Characterization and Use in Wheat Breeding of Leaf Rust Resistance Genes from Durable Varieties
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.1.1. Genetic Components of Leaf Rust Resistance in BM
2.1.2. Fine Mapping of APR Genes LrSV2 and LrcSV2
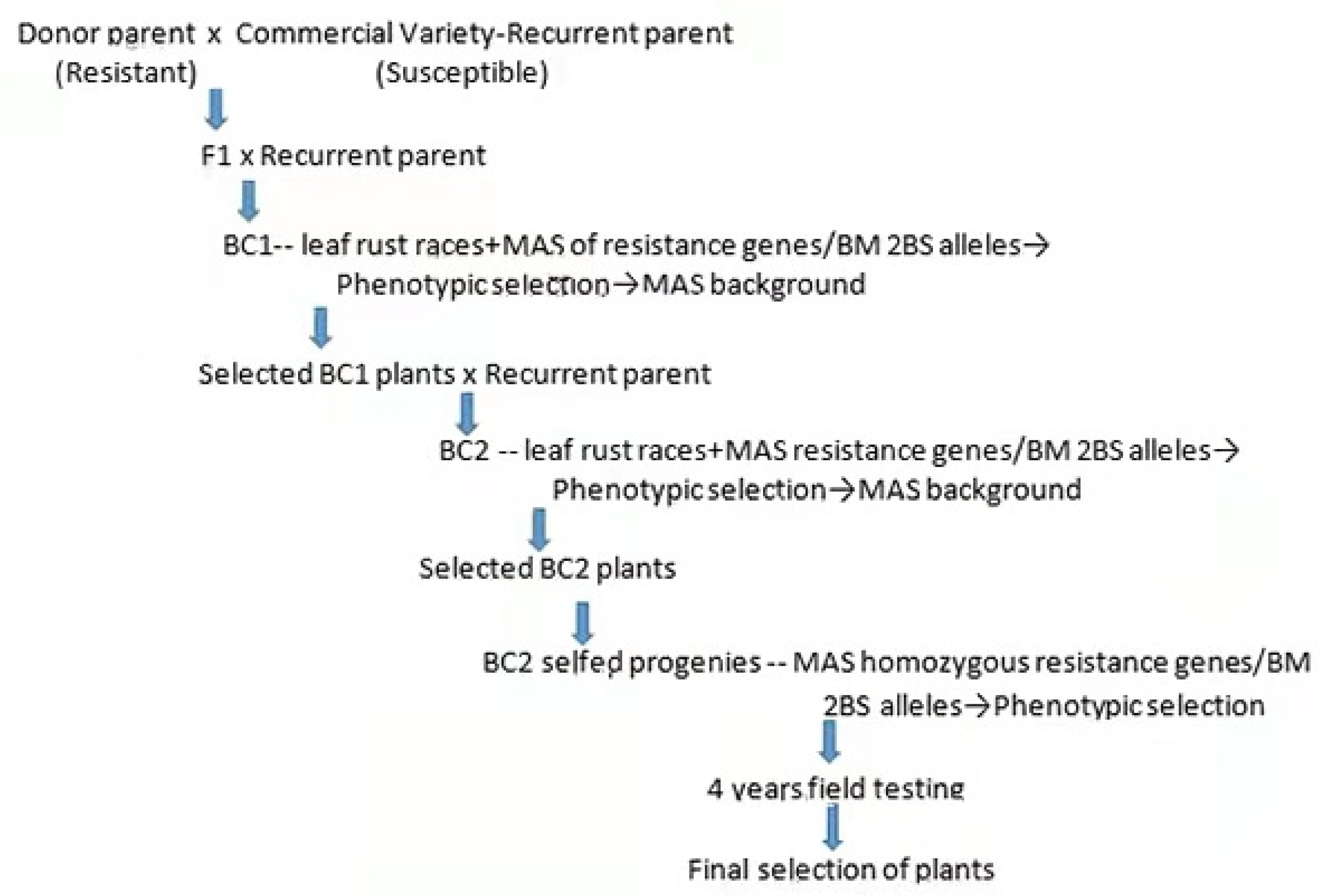
2.1.3. Introgression and Pyramiding of Resistance Genes for Leaf Rust from BM and SV in Seven Susceptible Commercial Wheat Varieties
2.2. Field Testing
2.2.1. Field Testing of F8 RIL Population from BM × P
2.2.2. Field Testing of Resistant Selected Lines
2.3. Rust Inoculations
2.4. DNA Markers
2.4.1. Mapping Resistance Genes in BM
2.4.2. Fine Mapping of Resistance Genes LrSV2 and LrcSV2
2.4.3. MAS for Leaf Rust Resistance Genes Introgression and Background MAS
2.4.4. Selection of Resistant Lines
2.5. Data Analysis
3. Results
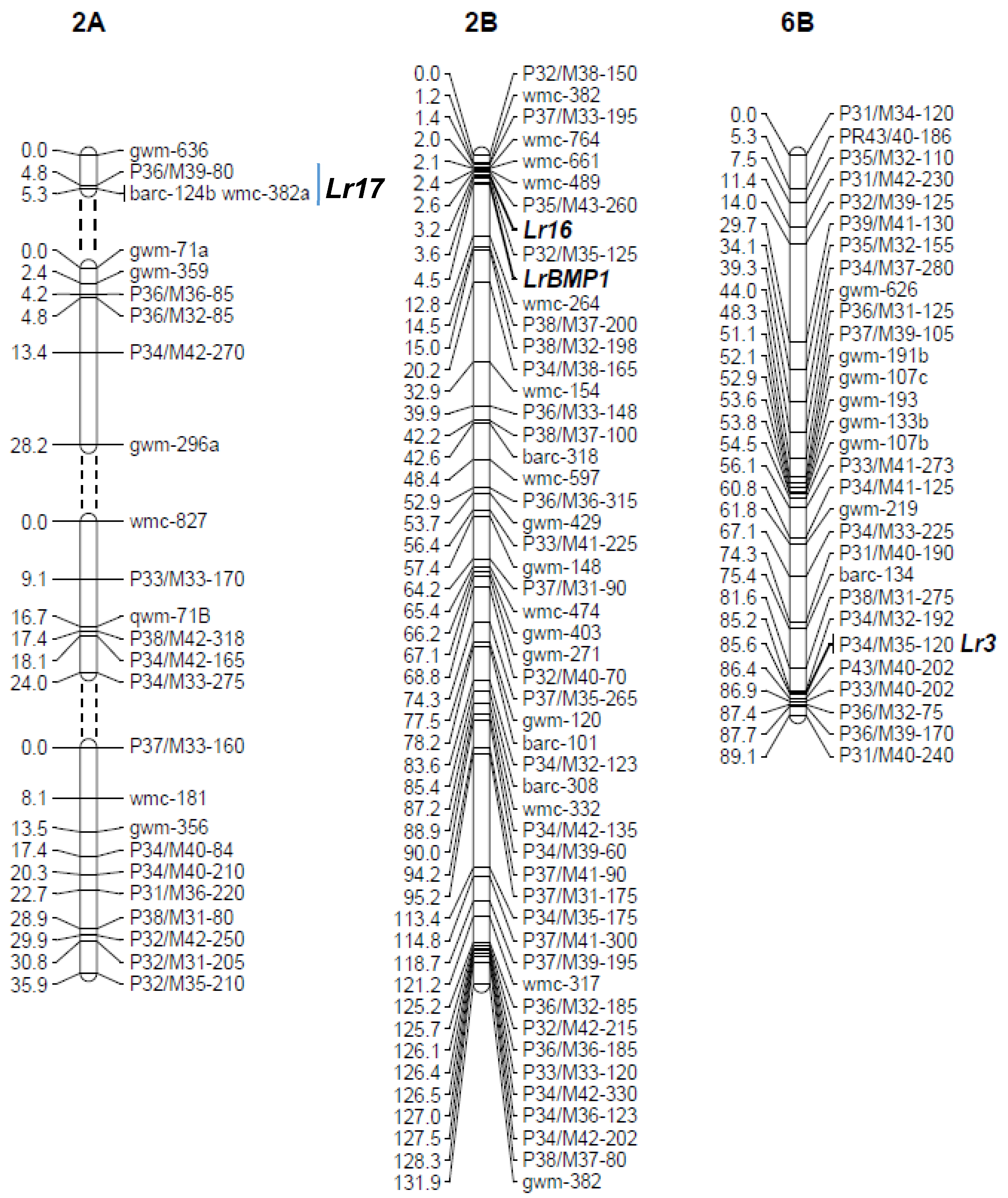
3.1. Genetic Mapping of Leaf Rust Resistance Genes in Buck Manantial
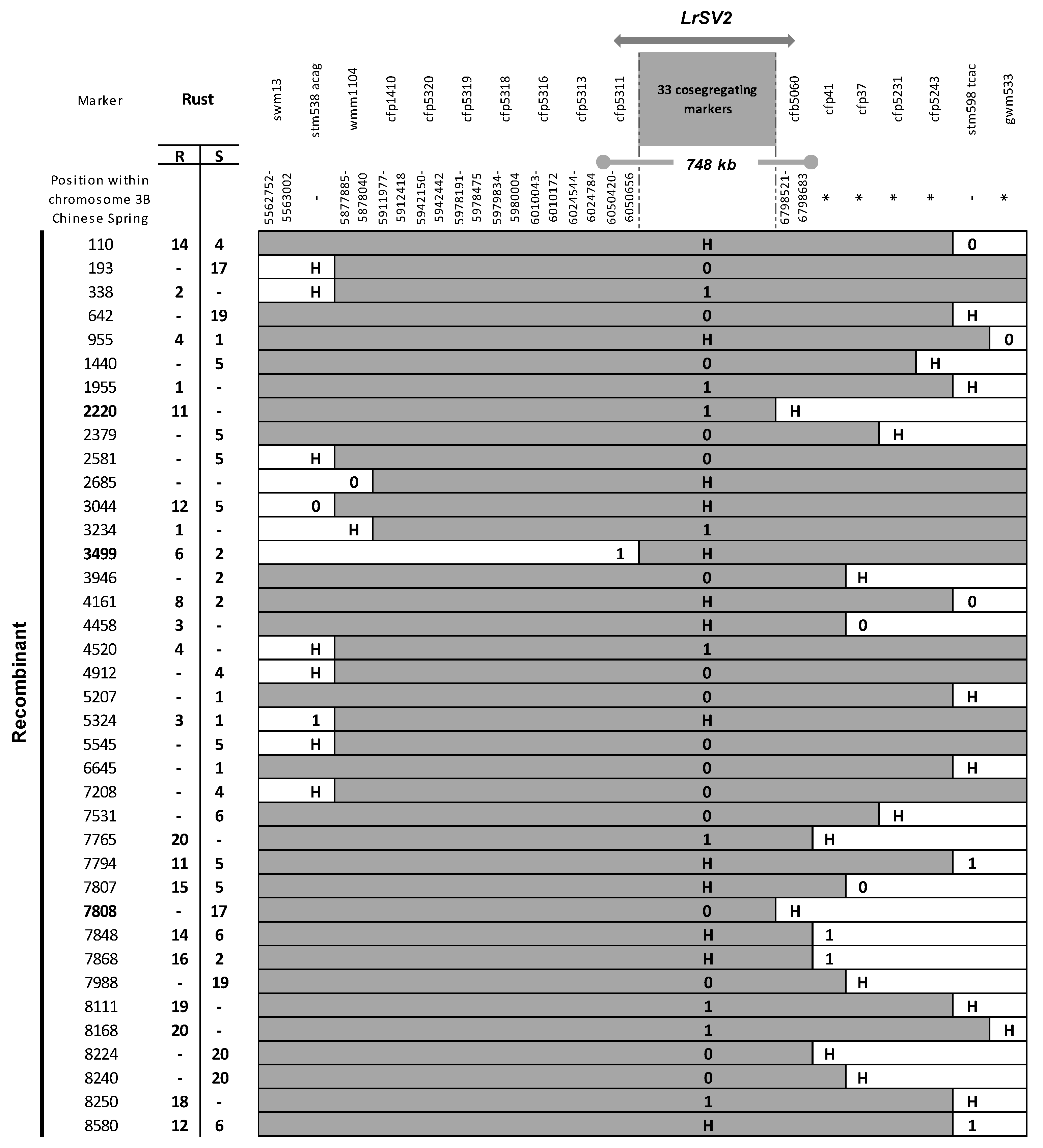
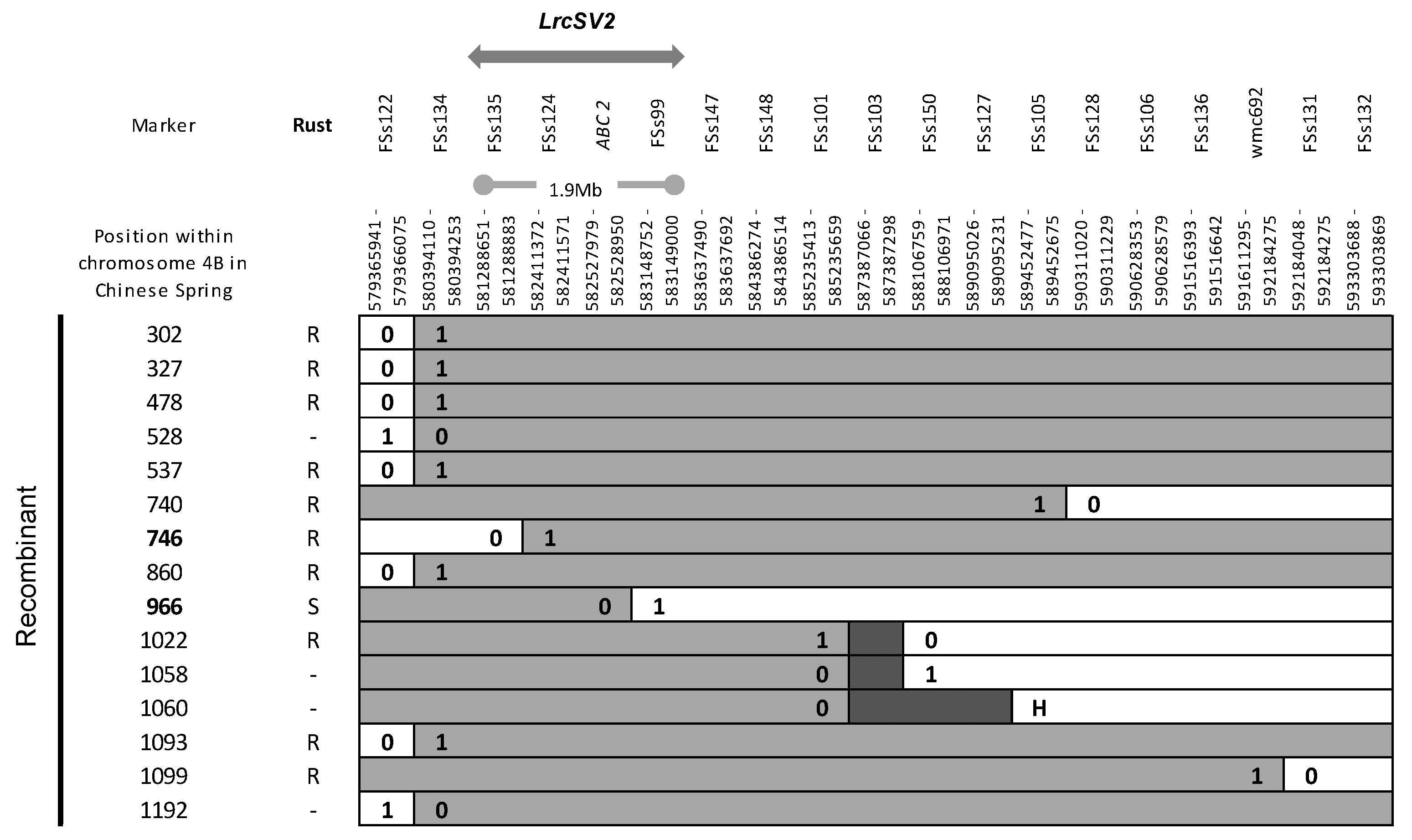
3.2. Fine Mapping of LrSV2 and LrcSV2
3.3. Introgression and Pyramiding of Leaf Rust Resistance Genes
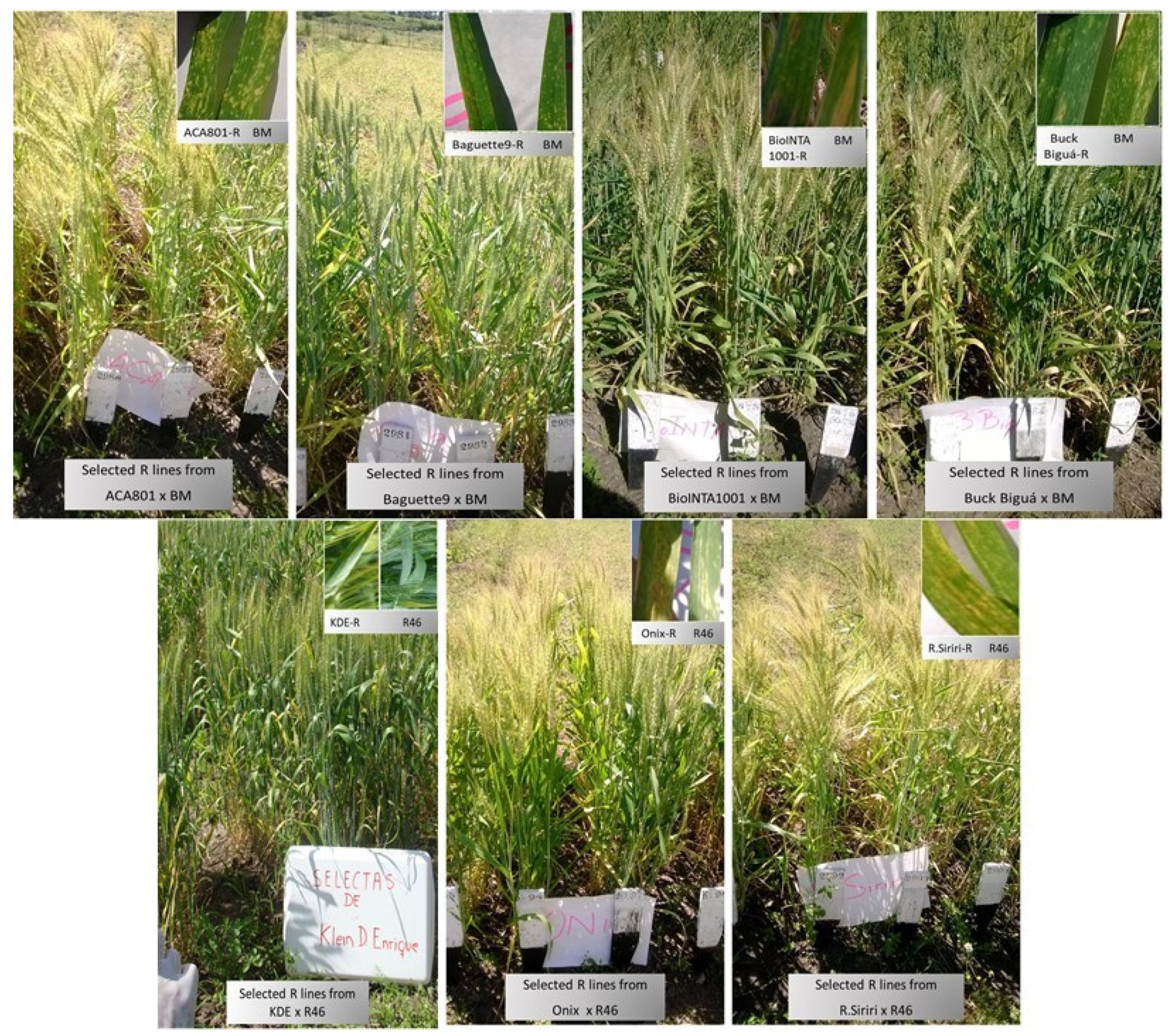
3.4. Field Testing of Introgressed Resistant Lines
4. Discusion
4.1. Identification and Introgression of Genetic Components of Durable Leaf Rust Resistance
4.2. Fine Mapping of Leaf Rust Resistance Genes Identified in Durable Varieties
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Huerta Espino, J.; Singh, R.P.; Germán, S.; McCallum, B.D.; Park, R.F.; Chen, W.Q.; Bhardwaj, S.C.; Goyeau, H. Global status of wheat leaf rust caused by Puccinia triticina. Euphytica 2011, 179, 143–160. [Google Scholar] [CrossRef]
- Rodriguez Amieva, P.J.; Tessi, J.L.; Frecha, J.H.; Vallega, J. Estimación de los daños producidos en la Argentina por las royas del tallo y de la hoja del trigo durante el período 1949–1958. Robigo 1961, 12, 18–22. [Google Scholar]
- Macagno, l.F.; Pizarro, J.B.; Cordone, G.E. Dirección Nacional Asistente de Planificación. Publicación Miscelánea n° 4; INTA: Buenos Aires, Argentina, 1993; pp. 15–28. [Google Scholar]
- Kolmer, J.A. Genetics of Resistance To Wheat Leaf Rust. Annu. Rev. Phytopathol. 1996, 34, 435–455. [Google Scholar] [CrossRef]
