, D.; Arend, D.; Mascher, M.; Stein, N.; Scholz, U.; Lange, M. BRIDGE—A visual analytics Web tool for barley genebank genomics. Front. Plant Sci. 2020, 11. [Google Scholar] [CrossRef]
- Dorofeev, V.F.; Filatenko, A.A.; Migushova, E.F.; Udachin, R.A.; Jakubziner, M.M. Flora of Cultivated Plants, Volume 1, Wheat; Brezhnev, D.D., Ed.; Kolos: Leningrad, Russia, 1979; p. 346. (In Russian) [Google Scholar]
- Hammer, K.; Gladis, T. Notes on infraspecific nomenclature and classifications of cultivated plants in Compositae, Cruciferae, Cucurbitaceae, Gramineae (with a remark on Triticum dicoccon Schrank) and Leguminosae. Genet. Resour. Crop Evol. 2014, 61, 1455–1467. [Google Scholar] [CrossRef]
- Key, J.M. A plant breeder’s perspective on taxonomy of cultivated plants. Biol. Zentralbl. 1988, 107, 369–379. [Google Scholar]
- Schiemann, E. Weizen Roggen Gerste. Systematik, Geschichte und Verwendung; Fischer: Jena, Germany, 1948; p. 102. [Google Scholar]
- Van Slageren, M.W. Wild Wheats: A Monograph of Aegilops L. and Amblyopyrum (Jaub. et Spach) Eig (Poaceae); Wageningen Agricultural University: Wageningen, The Netherlands; ICARDA: Aleppo, Syria, 1994; p. 514. [Google Scholar]
- Bowden, W.M. The taxonomy and nomenclature of the wheats, barleys and ryes and their relatives. Can. J. Bot. 1959, 37, 657–684. [Google Scholar] [CrossRef]
- Kimber, G.; Sears, E.R. Assignment of genome symbols in the Triticeae. In Proceedings of the 6th International Wheat Genetics Symposium, Kyoto, Japan, 28 November–3 December 1983; pp. 1195–1196. [Google Scholar]
- Chennaveeraiah, M.S. Karyomorphologic and cytotaxonomic studies in Aegilops. Acta Horti Gotobg. 1960, 23, 85–186. [Google Scholar]
- Eig, A. Monographisch-kritische Übersicht der Gattung Aegilops. Repert. Spec. Nov. Regni Veg. Beih. Berl. 1929, 55, 228. [Google Scholar]
- Hammer, K. Vorarbeiten zur monographischen Darstellung von Wildpflanzensortimenten: Aegilops L. Kulturpflanze 1980, 28, 33–180. [Google Scholar] [CrossRef]
- Hammer, K. Zur Taxonomie und Nomenklatur der Gattung Aegilops L. Feddes Rep. 1980, 91, 225–258. [Google Scholar] [CrossRef]
- Kihara, H. Considerations on the evolution and distribution of Aegilops species based on the analyser-method. Cytologia 1954, 19, 336–357. [Google Scholar] [CrossRef]
- Witcombe, J.R. A Guide to the Species of Aegilops L. Their Taxonomy, Morphology and Distribution; IBPGR Secretariat: Rome, Italy, 1983; p. 74. [Google Scholar]
- Zhukovsky, P.M. A critical-systematical survey of the species of the genus Aegilops L. Bull. Appl. Bot. Genet. Plant Breed 1928, 18, 417–609. [Google Scholar]
- Dvořák, J. Genome analysis in the Triticum-Aegilops alliance. In Proceedings of the 9th International Wheat Genetics Symposium, Saskatoon, SK, Canada, 2–7 August 1998; pp. 8–11. [Google Scholar]
- Kimber, G.; Tsunewaki, K. Genome symbols and plasma types in the wheat group. In Proceedings of the 7th International Wheat Genetics Symposium, Cambridge, UK, 13–19 July 1988; pp. 1209–1210. [Google Scholar]
- Edet, O.U.; Gorafi, Y.S.A.; Nasuda, S.; Tsujimoto, H. DArTseq-based analysis of genomic relationships among species of tribe Triticeae. Sci. Rep. 2018, 8, 16397. [Google Scholar] [CrossRef] [PubMed]
- Zeibig, F.; Kilian, B.; Frei, M. The grain quality of wheat wild relatives in the evolutionary context. Theor. Appl. Genet. 2021, in press. [Google Scholar]
- Bernhardt, N.; Brassac, J.; Dong, X.; Willing, E.-M.; Poskar, C.H.; Kilian, B.; Blattner, F.R. Genome-wide sequence information reveals recurrent hybridization among diploid wheat wild relatives. Plant J. 2020, 102, 493–506. [Google Scholar] [CrossRef] [PubMed]
- Bernhardt, N.; Brassac, J.; Kilian, B.; Blattner, F.R. Dated tribe-wide whole chloroplast genome phylogeny indicates recurrent hybridizations within Triticeae. BMC Evol. Biol. 2017, 17, 141. [Google Scholar] [CrossRef] [PubMed]
- Glémin, S.; Scornavacca, C.; Dainat, J.; Burgarella, C.; Viader, V.; Ardisson, M.; Sarah, G.; Santoni, S.; David, J.; Ranwez, V. Pervasive hybridizations in the history of wheat relatives. Sci. Adv. 2019, 5, eaav9188. [Google Scholar] [CrossRef] [PubMed]
- Li, L.-F.; Zhang, Z.-B.; Wang, Z.-H.; Li, N.; Sha, Y.; Wang, X.-F.; Ding, N.; Li, Y.; Zhao, J.; Wu, Y.; et al. Genome sequences of the five Sitopsis species of Aegilops and the origin of polyploid wheat B-subgenome. bioRxiv 2021. [Google Scholar] [CrossRef]
- Marcussen, T.; Sandve, S.R.; Heier, L.; Spannagl, M.; Pfeifer, M.; Jakobsen, K.S.; Wulff, B.B.H.; Steuernagel, B.; Mayer, K.F.X.; Olsen, O.-A. Ancient hybridizations among the ancestral genomes of bread wheat. Science 2014, 345. [Google Scholar] [CrossRef]
- Walkowiak, S.; Gao, L.; Monat, C.; Haberer, G.; Kassa, M.T.; Brinton, J.; Ramirez-Gonzalez, R.H.; Kolodziej, M.C.; Delorean, E.; Thambugala, D.; et al. Multiple wheat genomes reveal global variation in modern breeding. Nature 2020, 588, 277–283. [Google Scholar] [CrossRef]
- Feldman, M.; Levy, A.A. Origin and evolution of wheat and related Triticeae species. In Alien Introgression in Wheat: Cytogenetics, Molecular Biology, and Genomics; Molnár-Láng, M., Ceoloni, C., Doležel, J., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 21–76. [Google Scholar]
- Lilienfeld, F.A. H. Kihara: Genome-analysis in Triticum and Aegilops. X. Concluding review. Cytologia 1951, 16, 101–123. [Google Scholar] [CrossRef]
- Tsunewaki, K. Plasmon analysis in the Triticum-Aegilops complex. Breed. Sci. 2009, 59, 455–470. [Google Scholar] [CrossRef]
- Huang, S.; Sirikhachornkit, A.; Su, X.; Faris, J.; Gill, B.; Haselkorn, R.; Gornicki, P. Genes encoding plastid acetyl-CoA carboxylase and 3-phosphoglycerate kinase of the Triticum/Aegilops complex and the evolutionary history of polyploid wheat. Proc. Natl. Acad. Sci. USA 2002, 99, 8133–8138. [Google Scholar] [CrossRef]
- Middleton, C.P.; Senerchia, N.; Stein, N.; Akhunov, E.D.; Keller, B.; Wicker, T.; Kilian, B. Sequencing of chloroplast genomes from wheat, barley, rye and their relatives provides a detailed insight into the evolution of the Triticeae tribe. PLoS ONE 2014, 9, e85761. [Google Scholar] [CrossRef]
- Avni, R.; Lux, T.; Minz-Dub, A.; Millet, E.; Sela, H.; Distelfeld, A.; Deek, J.; Yu, G.; Steuernagel, B.; Pozniak, C.; et al. Genome sequences of Aegilops species of section Sitopsis reveal phylogenetic relationships and provide resources for wheat improvement. bioRxiv 2021. [Google Scholar] [CrossRef]
- Dvořák, J.; Zhang, H. Variation in repeated nucleotide sequences sheds light on the phylogeny of the wheat B and G genomes. Proc. Natl. Acad. Sci. USA 1990, 87, 9640–9644. [Google Scholar] [CrossRef] [PubMed]
- Ogihara, Y.; Tsunewaki, K. Diversity and evolution of chloroplast DNA in Triticum and Aegilops as revealed by restriction fragment analysis. Theor. Appl. Genet. 1988, 76, 321–332. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, P.; Stebbins, G.L. Morphological evidence concerning the origin of the B genome in wheat. Amer. J. Bot. 1956, 43, 297–304. [Google Scholar] [CrossRef]
- Gornicki, P.; Zhu, H.; Wang, J.; Challa, G.S.; Zhang, Z.; Gill, B.S.; Li, W. The chloroplast view of the evolution of polyploid wheat. New Phytol. 2014, 204, 704–714. [Google Scholar] [CrossRef]
- Dvořák, J.; Akhunov, E.D. Tempos of gene locus deletions and duplications and their relationship to recombination rate during diploid and polyploid evolution in the Aegilops-Triticum alliance. Genetics 2005, 171, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.C.; Yang, Z.L.; You, F.M.; Kawahara, T.; Waines, J.G.; Dvořák, J. The structure of wild and domesticated emmer wheat populations, gene flow between them, and the site of emmer domestication. Theor. Appl. Genet. 2007, 114, 947–959. [Google Scholar] [CrossRef] [PubMed]