- Annone, J.G.; García, R.; Botta, G.; Ivancovich, A. Yield losses caused by “Leaf Rust” and “Yellow Spot” of wheat: Estimates in the north of the Province of Buenos Aires. Agric. Technol. Mag. 2001, 6, 21–23. [Google Scholar]
- Germán, S.; Kohli, M.; Chaves, M.; Barcellos, A.; Nisi, J.; Annone, J.; Madariaga, R.; de Viedma, L. Breakdown of resistance of wheat cultivars and estimated losses caused by recent changes in the leaf rust population in South America. In Proceedings of the 11th International Cereal Rusts and Powdery Mildews Conference Abstract, Norwich, UK, 22–27 August 2004; p. A2.21. [Google Scholar]
- Flor, H.H. Host-parasite interactions in flax rust-its genetics and other implications. Phytopathology 1955, 45, 680–685. [Google Scholar]
- Samborski, D.J.; Dyck, P.L. Inheritance of virulence in wheat leaf rust on the standard differential wheat varieties. Can. J. Genet. Cytol. 1968, 10, 24–32. [Google Scholar] [CrossRef]
- McIntosh, R.A.; Welllings, C.R.; Park, R.F. Wheat Rusts: An Atlas of Resistance Genes; Plant Breeding Institute, The University of Sydney: Sydney, Australia, 1995. [Google Scholar]
- McIntosh, R.A.; Dubcovsky, J.; Rogers, J.M.; Xia, X.; Raupp, W.J. Catalogue of gene symbols for wheat: 2020 supplement. Annu. Wheat Newsl. 2020, 66, 109–128. [Google Scholar]
- Antonelli, E.F. La Roya Anaranjada (Puccinia triticina Erikss). Sobre la Efímera Resistencia Observada en la Última Década en Cultivares Comerciales de Trigo de Amplia Difusión en la Argentina; Grafos SRL: Necochea, Argentina, 2003; 22p. [Google Scholar]
- Johnson, R. Durable resistance: Definition of, genetic control, and attainment in plant breeding. Phytopathology 1981, 71, 567–568. [Google Scholar] [CrossRef]
- Messmer, M.M.; Seyfarth, R.; Keller, M.; Schachermayr, G.; Winzeler, M.; Zanetti, S.; Feuillet, C.; Keller, B. Genetic analysis of durable leaf rust resistance in winter wheat. Theor. Appl. Genet. 2000, 100, 419–431. [Google Scholar] [CrossRef]
- Schnurbusch, T.; Paillard, S.; Schori, A.; Messmer, M.; Schachermayr, G.; Winzeler, M.; Keller, B. Dissection of quantitative and durable leaf rust resistance in Swiss winter wheat reveals a major resistance QTL in the Lr34 chromosomal region. Theor. Appl. Genet. 2003, 108, 477–484. [Google Scholar] [CrossRef]
- Sawhney, R.N.; Nayar, S.K.; Sharma, J.B.; Bedi, R. Mechanism of durable resistance: A new approach. Theor. Appl. Genet. 1989, 78, 229–232. [Google Scholar] [CrossRef] [PubMed]
- Pretorius, Z.A.; Roelfs, A.P. The role of Lr10, Lr13, and Lr34 in the expression of adult-plant resistance to leaf rust in the wheat cultivar Era. Plant Dis. 1996, 80, 199–202. [Google Scholar] [CrossRef]
- Ingala, L.; Lopez, M.; D2arino, M.; Pergolesi, M.F.; Diéguez, M.J.; Sacco, F. Genetic analysis of leaf rust resistance genes and associated markers in the durable resistant wheat cultivar Sinvalocho MA. Theor. Appl. Genet. 2012, 124, 1305–1314. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Phogat, B.S.; Vikas, V.K.; Sharma, A.K.; Saharan, M.S.; Singh, A.K.; Kumari, J.; Singh, R.; Jacob, S.R.; Singh, G.P.; et al. Mining of Indian wheat germplasm collection for adult plant resistance to leaf rust. PLoS ONE 2019, 14, e0213468. [Google Scholar] [CrossRef] [Green Version]
- Favret, E.A.; Saione, H.A.; Franzone, P.M. New approaches in breeding for disease resistance. In Proceedings of the Cereal Breeding and Production Symposium, Special Report 718, Oregon State University, Marcos Juarez, Argentina, 7–12 November 1983; pp. 397–411. [Google Scholar]
- Samborski, D.J. The Cereal Rusts; Roelfs, A., Bushnell, W., Eds.; Academic Press Inc. (Harcourt Brace Jovanovich, Publishers): New York, NY, USA, 1985; Volume II, pp. 39–59. [Google Scholar]
- Dyck, P.L. The inheritance of leaf rust resistance in wheat cultivars kenyon and buck manantial. Can. J. Plant Sci. 1989, 69, 1113–1117. [Google Scholar] [CrossRef] [Green Version]
- Dyck, P.L. Genetics of adult-plant leaf rust resistance in ‘Chinese Spring’ and ‘Sturdy’ wheats. Crop Sci. 1991, 31, 309–311. [Google Scholar] [CrossRef]
- Tsilo, T.J.; Kolmer, J.A.; Anderson, J.A. Molecular Mapping and Improvement of Leaf Rust Resistance in Wheat Breeding Lines. Phytopathology 2014, 104, 865–870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lan, C.; Hale Lago, L.; Herrera-Foessel, S.A.; Basnet Bhoja, R.; Randhawa, M.S.; Huerta-Espino, J.; Dubcovsky, J.; Singh, R.P. Characterization and Mapping of Leaf Rust and Stripe Rust Resistance Loci in Hexaploid Wheat Lines UC1110 and PI610750 under Mexican Environments. Front. Plant Sci. 2017, 8, 1450. [Google Scholar] [CrossRef] [Green Version]