- Mori, N.; Ishii, T.; Ishido, T.; Hirosawa, S.; Watatani, H.; Kawahara; Nesbitt, M.; Belay, G.; Takumi, S.; Ogihara, Y.; et al. Origin of domesticated emmer and common wheat inferred from chloroplast DNA fingerprinting. In Proceedings of the Proceedings of the 10th International Wheat Genetic Symposium, Paestum, Italy, 1–6 September 2003; pp. 25–28. [Google Scholar]
- Özkan, H.; Brandolini, A.; Pozzi, C.; Effgen, S.; Wunder, J.; Salamini, F. A reconsideration of the domestication geography of tetraploid wheats. Theor. Appl. Genet. 2005, 110, 1052–1060. [Google Scholar] [CrossRef] [PubMed]
- Özkan, H.; Willcox, G.; Graner, A.; Salamini, F.; Kilian, B. Geographic distribution and domestication of wild emmer wheat (Triticum dicoccoides). Genet. Resour. Crop Evol. 2011, 58, 11–53. [Google Scholar] [CrossRef]
- Civáň, P.; Ivaničová, Z.; Brown, T.A. Reticulated origin of domesticated emmer wheat supports a dynamic model for the emergence of agriculture in the Fertile Crescent. PLoS ONE 2013, 8, e81955. [Google Scholar] [CrossRef]
- Maccaferri, M.; Harris, N.S.; Twardziok, S.O.; Pasam, R.K.; Gundlach, H.; Spannagl, M.; Ormanbekova, D.; Lux, T.; Prade, V.M.; Milner, S.G.; et al. Durum wheat genome highlights past domestication signatures and future improvement targets. Nat. Genet. 2019, 51, 885–895. [Google Scholar] [CrossRef]
- Pont, C.; Leroy, T.; Seidel, M.; Tondelli, A.; Duchemin, W.; Armisen, D.; Lang, D.; Bustos-Korts, D.; Goué, N.; Balfourier, F.; et al. Tracing the ancestry of modern bread wheats. Nat. Genet. 2019, 51, 905–911. [Google Scholar] [CrossRef]
- Gioia, T.; Nagel, K.A.; Beleggia, R.; Fragasso, M.; Ficco, D.B.; Pieruschka, R.; De Vita, P.; Fiorani, F.; Papa, R. Impact of domestication on the phenotypic architecture of durum wheat under contrasting nitrogen fertilization. J. Exp. Bot. 2015, 66, 5519–5530. [Google Scholar] [CrossRef]
- Vavilov, N.I. The origin, variation, immunity and breeding of cultivated plants. Soil Sci. 1951, 72, 482. [Google Scholar] [CrossRef]
- Kabbaj, H.; Sall, A.T.; Al-Abdallat, A.; Geleta, M.; Amri, A.; Filali-Maltouf, A.; Belkadi, B.; Ortiz, R.; Bassi, F.M. Genetic diversity within a global panel of durum wheat (Triticum durum) landraces and modern germplasm reveals the history of alleles exchange. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef]
- Martínez-Moreno, F.; Solís, I.; Noguero, D.; Blanco, A.; Özberk, İ.; Nsarellah, N.; Elias, E.; Mylonas, I.; Soriano, J.M. Durum wheat in the Mediterranean Rim: Historical evolution and genetic resources. Genet. Resour. Crop Evol. 2020, 67, 1415–1436. [Google Scholar] [CrossRef]
- Mazzucotelli, E.; Sciara, G.; Mastrangelo, A.M.; Desiderio, F.; Xu, S.S.; Faris, J.; Hayden, M.J.; Tricker, P.J.; Özkan, H.; Echenique, V.; et al. The Global Durum Wheat Panel (GDP): An international platform to identify and exchange beneficial alleles. Front. Plant Sci. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Mengistu, D.K.; Kidane, Y.G.; Catellani, M.; Frascaroli, E.; Fadda, C.; Pè, M.E.; Dell’Acqua, M. High-density molecular characterization and association mapping in Ethiopian durum wheat landraces reveals high diversity and potential for wheat breeding. Plant Biotech. J. 2016, 14, 1800–1812. [Google Scholar] [CrossRef] [PubMed]
- Mengistu, D.K.; Kiros, A.Y.; Pè, M.E. Phenotypic diversity in Ethiopian durum wheat (Triticum turgidum var. durum) landraces. Crop J. 2015, 3, 190–199. [Google Scholar] [CrossRef]
- Badaeva, E.D.; Badaev, N.S.; Gill, B.S.; Filatenko, A.A. Intraspecific karyotype divergence in Triticum araraticum (Poaceae). Plant Syst. Evol. 1994, 192, 117–145. [Google Scholar] [CrossRef]
- Nave, M.; Taş, M.; Raupp, J.; Tiwari, V.K.; Özkan, H.; Poland, J.; Hale, I.; Komatsuda, T.; Distelfeld, A. The independent domestication of Timopheev’s wheat: Insights from haplotype analysis of the Brittle rachis 1 (BTR1-A) gene. Genes 2021, 12, 338. [Google Scholar] [CrossRef] [PubMed]
- Badaeva, E.D.; Konovalov, F.A.; Knüpffer, H.; Fricano, A.; Ruban, A.S.; Kehel, Z.; Zoshchuk, S.A.; Surzhikov, S.A.; Neumann, K.; Graner, A.; et al. Genetic diversity, distribution and domestication history of the neglected GGAtAt genepool of wheat. Theor. Appl. Genet. 2021. [Google Scholar] [CrossRef]
- Kihara, H. Die Entdeckung des DD-Analysators beim Weizen. Agric. Hortic. Tokio 1944, 19, 889–890. [Google Scholar]
- McFadden, E.S.; Sears, E.R. The origin of Triticum spelta and its free-threshing hexaploid relatives. J. Hered. 1946, 37, 81–107. [Google Scholar] [CrossRef]
- Singh, N.; Wu, S.; Tiwari, V.; Sehgal, S.; Raupp, J.; Wilson, D.; Abbasov, M.; Gill, B.; Poland, J. Genomic analysis confirms population structure and identifies inter-lineage hybrids in Aegilops tauschii. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef]
- Dvořák, J.; Luo, M.C.; Yang, Z.L.; Zhang, H.B. The structure of the Aegilops tauschii genepool and the evolution of hexaploid wheat. Theor. Appl. Genet. 1998, 97, 657–670. [Google Scholar] [CrossRef]
- Wang, J.; Luo, M.-C.; Chen, Z.; You, F.M.; Wei, Y.; Zheng, Y.; Dvořák, J. Aegilops tauschii single nucleotide polymorphisms shed light on the origins of wheat D-genome genetic diversity and pinpoint the geographic origin of hexaploid wheat. New Phytol. 2013, 198, 925–937. [Google Scholar] [CrossRef]
- Badaeva, E.D.; Fisenko, A.V.; Surzhikov, S.A.; Yankovskaya, A.A.; Chikida, N.N.; Zoshchuk, S.A.; Belousova, M.K.; Dragovich, A.Y. Genetic heterogeneity of a diploid grass Aegilops tauschii revealed by chromosome banding methods and electrophoretic analysis of the seed storage proteins (gliadins). Russ. J. Genet. 2019, 55, 1315–1329. [Google Scholar] [CrossRef]
- Zhao, L.; Ning, S.; Yi, Y.; Zhang, L.; Yuan, Z.; Wang, J.; Zheng, Y.; Hao, M.; Liu, D. Fluorescence in situ hybridization karyotyping reveals the presence of two distinct genomes in the taxon Aegilops tauschii. BMC Genom. 2018, 19, 3. [Google Scholar] [CrossRef]
- Petersen, G.; Seberg, O.; Yde, M.; Berthelsen, K. Phylogenetic relationships of Triticum and Aegilops and evidence for the origin of the A, B, and D genomes of common wheat (Triticum aestivum). Mol. Phyl. Evol. 2006, 39, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Renfrew, J.M. Palaeoethnobotany—The Prehistoric Food Plants of the Near East and Europe; Columbia Univ. Pr.: London, UK, 1973; p. 248. [Google Scholar]
- Badaeva, E.D.; Ruban, A.S.; Zoshchuk, S.A.; Surzhikov, S.A.; Knüpffer, H.; Kilian, B. Molecular cytogenetic characterization of Triticum timopheevii chromosomes provides new insight on genome evolution of T. zhukovskyi. Plant Syst. Evol. 2016, 302, 943–956. [Google Scholar] [CrossRef]
- Menabde, V.L.; Ericzjan, A.A. To the investigation of Georgian wheat Zanduri. Proc. Georg. Acad. Sci. 1960, 25, 731–732. (In Georgian) [Google Scholar]
- Mitrofanova, O.; Badaeva, E.; Salina, E. Triticum timopheevii, T. araraticum and T. zhukovskyi, bread and durum wheat relatives carrying the G genome. In The World Wheat Book: A History of Wheat Breeding; Bonjean, A., Angus, W., van Ginkel, J.M., Eds.; Lavoisier Tec & Doc: Paris, France, 2016; Volume 3, pp. 1167–1228. [Google Scholar]
- Genesys. Available online: https://www.genesys-pgr.org/a/v2R2VmqW782 (accessed on 21 August 2021).
- Kilian, B.; Özkan, H.; Pozzi, C.; Salamini, F. Domestication of the Triticeae in the Fertile Crescent. In Genetics and Genomics of the Triticeae, 1st ed.; Muehlbauer, G.J., Feuillet, C., Eds.; Plant Genetics and Genomics: Crops and Models; Springer: New York, NY, USA, 2009; Volume 7, part 1 (Genetics of the Triticeae); pp. 81–119. [Google Scholar]
- Peng, J.H.; Sun, D.; Nevo, E. Domestication evolution, genetics and genomics in wheat. Mol. Biol. Evol. 2011, 28, 281. [Google Scholar] [CrossRef]
- Shewry, P.R. Wheat. J. Exp. Bot. 2009, 60, 1537–1553. [Google Scholar] [CrossRef]
- Zohary, D.; Hopf, M.; Weiss, E. Domestication of plants in the Old World, 4th ed.; Oxford Univ. Press: Oxford, UK, 2012; p. 264. [Google Scholar]
- Harlan, J.R.; de Wet, J.M.J. Toward a rational classification of cultivated plants. Taxon 1971, 20, 509–517. [Google Scholar] [CrossRef]
- Feldman, M.; Sears, E.R. The wild gene resources of wheat. Sci. Am. 1981, 244, 102–113. [Google Scholar] [CrossRef]
- Jauhar, P.P. Alien gene transfer and genetic enrichment of wheat. In Biodiversity and Wheat Improvement; Damania, A.B., Ed.; John Wiley and Sons: Chichester, UK, 1993; pp. 103–119. [Google Scholar]
- Kihara, H. Cytologische und genetische Studien bei wichtigen Getreidearten mit besonderer Rücksicht auf das Verhalten der Chromosomen und die Sterilität in den Bastarden. Mem. Cell. Sci. Kyoto Imp. Univ. Ser. B 1924, 1, 1–200. [Google Scholar]
- Molnár-Láng, M. The crossability of wheat with rye and other related species. In Alien Introgression in Wheat; Molnár-Láng, M., Ceoloni, C., Doležel, J., Eds.; Springer: Cham, Switzerland, 2015; pp. 103–120. [Google Scholar]
- Molnár-Láng, M.; Molnár, I.; Szakács, É.; Linc, G.; Bedö, Z. Production and molecular cytogenetic identification of wheat-alien hybrids and introgression lines. In Genomics of Plant Genetic Resources. Volume 1. Managing, Sequencing and Mining Genetic Resources; Tuberosa, R., Graner, A., Frison, E., Eds.; Genomics of plant genetic resources; Springer: New York, NY, USA; Heidelberg, Germany; Dordrecht, The Netherlands; London, UK, 2014; Volume 1, pp. 255–284. [Google Scholar]
- Moore, G. The control of recombination in wheat by Ph1 and its use in breeding. Methods Mol. Biol. 2014, 1145, 143–153. [Google Scholar] [CrossRef]
- Mujeeb-Kazi, A.; Gilchrist, L.I.; Fuentes-Davila, G.; Delgado, R. Production and utilization of D-genome synthetic hexaploids in wheat improvement. In Proceedings of the 3rd International Triticeae Symposium, ICARDA, Aleppo, Syria, 4-8 May 1997; Jaradat, A.A., Ed.; Science Publishers: New York, NY, USA, 1997; pp. 369–374. [Google Scholar]
- Riley, R.; Unrau, J.; Chapman, V. Evidence on the origin of the B genome of wheat. J. Hered. 1958, 49, 91–98. [Google Scholar] [CrossRef]
- Sharma, H.C.; Gill, B.S. Current status of wide hybridization in wheat. Euphytica 1983, 32, 17–31. [Google Scholar] [CrossRef]
- Salvi, S.; Porfiri, O.; Ceccarelli, S. Nazareno Strampelli, the ‘Prophet’ of the green revolution. J. Agric. Sci. 2012, 151, 1–5. [Google Scholar] [CrossRef]
- Borojevic, K.; Borojevic, K. Historic role of the wheat variety Akakomugi in southern and central European wheat breeding programs. Breed. Sci. 2005, 55, 253–256. [Google Scholar] [CrossRef][Green Version]
- Dreisigacker, S.; Zhang, P.; Warburton, M.L.; Skovmand, B.; Hoisington, D.; Melchinger, A.E. Genetic diversity among and within CIMMYT wheat landrace accessions investigated with SSRs and implications for plant genetic resources management. Crop Sci. 2005, 45, 653–661. [Google Scholar] [CrossRef]
- Matsumoto, T. Norin 10—A dwarf winter wheat variety. Jap. Agric. Res. Quart. 1968, 3, 22–26. [Google Scholar]
- Reitz, L.P.; Salmon, S.C. Origin, history, and use of Norin 10 wheat. Crop Sci. 1968, 8, 686–689. [Google Scholar] [CrossRef]
- Lopes, M.S.; El-Basyoni, I.; Baenziger, P.S.; Singh, S.; Royo, C.; Ozbek, K.; Aktas, H.; Ozer, E.; Ozdemir, F.; Manickavelu, A.; et al. Exploiting genetic diversity from landraces in wheat breeding for adaptation to climate change. J. Exp. Bot. 2015, 66, 3477–3486. [Google Scholar] [CrossRef]
- Hede, A.R.; Skovmand, B.; Reynolds, M.P.; Crossa, J.; Vilhelmsen, A.L.; Stolen, O. Evaluating genetic diversity for heat tolerance traits in Mexican wheat landraces. Genet. Resour. Crop Evol. 1999, 46, 37–45. [Google Scholar] [CrossRef]
- Vikram, P.; Franco, J.; Burgueno-Ferreira, J.; Li, H.H.; Sehgal, D.; Saint Pierre, C.; Ortiz, C.; Sneller, C.; Tattaris, M.; Guzman, C.; et al. Unlocking the genetic diversity of Creole wheats. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Royo, C.; Briceño-Félix, G.A. Spanish wheat pool. In The World Wheat Book: A History of Wheat Breeding; Bonjean, A.P., Angus, W.J., van Ginkel, M., Eds.; Lavoisier Publishing Inc.: Paris, France, 2011; Volume 2, pp. 121–154. [Google Scholar]
- Taranto, F.; D’Agostino, N.; Rodriguez, M.; Pavan, S.; Minervini, A.P.; Pecchioni, N.; Papa, R.; De Vita, P. Whole genome scan reveals molecular signatures of divergence and selection related to important traits in durum wheat germplasm. Front. Genet. 2020, 11. [Google Scholar] [CrossRef]
- Bieńkowska, T.; Suchowilska, E.; Wiwart, M. Triticum polonicum L. as promising source material for breeding new wheat cultivars. J. Elem. 2020, 25, 237–248. [Google Scholar]
- Bienkowska, T.; Suchowilska, E.; Kandler, W.; Krska, R.; Wiwart, M. Triticum polonicum L. as potential source material for the biofortification of wheat with essential micronutrients. Plant Genet. Resour.-Charact. Util. 2019, 17, 213–220. [Google Scholar] [CrossRef]
- Chen, Q.; Kang, H.-Y.; Fan, X.; Wang, Y.; Sha, L.-N.; Zhang, H.-Q.; Zhong, M.-Y.; Xu, L.-L.; Zeng, J.; Yang, R.-W.; et al. Evolutionary history of Triticum petropavlovskyi Udacz. et Migusch. inferred from the sequences of the 3-phosphoglycerate kinase gene. PLoS ONE 2013, 8, e71139. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.Y.; Fan, X.; Zhang, H.Q.; Sha, L.N.; Sun, G.L.; Zhou, Y.H. The origin of Triticum petropavlovskyi Udacz. et Migusch.: Demonstration of the utility of the genes encoding plastid acetyl-CoA carboxylase sequence. Mol. Biol. Evol. 2010, 25, 381–395. [Google Scholar] [CrossRef]
- Akond, A.S.M.G.M.; Watanabe, N. Genetic variation among Portuguese landraces of ‘Arrancada’ wheat and Triticum petropavlovskyi by AFLP-based assessment. Genet. Resour. Crop Evol. 2005, 52, 619–628. [Google Scholar] [CrossRef]
- Martynov, S.P.; Dobrotvorskaya, T.V.; Krupnov, V.A. Analysis of the distribution of Triticum timopheevii Zhuk. genetic material in common wheat varieties (Triticum aestivum L.). Russ. J. Genet. 2018, 54, 166–175. [Google Scholar] [CrossRef]
- McIntosh, R.A.; Yamazaki, Y.; Dubkovsky, G.; Rogers, J.; Morris, C.F.; Appels, R.; Xia, X.C. Catalogue of gene symbols for wheat. In Proceedings of the 12th International Wheat Genetics Symposium, Yokohama, Japan, 8–13 September 2013; p. 395. [Google Scholar]
- Maan, S.S.; Lucken, K.A. Interacting male sterility-male fertility restoration systems for hybrid wheat research. Crop Sci. 1972, 12, 360–364. [Google Scholar] [CrossRef]
- Mikó, P.; Megyeri, M.; Kovács, G. Characterization of Triticum timopheevii Zhuk. gene bank accessions to gain useful materials for organic wheat breeding. In Proceedings of the Agrisafe Final Conference, Climate Change: Challenges and Opportunities in Agriculture, Budapest, Hungary, 21–23 March 2011; pp. 90–93. [Google Scholar]
- Wurschum, T.; Leiser, W.L.; Weissmann, S.; Maurer, H.P. Genetic architecture of male fertility restoration of Triticum timopheevii cytoplasm and fine-mapping of the major restorer locus Rf3 on chromosome 1B. Theor. Appl. Genet. 2017, 130, 1253–1266. [Google Scholar] [CrossRef]