- Rollar, S.; Serfling, A.; Geyer, M.; Hartl, L.; Mohler, V.; Ordon, F. QTL mapping of adult plant and seedling resistance to leaf rust (Puccinia triticina Eriks.) in a multiparent advanced generation intercross (MAGIC) wheat population. Theor. Appl. Genet. 2021, 134, 37–51. [Google Scholar] [CrossRef]
- Diéguez, M.J.; Petignat, C.; Ferella, L.; Fiorentino, G.; Silva, M.; Dabove, M.A.; Yanez, G.R.; López, M.; Pergolesi, M.F.; Ingala, L.; et al. Mapping a gene on wheat chromosome 4BL involved in a complementary interaction with adult plant leaf rust resistance gene LrSV2. Theor. Appl. Genet. 2018, 131, 2333–2344. [Google Scholar] [CrossRef]
- Saione, H.A.; Favret, E.A.; Franzone, P.M.; Sacco, F. Host genetic analysis by using Puccinia recondita tritici induced mutants for in-creased virulence. J. Phytopathol. 1993, 138, 225–232. [Google Scholar] [CrossRef]
- Tanksley, S.D.; Young, N.; Paterson, A.H.; Bonierbale, M.W. RFLP Mapping in Plant Breeding: New Tools for an Old Science. Nat. Biotechnol. 1989, 7, 257–264. [Google Scholar] [CrossRef]
- Ragot, M.; Biasiolli, M.; Delbut, M.F.; Dell’Orco, A.; Malgarini, L.; Thevenin, P.; Vernoy, J.; Vivant, J.; Zimmermann, R.; Gay, G. Marker-assisted backcrossing: A practical example. In Techniques Et Utilisations Des Marqueurs Moléculaires; Les Colloques, No. 72; INRA Editions: Montpellier, France, 1994; pp. 45–56. [Google Scholar]
- Johnson, G.R.; Mumm, R.H. Marker assisted maize breeding. In Proceedings of the 51st Corn and Sorghum Conference, Chicago, IL, USA, 11–12 December 1996; pp. 75–84. [Google Scholar]
- Leonova, I.N.; Skolotneva, E.S.; Salina, E.A. Genome-wide association study of leaf rust resistance in Russian spring wheat varieties. BMC Plant Biol. 2020, 20, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Yadava, P.S.; Mishraa, V.K.; Aruna, B.; Chandb Vishwakarmaa, R.M.; Vasisthaa, N.K.; Mishrac, A.N.; Kalappanavard, I.K.; Joshi, A.K. Enhanced resistance in wheat against stem rust achieved by marker assisted backcrossing involving three independent Sr genes. Curr. Plant Biol. 2015, 2, 25–33. [Google Scholar] [CrossRef] [Green Version]