- Villareal, R.L.; Rajaram, S.; Mujeeb-Kazi, A.; Del Toro, E. The effect of chromosome 1B/1R translocation on the yield potential of certain spring wheats (Triticum aestivum L.). Plant Breed. 1991, 106, 77–81. [Google Scholar] [CrossRef]
- Schlegel, G.; Schlegel, R. A compendium of reciprocal intervarietal translocations in hexaploid wheat. Genet. Resour. Crop Evol. 1989, 37, 163–176. [Google Scholar] [CrossRef]
- Friebe, B.; Jiang, J.; Raupp, W.J.; McIntosh, R.A.; Gill, B.S. Characterization of wheat-alien translocations conferring resistance to diseases and pests: Current status. Euphytica 1996, 91, 59–87. [Google Scholar] [CrossRef]
- Rabinovich, S.V. Importance of wheat-rye translocations for breeding modern cultivar of Triticum aestivum L. Euphytica 1998, 100, 323–340. [Google Scholar] [CrossRef]
- Badaeva, E.D.; Dedkova, O.S.; Gay, G.; Pukhalskyi, V.A.; Zelenin, A.V.; Bernard, S.; Bernard, M. Chromosomal rearrangements in wheat: Their types and distribution. Genome 2007, 50, 907–926. [Google Scholar] [CrossRef] [PubMed]
- Ren, T.H.; Chen, F.; Yan, B.J.; Zhang, H.Q.; Ren, Z.L. Genetic diversity of wheat-rye 1BL.1RS translocation lines derived from different wheat and rye sources. Euphytica 2010, 183, 133–146. [Google Scholar] [CrossRef]
- Skovmand, B.; Villareal, R.L.; van Ginkel, M.; Rajaram, S.; Ferrara, G.O. Semidwarf Bread Wheats: Names, Parentages, Pedigrees and Origins; CIMMYT: Ciudad de México, Mexico, 1997; p. 77. [Google Scholar]
- Doussinault, G.; Delibes, A.; Sanchez-Monge, R.; Garcia-Olmedo, F. Transfer of a dominant gene for resistance to eyespot disease from a wild grass to hexaploid wheat. Nature 1983, 303, 698–700. [Google Scholar] [CrossRef]
- Bariana, H.S.; McIntosh, R.A. Characterisation and origin of rust and powdery mildew resistance genes in VPM1 wheat. Euphytica 1994, 76, 53–61. [Google Scholar] [CrossRef]
- Cruz, C.D.; Peterson, G.L.; Bockus, W.W.; Kankanala, P.; Dubcovsky, J.; Jordan, K.W.; Akhunov, E.; Chumley, F.; Baldelomar, F.D.; Valent, B. The 2NS translocation from Aegilops ventricosa confers resistance to the Triticum pathotype of Magnaporthe oryzae. Crop Sci. 2016, 56, 990–1000. [Google Scholar] [CrossRef] [PubMed]
- Cruz, C.D.; Valent, B. Wheat blast disease: Danger on the move. Trop. Plant Pathol. 2017, 42, 210–222. [Google Scholar] [CrossRef]
- Gao, L.; Koo, D.-H.; Juliana, P.; Rife, T.; Singh, D.; Lemes da Silva, C.; Lux, T.; Dorn, K.M.; Clinesmith, M.; Silva, P.; et al. The Aegilops ventricosa 2NvS segment in bread wheat: Cytology, genomics and breeding. Theor. Appl. Genet. 2021, 134, 529–542. [Google Scholar] [CrossRef]
- Singh, D.; Wang, X.; Kumar, U.; Gao, L.; Noor, M.; Imtiaz, M.; Singh, R.P.; Poland, J. High-throughput phenotyping enabled genetic dissection of crop lodging in wheat. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef]
- Schneider, A.; Molnár, I.; Molnár-Láng, M. Utilisation of Aegilops (goatgrass) species to widen the genetic diversity of cultivated wheat. Euphytica 2008, 163, 1–19. [Google Scholar] [CrossRef]
- Olivera, P.D.; Rouse, M.N.; Jin, Y. Identification of new sources of resistance to wheat stem rust in Aegilops spp. in the tertiary genepool of wheat. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Gill, B.S.; Sharma, C.; Raupp, W.J.; Browder, L.E.; Heachett, J.H.; Harvey, T.L.; Moseman, J.G.; Waines, J.G. Evaluation of Aegilops species for resistance to wheat powdery mildew, wheat leaf rust, Hessian fly, and greenbug. Plant Dis. 1985, 69, 314–316. [Google Scholar]
- Rawat, N.; Schoen, A.; Singh, L.; Mahlandt, A.; Wilson, D.L.; Liu, S.; Lin, G.; Gill, B.S.; Tiwari, V.K. TILL-D: An Aegilops tauschii TILLING resource for wheat improvement. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Suneja, Y.; Gupta, A.K.; Bains, N.S. Stress adaptive plasticity: Aegilops tauschii and Triticum dicoccoides as potential donors of drought associated morpho-physiological traits in wheat. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef]
- Feuillet, C.; Langridge, P.; Waugh, R. Cereal breeding takes a walk on the wild side. Trends Genet. 2008, 24, 24–32. [Google Scholar] [CrossRef]
- Martín, A.C.; Rey, M.D.; Shaw, P.; Moore, G. Dual effect of the wheat Ph1 locus on chromosome synapsis and crossover. Chromosoma 2017, 126, 669–680. [Google Scholar] [CrossRef]
- Rey, M.D.; Martín, A.C.; Higgins, J.; Swarbreck, D.; Uauy, C.; Shaw, P.; Moore, G. Exploiting the ZIP4 homologue within the wheat Ph1 locus has identified two lines exhibiting homoeologous crossover in wheat-wild relative hybrids. Mol. Breed. New Strateg. Plant Improv. 2017, 37, 95. [Google Scholar] [CrossRef]
- Baker, L.; Grewal, S.; Yang, C.-Y.; Hubbart-Edwards, S.; Scholefield, D.; Ashling, S.; Burridge, A.J.; Przewieslik-Allen, A.M.; Wilkinson, P.A.; King, I.P.; et al. Exploiting the genome of Thinopyrum elongatum to expand the gene pool of hexaploid wheat. Theor. Appl. Genet. 2020, 133, 2213–2226. [Google Scholar] [CrossRef]
- Chen, P.D.; Tsujimoto, H.; Gill, B.S. Transfer of Ph(I) genes promoting homoeologous pairing from Triticum speltoides to common wheat. Theor. Appl. Genet. 1994, 88, 97–101. [Google Scholar] [CrossRef]
- Chhuneja, P.; Kaur, S.; Goel, R.K.; Aghaee-Sarbarzeh, M.; Prashar, M.; Dhaliwal, H.S. Transfer of leaf rust and stripe rust resistance from Aegilops umbellulata Zhuk. to bread wheat (Triticum aestivum L.). Genet. Resour. Crop Evol. 2008, 55, 849–859. [Google Scholar] [CrossRef]
- Aghaee-Sarbarzeh, M.; Ferrahi, M.; Singh, S.; Singh, H.; Friebe, B.; Gill, B.S.; Dhaliwal, H.S. Ph-I-induced transfer of leaf and stripe rust-resistance genes from Aegilops triuncialis and Ae. geniculata to bread wheat. Euphytica 2002, 127, 377–382. [Google Scholar] [CrossRef]
- Aberkane, H.; Payne, T.; Kishi, M.; Smale, M.; Amri, A.; Jamora, N. Transferring diversity of goat grass to farmers’ fields through the development of synthetic hexaploid wheat. Food Sec. 2020, 12, 1017–1033. [Google Scholar] [CrossRef]
- Ma’Arup, R.; Trethowan, R.M.; Ahmed, N.U.; Bramley, H.; Sharp, P.J. Emmer wheat (Triticum dicoccon Schrank) improves water use efficiency and yield of hexaploid bread wheat. Plant Sci. 2020, 295, 110212. [Google Scholar] [CrossRef] [PubMed]
- Ullah, S.; Bramley, H.; Mahmood, T.; Trethowan, R. The impact of emmer genetic diversity on grain protein content and test weight of hexaploid wheat under high temperature stress. J. Cereal Sci. 2020, 95, 103052. [Google Scholar] [CrossRef]
- Al Hakimi, A.; Monneveaux, P.; Nachit, M.M. Direct and indirect selection for drought tolerance in alien tetraploid wheat x durum wheat crosses. In Wheat: Prospects for Global Improvement. Developments in Plant Breeding; Braun, H., Altay, F., Kronstad, W.E., Beniwal, S.P.S., McNab, A., Eds.; Springer: Dordrecht, The Netherlands, 1997; Volume 6, pp. 353–360. [Google Scholar]
- Bassi, F.M.; Brahmi, H.; Sabraoui, A.; Amri, A.; Nsarellah, N.; Nachit, M.M.; Al-Abdallat, A.; Chen, M.S.; Lazraq, A.; El Bouhssini, M. Genetic identification of loci for Hessian fly resistance in durum wheat. Mol. Breed. 2019, 39, 24. [Google Scholar] [CrossRef]
- Liu, X.; Brown-Guedira, G.; Hatchett, J.; Owuoche, J.; Chen, M. Genetic characterization and molecular mapping of a Hessian fly-resistance gene transferred from T. turgidum ssp. dicoccum to common wheat. Theor. Appl. Genet. 2005, 111, 1308–1315. [Google Scholar] [CrossRef] [PubMed]
- Rosyara, U.; Kishii, M.; Payne, T.; Sansaloni, C.P.; Singh, R.P.; Braun, H.J.; Dreisigacker, S. Genetic contribution of synthetic hexaploid wheat to CIMMYT’s spring bread wheat breeding germplasm. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef]
- Kaur, A.; Chhuneja, P.; Srivastava, P.; Singh, K.; Kaur, S. Evaluation of Triticum durum–Aegilops tauschii derived primary synthetics as potential sources of heat stress tolerance for wheat improvement. Plant Genet. Resour. 2021, 19, 74–89. [Google Scholar] [CrossRef]
- Börner, A.; Ogbonnaya, F.C.; Röder, M.S.; Rasheed, A.; Periyannan, S.; Lagudah, E.S. Aegilops tauschii introgressions in wheat. In Alien Introgression in Wheat: Cytogenetics, Molecular Biology, and Genomics; Molnár-Láng, M., Ceoloni, C., Doležel, J., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 245–271. [Google Scholar]
- Gill, B.S.; Raupp, W.J.; Sharma, H.C.; Browder, L.E.; Hatchett, J.H.; Harvey, T.L.; Moseman, J.G.; Waines, J.G. Resistance in Aegilops squarrosa to wheat leaf rust, wheat powdery mildew, greenbug, and Hessian fly. Plant Dis. 1986, 70, 553–556. [Google Scholar] [CrossRef]
- Röder, M.S.; Huang, X.-Q.; Börner, A. Fine mapping of the region on wheat chromosome 7D controlling grain weight. Funct. Int. Genom. 2008, 8, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Maes, B.; Trethowan, R.M.; Reynolds, M.P.; van Ginkel, M.; Skovmand, B. The influence of glume pubescence on spikelet temperature of wheat under freezing conditions. Austr. J. Plant Pathol. 2001, 28, 141–148. [Google Scholar] [CrossRef]
- Reynolds, M.P.; Saint Pierre, C.; Saad, A.S.I.; Vargas, M.; Condon, A.G. Evaluating potential genetic gains in wheat associated with stress-adaptive trait expression in elite genetic resources under drought and heat stress. Crop Sci. 2007, 47, S172–S189. [Google Scholar] [CrossRef]
- Szabo-Hever, A.; Zhang, Q.; Friesen, T.L.; Zhong, S.; Elias, E.M.; Cai, X.; Jin, Y.; Faris, J.D.; Chao, S.; Xu, S.S. Genetic diversity and resistance to Fusarium head blight in synthetic hexaploid wheat derived from Aegilops tauschii and diverse Triticum turgidum subspecies. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef]
- Villareal, R.; Sayre, K.; Banuelos, O.; Mujeeb-Kazi, A. Registration of four synthetic hexaploid wheat germplasm lines tolerant to waterlogging. Crop Sci. 2001, 41, 274. [Google Scholar] [CrossRef]
- Yang, J.; Sears, R.G.; Gill, B.S.; Paulsen, G.M. Growth and senescence characteristics associated with tolerance of wheat-alien amphiploids to high temperature under controlled conditions. Euphytica 2002, 126, 185–193. [Google Scholar] [CrossRef]
- Loughman, R.; Lagudah, E.S.; Trottet, M.; Wilson, R.E.; Mathews, A. Septoria nodorum blotch resistance in Aegilops tauschii and its expression in synthetic amphiploids. Austral. J. Agric. Res. 2001, 52, 1393–1402. [Google Scholar] [CrossRef]
- Nicholson, P.; Rezanoor, H.N.; Worland, A.J. Chromosomal location of resistance to Septoria nodorum in a synthetic hexaploid wheat determined by the study of chromosomal substitution lines in ‘Chinese Spring’ wheat. Plant Breed. 1993, 110, 177–184. [Google Scholar] [CrossRef]
- Lage, J.; Skovmand, B.; Andersen, S.B. Field evaluation of emmer wheat-derived synthetic hexaploid wheat for resistance to Russian wheat aphid (Homoptera: Aphididae). J. Econ. Entomol. 2004, 97, 1065–1070. [Google Scholar] [CrossRef]
- Lage, J.; Skovmand, B.; Andersen, S.B. Expression and suppression of resistance to greenbug (Homoptera: Aphididae) in synthetic hexaploid wheats derived from Triticum dicoccum x Aegilops tauschii crosses. J. Econ. Entomol. 2003, 96, 202–206. [Google Scholar] [CrossRef]
- King, J.; Grewal, S.; Yang, C.-Y.; Hubbart Edwards, S.; Scholefield, D.; Ashling, S.; Harper, J.A.; Allen, A.M.; Edwards, K.J.; Burridge, A.J.; et al. Introgression of Aegilops speltoides segments in Triticum aestivum and the effect of the gametocidal genes. Ann. Bot. 2018, 121, 229–240. [Google Scholar] [CrossRef]
- King, J.; Grewal, S.; Yang, C.-Y.; Hubbart, S.; Scholefield, D.; Ashling, S.; Edwards, K.J.; Allen, A.M.; Burridge, A.; Bloor, C.; et al. A step change in the transfer of interspecific variation into wheat from Amblyopyrum muticum. Plant Biotechnol. J. 2017, 15, 217–226. [Google Scholar] [CrossRef]
- Iefimenko, T.S.; Fedak, Y.G.; Antonyuk, M.Z.; Ternovska, T.K. Microsatellite analysis of chromosomes from the fifth homoeologous group in the introgressive Triticum aestivum/Amblyopyrum muticum wheat lines. Cytol. Genet. 2015, 49, 183–191. [Google Scholar] [CrossRef]
- King, J.; Newell, C.; Grewal, S.; Hubbart-Edwards, S.; Yang, C.-Y.; Scholefield, D.; Ashling, S.; Stride, A.; King, I.P. Development of stable homozygous wheat/Amblyopyrum muticum (Aegilops mutica) introgression lines and their cytogenetic and molecular characterization. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef]
- Sansaloni, C.; Franco, J.; Santos, B.; Percival-Alwyn, L.; Singh, S.; Petroli, C.; Campos, J.; Dreher, K.; Payne, T.; Marshall, D.; et al. Diversity analysis of 80,000 wheat accessions reveals consequences and opportunities of selection footprints. Nat. Commun. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Keilwagen, J.; Lehnert, H.; Berner, T.; Badaeva, E.; Himmelbach, A.; Börner, A.; Kilian, B. Detecting major introgressions in wheat and their putative origin using coverage analysis. Scientific Rep. 2021, in press. [Google Scholar] [CrossRef]
- Sharma, S.; Upadhyaya, H.; Varshney, R.; Gowda, C. Pre-breeding for diversification of primary gene pool and genetic enhancement of grain legumes. Front. Plant Sci. 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Olivera, P.; Newcomb, M.; Szabo, L.J.; Rouse, M.; Johnson, J.; Gale, S.; Luster, D.G.; Hodson, D.; Cox, J.A.; Burgin, L.; et al. Phenotypic and genotypic characterization of race TKTTF of Puccinia graminis f. sp. tritici that caused a wheat stem rust epidemic in southern Ethiopia in 2013–14. Phytopathology® 2015, 105, 917–928. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S. Deadly new wheat disease threatens Europe’s crops. Nature 2017, 542, 145–146. [Google Scholar] [CrossRef]
- Lewis, C.M.; Persoons, A.; Bebber, D.P.; Kigathi, R.N.; Maintz, J.; Findlay, K.; Bueno-Sancho, V.; Corredor-Moreno, P.; Harrington, S.A.; Kangara, N.; et al. Potential for re-emergence of wheat stem rust in the United Kingdom. Commun. Biol. 2018, 1, 13. [Google Scholar] [CrossRef] [PubMed]
- Olivera Firpo, P.D.; Newcomb, M.; Flath, K.; Sommerfeldt-Impe, N.; Szabo, L.J.; Carter, M.; Luster, D.G.; Jin, Y. Characterization of Puccinia graminis f. sp. tritici isolates derived from an unusual wheat stem rust outbreak in Germany in 2013. Plant Pathol. 2017, 66, 1258–1266. [Google Scholar] [CrossRef]
- Shamanin, V.; Salina, E.; Zelenskiy, Y.; Kokhmetova, A.; Patpour, M.; Hovmøller, M.; Olivera, P.; Szabo, L.; Yue, J.; Meyer, M.; et al. Large scale wheat stem rust outbreaks in Western Siberia/Northern Kazakhstan in 2015-2017. In Proceedings of the Borlaug Global Rust Initiative: Technical Workshop, Marrakech, Morocco, 14–17 April 2018. [Google Scholar]
- Kildea, S.; Sheppard, L.; Cucak, M.; Hutton, F. Detection of virulence to Septoria tritici blotch (STB) resistance conferred by the winter wheat cultivar Cougar in the Irish Zymoseptoria tritici population and potential implications for STB control. Plant Pathol. 2021. [Google Scholar] [CrossRef]
- Wang, X.; Liu, Y.; Chen, L.; Zhao, D.; Wang, X.; Zhang, Z. Wheat resistome in response to barley yellow dwarf virus infection. Funct. Int. Genom. 2013, 13, 155–165. [Google Scholar] [CrossRef]
- Hussain, B.; Akpınar, B.A.; Alaux, M.; Algharib, A.M.; Sehgal, D.; Ali, Z.; Appels, R.; Aradottir, G.I.; Batley, J.; Bellec, A.; et al. Wheat genomics and breeding: Bridging the gap. AgriRxiv 2021. [Google Scholar] [CrossRef]
- Hoffmann, B.; Burucs, Z. Adaptation of wheat (Triticum aestivum L.) genotypes and related species to water deficiency. Cereal Res. Commun. 2005, 33, 681–687. [Google Scholar] [CrossRef]