- Vrana, J.; Kubaláková, M.; Simková, H.; Číhalíkovái, J.; Lysak, M.; Dolezel, J. Flow Sorting of Mitotic Chromosomes in Common Wheat (Triticum aestivum L.). Genetics 2000, 156, 2033–2041. [Google Scholar] [CrossRef] [PubMed]
- Choulet, F.; Alberti, A.; Theil, S.; Glover, N.; Barbe, V.; Daron, J.; Pingault, L.; Sourdille, P.; Couloux, A.; Paux, E.; et al. Structural and functional partitioning of bread wheat chromosome 3B. Science 2014, 345, 1249721. [Google Scholar] [CrossRef]
- IWGSC—International Wheat Genome Sequencing Consortium. A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 2014, 345, 1251788. [Google Scholar] [CrossRef]
- Diéguez, M.J.; Pergolesi, M.F.; Velasquez, S.M.; Ingala, L.; López, M.; Darino, M.; Paux, E.; Feuillet, C.; Sacco, F. Fine mapping of LrSV2, a race-specific adult plant leaf rust resistance gene on wheat chromosome 3BS. Theor. Appl. Genet. 2014, 127, 1133–1141. [Google Scholar] [CrossRef] [PubMed]
- Favret, E.A.; Cenoz, H.P.; Sikvero Sanz, O.I.; Solari, R.; Mujica, F.L. Efecto de Posición inducido en trigo para reacción a las royas. In Plants; Symp.IAEA/FAO: Pullman, WA, USA, 1969; pp. 123–133. [Google Scholar]
- Germán, S.E.; Kolmer, J.A. Virulence phenotypes of Puccinia recondita f. sp. tritici in Uruguay. Plant Dis. 1994, 78, 1139–1141. [Google Scholar] [CrossRef]
- Mains, E.B.; Jackson, H.S. Physiologic specialization in the leaf rust of wheat; Puccinia triticina Erikss. Phytopathology 1926, 16, 89–120. [Google Scholar]
- Long, D.L.; Kolmer, J.A. A North American system of nomenclature for Puccinia recondita f. sp. tritici. Phytopathology 1989, 79, 525–529. [Google Scholar] [CrossRef]
- Sacco, F.; Suárez, E.Y.; Naranjo, T. Mapping of the leaf rust resistance gene Lr3 on chromosome 6B of Sinvalocho MA wheat. Genome 1998, 41, 686–690. [Google Scholar] [CrossRef]
- Diéguez, M.J.; Altieri, E.; Ingala, L.R.; Perera, E.; Sacco, F.; Naranjo, T. Physical and genetic mapping of AFlPs and the leaf rust resistance Lr3 gene on chromosome 6Bl of wheat. Theor. Appl. Genet. 2006, 112, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Benbouza, H.; Jacquemin, J.M.; Baudoin, J.P.; Mergeai, G. Optimization of a reliable, fast, cheap and sensitive silver staining method to detect SSr markers in polyacrylamide gels. Biotechnol. Agron. Soc. Environ. 2006, 10, 77–81. [Google Scholar]