- Rajaram, S. Prospects and promise of wheat breeding in the 21st century. Euphytica 2001, 119, 3–15. [Google Scholar] [CrossRef]
- Sallam, A.; Alqudah, A.M.; Dawood, M.F.A.; Baenziger, P.S.; Börner, A. Drought stress tolerance in wheat and barley: Advances in physiology, breeding and genetics research. Int. J. Mol. Sci. 2019, 20, 3137. [Google Scholar] [CrossRef]
- Damania, A.B.; Altunji, H.; Dhaliwal, H.S. Evaluation of Aegilops spp. for Drought and Frost Tolerance; Genetics Research Unit Annual Report 1992; ICARDA: Aleppo, Syria, 1992; pp. 45–46. [Google Scholar]
- Jaradat, A.A. Ecogeography, genetic diversity, and breeding value of wild emmer wheat (Triticum dicoccoides Körn. ex Asch. & Graebn.) Thell. Austral. J. Agric. Res. 2011, 5, 1072–1086. [Google Scholar]
- Nevo, E.; Chen, G. Drought and salt tolerances in wild relatives for wheat and barley improvement. Plant Cell Environ. 2010, 33, 670–685. [Google Scholar] [CrossRef]
- Dixon, J.; Braun, H.-J.; Kosina, P.; Crouch, J.H. Wheat Facts and Futures; CIMMYT: Ciudad de México, Mexico, 2009; p. 95. [Google Scholar]
- Zaharieva, M.; Ayana, N.; Hakimi, A.; Misra, S.; Monneveux, P. Cultivated emmer wheat (Triticum dicoccon Schrank), an old crop with promising future: A review. Genet. Resour. Crop Evol. 2010, 57, 937–962. [Google Scholar] [CrossRef]
- Zaharieva, M.; Monneveux, P. Cultivated einkorn wheat (Triticum monococcum L. subsp. monococcum): The long life of a founder crop of agriculture. Genet. Resour. Crop Evol. 2014, 61, 677–706. [Google Scholar] [CrossRef]
- Oliver, R.E.; Cai, X.; Friesen, T.L.; Halley, S.; Stack, R.W.; Xu, S.S. Evaluation of Fusarium head blight resistance in tetraploid wheat (Triticum turgidum L.). Crop Sci. 2008, 48, 213–222. [Google Scholar] [CrossRef]
- Saleh, M.M. Stress breeding of neglected tetraploid primitive wheat (Triticum dicoccum, Triticum carthlicum and Triticum polonicum). Curr. Bot. 2020, 11, 99–110. [Google Scholar] [CrossRef]
- Bencze, S.; Makádi, M.; Aranyos, T.J.; Földi, M.; Hertelendy, P.; Mikó, P.; Bosi, S.; Negri, L.; Drexler, D. Re-introduction of ancient wheat cultivars into organic agriculture—Emmer and einkorn cultivation experiences under marginal conditions. Sustainability 2020, 12, 1584. [Google Scholar] [CrossRef]
- Bonafaccia, G.; Galli, V.; Francisci, R.; Mair, V.; Skrabanja, V.; Kreft, I. Characteristics of spelt wheat products and nutritional value of spelt wheat-based bread. Food Chem. 2000, 68, 437–441. [Google Scholar] [CrossRef]
- Konvalina, P.; Stehno, Z.; Capouchová, I.; Moudrý jr., J.; Jůza, M.; Moudrý, J. Emmer wheat using and growing in the Czech Republic. Lucr. Ştiinţifice Ser. Agron. 2010, 53, 1–5. [Google Scholar]
- Lacko-Bartošová, M.; Čurná, V.; Lacko-Bartošová, L. Emmer—Ancient wheat suitable for ecological farming. Res. J. Agric. Sci. 2015, 47, 3–10. [Google Scholar]
- Stagnari, F.; Codianni, P.; Pisante, M. Agronomic and kernel quality of ancient wheats grown in central and southern Italy. Cereal Res. Commun. 2008, 36, 313–326. [Google Scholar] [CrossRef]
- Escarnot, E.; Jacquemin, J.M.; Agneessens, R.; Paquot, M. Comparative study of the content and profiles of macronutrients in spelt and wheat, a review. Biotech. Agron. Soc. Environ. 2012, 16, 243–256. [Google Scholar]
- Yenagi, N.; Hanchinal, R.R.; Patil, C.S.; Koppikar, V.; Halagi, M. Glycemic and lipidemic response to dicoccum wheat (Triticum dicoccum) in the diet of diabetic patients. Int. J. Diabetes Dev. Ctries. 2001, 21, 153–155. Available online: https://www.semanticscholar.org/paper/GLYCEMIC-AND-LIPIDEMIC-RESPONSE-TO-DICOCCUM-WHEAT-Yenagi-Hanchinal/d00cd8d4bab7b02e0bfe451047aed9536cf377ad (accessed on 21 September 2021).
- Hidalgo, A.; Brandolini, A. Nutritional properties of einkorn wheat (Triticum monococcum L.). J. Sci. Food Agric. 2014, 94, 601–612. [Google Scholar] [CrossRef] [PubMed]
- Wiwart, M.; Suchowilska, E.; Kandler, W.; Sulyok, M.; Groenwald, P.; Krska, R. Can Polish wheat (Triticum polonicum L.) be an interesting gene source for breeding wheat cultivars with increased resistance to Fusarium head blight? Genet. Resour. Crop Evol. 2013, 60, 2359–2373. [Google Scholar] [CrossRef]
- Cooper, R. Re-discovering ancient wheat varieties as functional foods. J. Tradit. Complement. Med. 2015, 5, 138–143. [Google Scholar] [CrossRef] [PubMed]
- Stallknecht, G.F.; Gilbertson, K.M.; Ranney, J.E. Alternative wheat cereals as food grains: Einkorn, emmer, spelt, kamut, and triticale. In Progress in New Crops; Janick, J., Ed.; ASHS Press: Alexandria, VA, USA, 1996; pp. 156–170. [Google Scholar]
- Bond, A. Discovering einkorn in Haute Provence, France. Cerealist 1989, 1, 6–7. [Google Scholar]
- Galterio, G.; Codianni, P.; Giusti, A.M.; Pezzarossa, B.; Cannella, C. Assessment of the agronomic and technological characteristics of Triticum turgidum ssp. dicoccum Schrank and T. spelta L. Nahrung 2003, 47, 54–59. [Google Scholar] [CrossRef]
- Keskin Şan, S.; Özbek, Ö.; Eser, V.; Göçmen Taşkin, B. Polymorphism in seed endosperm proteins (gliadins and glutenins) of Turkish cultivated einkorn wheat [Triticum monococcum ssp. monococcum] landraces. Cereal Res. Commun. 2015, 43, 108–122. [Google Scholar] [CrossRef][Green Version]
- Cakmak, I.; Torun, A.; Millet, E.; Feldman, M.; Fahima, T.; Korol, A.; Nevo, E.; Braun, H.J.; Özkan, H. Triticum dicoccoides: An important genetic resource for increasing zinc and iron concentration in modern cultivated wheat. Soil Sci. Plant Nut. 2004, 50, 1047–1054. [Google Scholar] [CrossRef]
- García, A.B.; Castellano, L.; Guzmán, C.; Alvarez, J.B. Potential use of wild einkorn wheat for wheat grain quality improvement: Evaluation and characterization of Glu-1, Wx and Ha loci. Agronomy 2021, 11, 816. [Google Scholar] [CrossRef]
- Liu, J.; Huang, L.; Li, T.; Liu, Y.; Yan, Z.; Tang, G.; Zheng, Y.; Liu, D.; Wu, B. Genome-wide association study for grain micronutrient concentrations in wheat advanced lines derived from wild emmer. Front. Plant Sci. 2021, 12. [Google Scholar] [CrossRef]
- Erba, D.; Hidalgo, A.; Bresciani, J.; Brandolini, A. Environmental and genotypic influences on trace element and mineral concentrations in whole meal flour of einkorn (Triticum monococcum L. subsp. monococcum). J. Cereal Sci. 2011, 54, 250–254. [Google Scholar] [CrossRef]
- Özkan, H.; Brandolini, A.; Torun, A.; Altintas, S.; Eker, S.; Kilian, B.; Braun, H.J.; Salamini, F.; Cakmak, I. Natural variation and identification of microelements content in seeds of einkorn wheat (Triticum monococcum). In Wheat Production in Stressed Environments; Buck, H.T., Nisi, J.E., Salomon, N., Eds.; Springer: Dordrecht, The Netherlands, 2007; Volume 12, pp. 455–462. [Google Scholar] [CrossRef]
- Chhuneja, P.; Dhaliwal, H.S.; Bains, N.S.; Singh, K. Aegilops kotschyi and Aegilops tauschii as sources for higher levels of grain iron and zinc. Plant Breed. 2006, 125, 529–531. [Google Scholar] [CrossRef]
- Kumar, A.; Kapoor, P.; Chunduri, V.; Sharma, S.; Garg, M. Potential of Aegilops sp. for improvement of grain processing and nutritional quality in wheat (Triticum aestivum). Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Rawat, N.; Tiwari, V.K.; Singh, N.; Randhawa, G.S.; Singh, K.; Chhuneja, P.; Dhaliwal, H.S. Evaluation and utilization of Aegilops and wild Triticum species for enhancing iron and zinc content in wheat. Genet. Resour. Crop Evol. 2009, 56. [Google Scholar] [CrossRef]
- Leonova, I.N.; Budashkina, E.B.; Kalinina, N.P.; Röder, M.S.; Börner, A.; Salina, E.A. Triticum aestivum-Triticum timopheevii introgression lines as a source of pathogen resistance genes. Czech J. Genet. Breed. 2011, 47, S49–S55. [Google Scholar] [CrossRef]
- EiB. Available online: https://excellenceinbreeding.org/ (accessed on 21 September 2021).