- Röder, M.S.; Korzun, V.; Wendehake, K.; Plaschke, J.; Tixier, M. A microsatellite map of wheat. Genetics 1998, 149, 2007–2023. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.J.; Shi, J.R.; Singh, S.; Fickus, W.; Costa, J.M.; Lewis, J.; Gill, B.S.; Ward, R.; Cregan, P.B. Development and mapping of microsatellite (SSR) markers in wheat. Theor. Appl. Genet. 2005, 110, 550–560. [Google Scholar] [CrossRef]
- Somers, D.J.; Isaac, P.; Edwards, K. A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 2004, 109, 1105–1114. [Google Scholar] [CrossRef]
- Sourdille, P.; Singh, S.; Cadalen, T.; Brown-Guedira, G.L.; Gay, G.; Qi, L.; Gill, B.S.; Dufour, P.; Murigneux, A.; Bernard, M. Microsatellite-based deletion bin system for the establishment of genetic-physical map relationships in wheat (Triticum aestivum L.). Funct. Integr. Genom. 2004, 4, 12–25. [Google Scholar] [CrossRef]
- Paux, E.; Roger, D.; Badaeva, E.; Gay, G.; Bernard, M.; Sourdille, P.; Feuillet, C. Characterizing the composition and evolution of homoe-ologous genomes in hexaploid wheat through BAC-end sequencing on chromosome 3B. Plant J. 2006, 48, 463–474. [Google Scholar] [CrossRef]
- Liu, S.; Anderson, J.A. Targeted molecular mapping of a major wheat QTl for Fusarium head blight resistance using wheat ESTs and synteny with rice. Genome 2003, 46, 817–823. [Google Scholar] [CrossRef] [PubMed]
- Hayden, M.J.; Stephenson, P.; Logojan, A.M.; Khatkar, D.; Rogers, C.; Elsden, J.; Koebner, R.M.D.; Snape, J.W.; Sharp, P.J. Development and genetic mapping of sequence-tagged microsatellites (STMs) in bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 2006, 113, 1271–1281. [Google Scholar] [CrossRef]
- Pestsova, E.; Ganal, M.W.; Röde, M.S. Isolation and mapping of microsatellite markers specific for the D genome of bread wheat. Genome 2000, 43, 689–697. [Google Scholar] [CrossRef] [PubMed]
- Bryan, G.J.; Collins, A.J.; Stephenson, P.; Orry, A.; Smith, J.B.; Gale, M.D. Isolation and characterisation of microsatellites from hexaploid bread wheat. Theor. Appl. Genet. 1997, 94, 557–563. [Google Scholar] [CrossRef]