- Jakob, S.S.; Rödder, D.; Engler, J.O.; Shaaf, S.; Özkan, H.; Blattner, F.R.; Kilian, B. Evolutionary history of wild barley (Hordeum vulgare subsp. spontaneum) analyzed using multilocus sequence data and paleodistribution modeling. Gen. Biol. Evol. 2014, 6, 685–702. [Google Scholar] [CrossRef]
- Singh, N.; Wu, S.; Raupp, W.J.; Sehgal, S.; Arora, S.; Tiwari, V.; Vikram, P.; Singh, S.; Chhuneja, P.; Gill, B.S.; et al. Efficient curation of genebanks using next generation sequencing reveals substantial duplication of germplasm accessions. Sci. Rep. 2019, 9, 650. [Google Scholar] [CrossRef]
- Brown, A.H.D. Core collections: A practical approach to genetic resources management. Genome 1989, 31, 818–824. [Google Scholar] [CrossRef]
- Upadhyaya, H.D.; Ortiz, R. A mini core subset for capturing diversity and promoting utilization of chickpea genetic resources in crop improvement. Theor. Appl. Genet. 2001, 102, 1292–1298. [Google Scholar] [CrossRef]
- Upadhyaya, H.D.; Dwivedi, S.L.; Baum, M.; Varshney, R.K.; Udupa, S.M.; Gowda, C.L.L.; Hoisington, D.; Singh, S. Genetic structure, diversity, and allelic richness in composite collection and reference set in chickpea (Cicer arietinum L.). BMC Plant Biol. 2008, 8, 106. [Google Scholar] [CrossRef]
- Glaszmann, J.C.; Kilian, B.; Upadhyaya, H.D.; Varshney, R.K. Accessing genetic diversity for crop improvement. Curr. Opin. Plant Biol. 2010, 13, 167–173. [Google Scholar] [CrossRef]
- Khazaei, H.; Street, K.; Bari, A.; Mackay, M.; Stoddard, F.L. The FIGS (Focused Identification of Germplasm Strategy) approach identifies traits related to drought adaptation in Vicia faba genetic resources. PLoS ONE 2013, 8, e63107. [Google Scholar] [CrossRef] [PubMed]
- Lande, R.; Thompson, R. Efficiency of marker-assisted selection in the improvement of quantitative traits. Genetics 1990, 124, 743. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, H.K.; Sethi, G.S.; Singh, S.; Pratap, A.; Sharma, S. Efficient haploid induction in wheat by using pollen of Imperata cylindrica. Plant Breed. 2005, 124, 96–98. [Google Scholar] [CrossRef]
- Ghosh, S.; Watson, A.; Gonzalez-Navarro, O.E.; Ramirez-Gonzalez, R.H.; Yanes, L.; Mendoza-Suárez, M.; Simmonds, J.; Wells, R.; Rayner, T.; Green, P.; et al. Speed breeding in growth chambers and glasshouses for crop breeding and model plant research. Nat. Protoc. 2018, 13, 2944–2963. [Google Scholar] [CrossRef]
- Sharma, S.; Sethi, G.S.; Chaudhary, H.K. Influence of winter and spring wheat genetic backgrounds on haploid induction parameters and trait correlations in the wheat × maize system. Euphytica 2005, 144, 199–205. [Google Scholar] [CrossRef]
- Arora, S.; Steuernagel, B.; Gaurav, K.; Chandramohan, S.; Long, Y.; Matny, O.; Johnson, R.; Enk, J.; Periyannan, S.; Singh, N.; et al. Resistance gene cloning from a wild crop relative by sequence capture and association genetics. Nat. Biotechnol. 2019, 37, 139–143. [Google Scholar] [CrossRef]
- Bai, G.; Kolb, F.L.; Shaner, G.; Domier, L.L. Amplified fragment length polymorphism markers linked to a major quantitative trait locus controlling scab resistance in wheat. Phytopathology® 1999, 89, 343–348. [Google Scholar] [CrossRef]
- Rawat, N.; Pumphrey, M.O.; Liu, S.; Zhang, X.; Tiwari, V.K.; Ando, K.; Trick, H.N.; Bockus, W.W.; Akhunov, E.; Anderson, J.A.; et al. Wheat Fhb1 encodes a chimeric lectin with agglutinin domains and a pore-forming toxin-like domain conferring resistance to Fusarium head blight. Nat. Genet. 2016, 48, 1576–1580. [Google Scholar] [CrossRef] [PubMed]
- Waldron, B.; Moreno-Sevilla, B.; Anderson, J.; Stack, R.; Frohberg, R. RFLP mapping of QTL for Fusarium head blight resistance in wheat. Crop Sci. 1999, 39, 805–811. [Google Scholar] [CrossRef]
- Bernardo, R. Genomewide selection for rapid introgression of exotic germplasm in maize. Crop Sci. 2009, 49, 419–425. [Google Scholar] [CrossRef]
- Meuwissen, T.H.; Hayes, B.J.; Goddard, M.E. Prediction of total genetic value using genome-wide dense marker maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [CrossRef] [PubMed]
- VanRaden, P.M. Efficient methods to compute genomic predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef] [PubMed]
- Arbelaez, J.D.; Moreno, L.T.; Singh, N.; Tung, C.-W.; Maron, L.G.; Ospina, Y.; Martinez, C.P.; Grenier, C.; Lorieux, M.; McCouch, S. Development and GBS-genotyping of introgression lines (ILs) using two wild species of rice, O. meridionalis and O. rufipogon, in a common recurrent parent, O. sativa cv. Curinga. Mol. Breed. 2015, 35, 81. [Google Scholar] [CrossRef] [PubMed]
- Nyine, M.; Adhikari, E.; Clinesmith, M.; Jordan, K.W.; Fritz, A.K.; Akhunov, E. Genomic patterns of introgression in interspecific populations created by crossing wheat with its wild relative. G3 Genes|Genomes|Genet. 2020, 10, 3651. [Google Scholar] [CrossRef] [PubMed]
- Schnell, F.W.; Utz, H.F. F1 Leistung und Elternwahl in der Züchtung von Selbstbefruchtern. In Bericht über die Arbeitstagung 1975 der Arbeitsgemeinschaft der Saatgutleiter; Bundesversuchsanstalt für Alpenländische Landwirtschaft Gumpenstein: Irdning, Austria, 1976; pp. 243–248. [Google Scholar]
- Becker, H. Pflanzenzüchtung; Verlag Eugen Ulmer: Stuttgart, Germany, 2011; Volume Nr 2. [Google Scholar]
- Endelman, J.B.; Atlin, G.N.; Beyene, Y.; Semagn, K.; Zhang, X.; Sorrells, M.E.; Jannink, J.-L. Optimal design of preliminary yield trials with genome-wide markers. Crop Sci. 2014, 54, 48–59. [Google Scholar] [CrossRef]
- Holland, J.B.; Nyquist, W.E.; Cervantes-Martínez, C.T. Estimating and interpreting heritability for plant breeding: An update. In Plant Breeding Reviews; Janick, J., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2003; Volume 22, pp. 9–111. [Google Scholar]
- Piepho, H.-P.; Möhring, J. Computing heritability and selection response from unbalanced plant breeding trials. Genetics 2007, 177, 1881–1888. [Google Scholar] [CrossRef]
- Allier, A.; Moreau, L.; Charcosset, A.; Teyssèdre, S.; Lehermeier, C. Usefulness criterion and post-selection parental contributions in multi-parental crosses: Application to polygenic trait introgression. G3 2019, 9, 1469–1479. [Google Scholar] [CrossRef]
- Lehermeier, C.; Teyssèdre, S.; Schön, C.C. Genetic gain increases by applying the usefulness criterion with improved variance prediction in selection of crosses. Genetics 2017, 207, 1651–1661. [Google Scholar] [CrossRef]
- Mrode, R.; Brotherstone, S.; White, I.; Swanson, G.; Coffey, M.; Jones, H.; Thompson, R. Random regression model for the genetic evaluation of production traits of dairy cattle in the UK. Interbull Bull. 2005, 33, 211–214. [Google Scholar]
- Bernardo, R. Genomewide selection of parental inbreds: Classes of loci and virtual biparental populations. Crop Sci. 2014, 54, 2586–2595. [Google Scholar] [CrossRef]
- Mohammadi, M.; Tiede, T.; Smith, K.P. PopVar: A genome-wide procedure for predicting genetic variance and correlated response in biparental breeding populations. Crop Sci. 2015, 55, 2068–2077. [Google Scholar] [CrossRef]
- Gianola, D.; Fernando, R.L.; Stella, A. Genomic-assisted prediction of genetic value with semiparametric procedures. Genetics 2006, 173, 1761–1776. [Google Scholar] [CrossRef] [PubMed]
- Gianola, D.; van Kaam, J.B.C.H.M. Reproducing kernel Hilbert spaces regression methods for genomic assisted prediction of quantitative traits. Genetics 2008, 178, 2289. [Google Scholar] [CrossRef]
- Jiang, Y.; Reif, J.C. Modeling epistasis in genomic selection. Genetics 2015, 201, 759–768. [Google Scholar] [CrossRef]
- Mullis, M.N.; Matsui, T.; Schell, R.; Foree, R.; Ehrenreich, I.M. The complex underpinnings of genetic background effects. Nat. Commun. 2018, 9, 3548. [Google Scholar] [CrossRef]
- Merchuk-Ovnat, L.; Fahima, T.; Ephrath, J.E.; Krugman, T.; Saranga, Y. Ancestral QTL alleles from wild emmer wheat enhance root development under drought in modern wheat. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef]
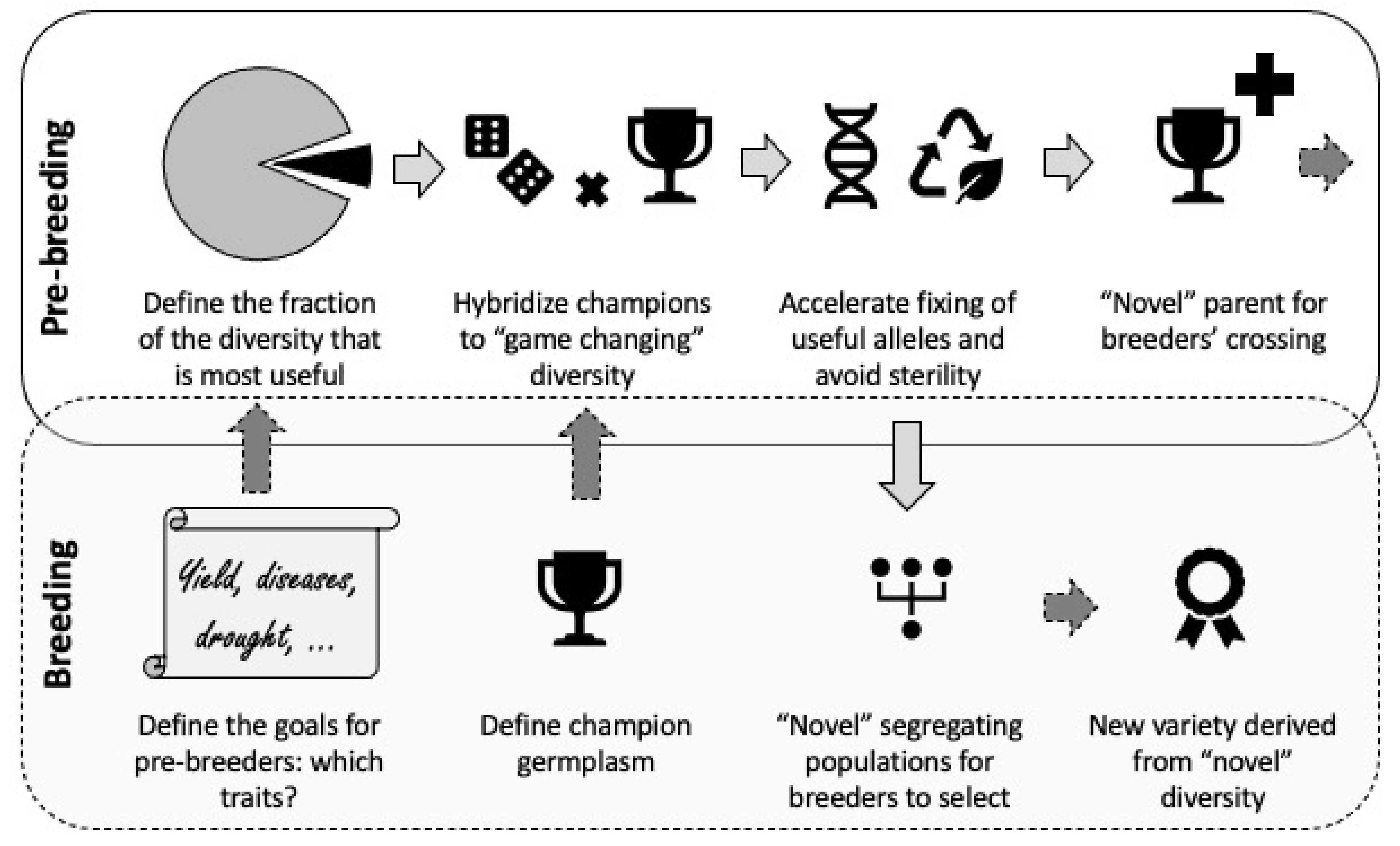
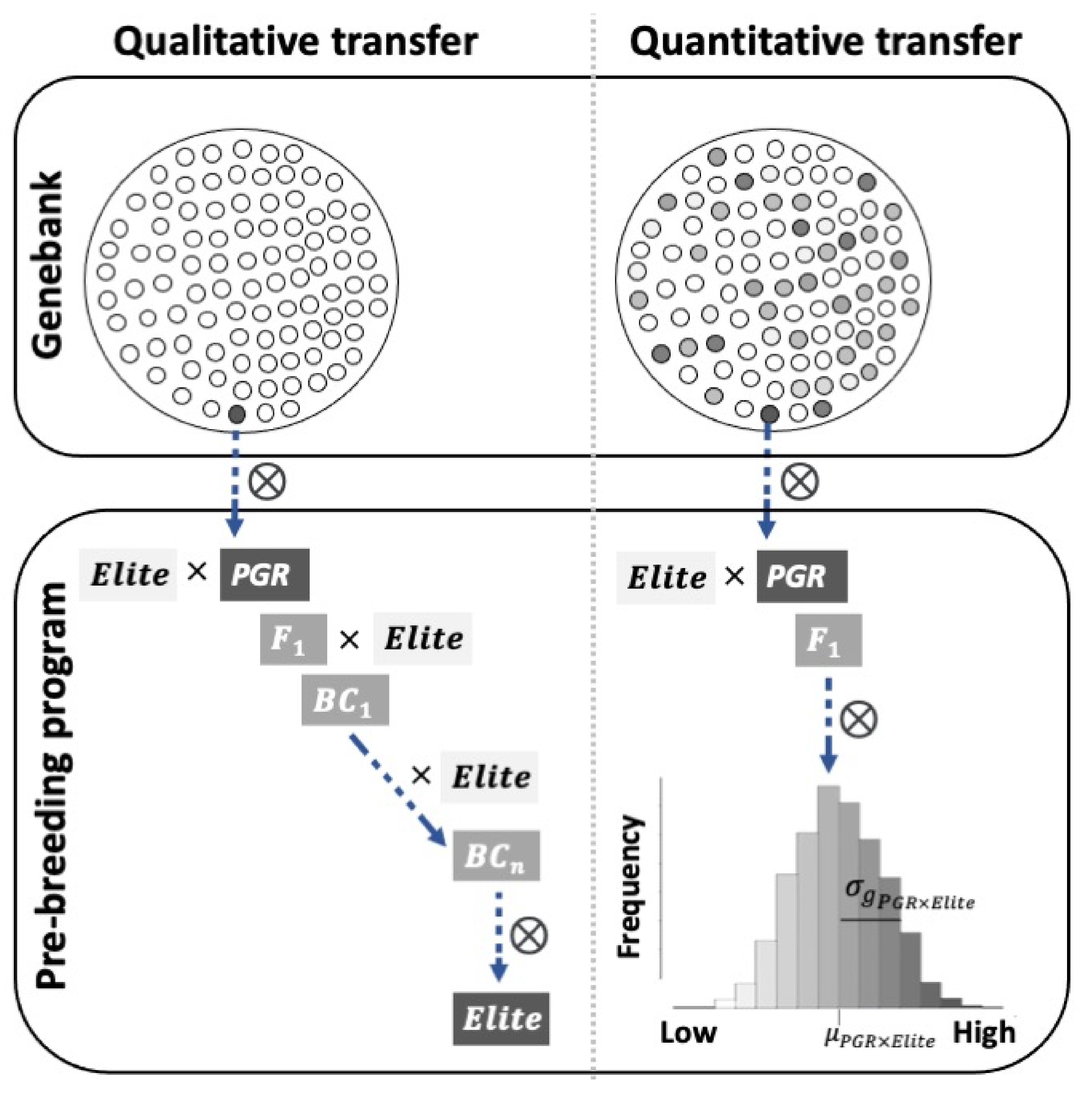
| Country | Institute | Institute Acronym (FAO WIEWS Institure Code) | Number of Accessions | Information Source |
|---|---|---|---|---|
| Triticum | ||||
| Mexico | International Maize and Wheat Improvement Center | CIMMYT (MEX002) | 142,484 | Genesys |
| United States of America | National Small Grains Germplasm Research Facility, USDA-ARS | NSGC (USA029) | 63,941 | Genesys |
| Australia | Australian Grains Genebank, Department of Economic Development Jobs Transport and Resources | AGG (AUS165) | 41,154 | Genesys |
| China | Institute of Crop Science, Chinese Academy of Agricultural Sciences | ICS-CAAS (CHN001) | 41,030 | FAO WIEWS |
| Lebanon | International Centre for Agricultural Research in Dry Areas | ICARDA (LBN002) | 38,897 | Genesys |
| Russian Federation | N.I. Vavilov All-Russian Scientific Research Institute of Plant Genetic Resources | VIR (RUS001) | 38,315 | Genesys |
| India | National Bureau of Plant Genetic Resources | NBPGR (IND001) | 35,889 | FAO WIEWS |
| Japan | Department of Genetic Resources I, National Institute of Agrobiological Sciences | NIAS (JPN003) | 34,652 | FAO WIEWS |
| Italy | Istituto di Bioscienze e Biorisorse, Consiglio Nazionale delle Ricerche, Bari | IBBR-CNR (ITA436) | 29,680 | https://ibbr.cnr.it/mgd accessed on 23 August 2021 |
| Germany | Genebank, Leibniz Institute of Plant Genetics and Crop Plant Research | IPK (DEU146) | 27,442 | Genesys, EURISCO |
| Aegilops | ||||
| Israel | Lieberman Germplasm Bank, Institute for Cereal Crops Improvement, Tel-Aviv University | ICCI-TELAVUN (ISR003) | 7520 | Genesys |
| Lebanon | International Centre for Agricultural Research in Dry Areas | ICARDA (LBN002) | 5081 | Genesys |
| Russian Federation | N.I. Vavilov All-Russian Scientific Research Institute of Plant Genetic Resources | VIR (RUS001) | 3362 | Genesys |
| Islamic Republic of Iran | National Plant Gene Bank of Iran, Seed and Plant Improvement Institute | NPGBI-SPII (IRN029) | 2653 | FAO WIEWS |
| Japan | Department of Genetic Resources I, National Institute of Agrobiological Sciences | NIAS (JPN003) | 2433 | FAO WIEWS |
| Japan | Plant Germplasm Institute, Faculty of Agriculture, Kyoto University | KYOPGI (JPN001) | 2396 | FAO WIEWS |
| United States of America | National Small Grains Germplasm Research Facility, USDA-ARS | NSGC (USA029) | 2245 | Genesys |
| Mexico | International Maize and Wheat Improvement Center | CIMMYT (MEX002) | 2203 | Genesys |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharma, S.; Schulthess, A.W.; Bassi, F.M.; Badaeva, E.D.; Neumann, K.; Graner, A.; Özkan, H.; Werner, P.; Knüpffer, H.; Kilian, B. Introducing Beneficial Alleles from Plant Genetic Resources into the Wheat Germplasm. Biology 2021, 10, 982. https://doi.org/10.3390/biology10100982
Sharma S, Schulthess AW, Bassi FM, Badaeva ED, Neumann K, Graner A, Özkan H, Werner P, Knüpffer H, Kilian B. Introducing Beneficial Alleles from Plant Genetic Resources into the Wheat Germplasm. Biology. 2021; 10(10):982. https://doi.org/10.3390/biology10100982
Chicago/Turabian StyleSharma, Shivali, Albert W. Schulthess, Filippo M. Bassi, Ekaterina D. Badaeva, Kerstin Neumann, Andreas Graner, Hakan Özkan, Peter Werner, Helmut Knüpffer, and Benjamin Kilian. 2021. "Introducing Beneficial Alleles from Plant Genetic Resources into the Wheat Germplasm" Biology 10, no. 10: 982. https://doi.org/10.3390/biology10100982
APA StyleSharma, S., Schulthess, A. W., Bassi, F. M., Badaeva, E. D., Neumann, K., Graner, A., Özkan, H., Werner, P., Knüpffer, H., & Kilian, B. (2021). Introducing Beneficial Alleles from Plant Genetic Resources into the Wheat Germplasm. Biology, 10(10), 982. https://doi.org/10.3390/biology10100982