- Bossolini, E.; Krattinger, S.G.; Keller, B. Development of simple sequence repeat markers specific for the Lr34 resistance region of wheat using sequence information from rice and Aegilops tauschii. Theor. Appl. Genet. 2006, 113, 1049–1062. [Google Scholar] [CrossRef] [PubMed]
- Van Ooijen, J.W.; Voorrips, R.E. JoinMap®, Version 3.0, Software for the Calculation of Genetic Linkage Maps; Plant Re-Search International: Wageningen, The Netherlands, 2001. [Google Scholar]
- Paux, E.; Faure, S.; Choulet, F.; Roger, D.; Gauthier, V.; Martinant, J.P.; Sourdille, P.; Balfourier, F.; Le-Paslier, M.C.; Chauveau, A.; et al. Insertion site-based polymorphism markers open new perspectives for genome saturation and marker-assisted se-lection in wheat. Plant J. 2010, 8, 196–210. [Google Scholar]
- Martins, W.S.; Lucas, D.C.S.; Neves, K.F.D.S.; Bertioli, D.J. WebSat—A Web Software for MicroSatellite marker development. Bioinformation 2009, 3, 282–283. [Google Scholar] [CrossRef] [Green Version]
- Benchimol, L.L.; Souza, C.L.D.; Souza, A.P.D. Microsatellite assisted backcross selection in maize. Genet. Mol. Biol. 2005, 28, 789–797. [Google Scholar] [CrossRef]
- McCartney, C.A.; Somers, D.J.; McCallum, B.D.; Thomas, J.; Humphreys, D.G.; Menzies, J.G.; Brown, P.D. Microsatellite tagging of the leaf rust resistance gene Lr16 on wheat chromosome 2BSc. Mol. Breed. 2005, 15, 329–337. [Google Scholar] [CrossRef]
- Choulet, F.; Wicker Rustenholz, C.; Paux, E.; Salse, J.; Leroy, P.; Schlub, S.; Le Paslier, M.C.; Magdelenat, G.; Gonthier, C.; Couloux, A.; et al. Megabase level sequencing reveals contrasted organization and evolution patterns of the wheat gene and transposable element spaces. Plant Cell 2010, 22, 1686–1701. [Google Scholar] [CrossRef] [Green Version]
- Paux, E.; Sourdille, P.; Salse, J.; Saintenac, C.; Choulet, F.; Leroy, P.; Korol, A.; Michalak, M.; Kianian, S.; Spielmeyer, W.; et al. A Physical Map of the 1-Gigabase Bread Wheat Chromosome 3B. Science 2008, 322, 101–104. [Google Scholar] [CrossRef]
- Qiu, L.; Wang, H.; Li, Y.; Wang, W.; Liu, Y.; Mu, J.; Geng, M.; Guo, W.; Hu, Z.; Ma, J.; et al. Fine Mapping of the Wheat Leaf Rust Resistance Gene LrLC10 (Lr13) and Validation of Its Co-segregation Markers. Front. Plant Sci. 2020, 11, 470. [Google Scholar] [CrossRef]
- Zeller, F.J. 1B/1R wheat-rye chromosome substitutions and translocations. In Proceedings of the 4th International Wheat Genetics Symposium, Colombia, MO, USA, 6–11 August 1973; Alien Genetic Material. pp. 209–221.14. [Google Scholar]
- Das, B.K.; Saini, A.; Bhagwat, S.G.; Jawali, N. Development of SCAR markers for identification of stem rust resistance gene Sr31 in the homozygous or heterozygous condition in bread wheat. Plant Breed. 2006, 125, 544–549. [Google Scholar] [CrossRef]
- Ellis, J.G.; Lagudah, E.; Spielmeyer, W.; Dodds, P. The past, present and future of breeding rust resistant wheat. Front. Plant Sci. 2014, 5, 641. [Google Scholar] [CrossRef] [Green Version]
- Kolmer, J.A.; Bernardo, A.; Bai, G.; Hayden, M.J.; Chao, S. Adult Plant Leaf Rust Resistance Derived from Toropi Wheat is Conditioned by Lr78 and Three Minor QTL. Phytopathology 2018, 108, 246–253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Figlan, S.; Ntushelo, K.; Mwadzingeni, L.; Terefe, T.; Tsilo Toi, J.; Shimelis, H. Breeding Wheat for Durable Leaf Rust Resistance in Southern Africa: Variability, Distribution, Current Control Strategies, Challenges and Future Prospects. Front. Plant Sci. 2020, 11, 549. [Google Scholar] [CrossRef]
- Roelfs, A.P. Breeding Strategies for Resistance to the Rusts of Wheat; DF CIMMYt: Mexico City, Mexico, 1988; pp. 10–22. [Google Scholar]
- German, S.E.; Kolmer, J.A. Effect of gene Lr34 in the enhancement of resistance to leaf rust of wheat. Theor. Appl. Genet. 1992, 84, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Vanzetti, L.S.; Brevis, J.C.; Dubcovsky, J.; Helguera, M. Identification of microsatellites linked to Lr47. Electron. J. Biotechnol. 2006, 9, 267–271. [Google Scholar] [CrossRef] [Green Version]
- Helguera, M.; Khan, I.; Dubcovsky, J. Development of PCR markers for the wheat leaf rust resistance gene Lr47. Theor. Appl. Genet 2000, 100, 1137–1143. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.; Knox, R.E.; DePauw, R.M.; Singh, A.K.; Cuthbert, R.D.; Campbell, H.L.; Shorter, S.; Bhavani, S. Stripe rust and leaf rust resistance QTL mapping, epistatic interactions, and co-localization with stem rust resistance loci in spring wheat evaluated over three continents. Theor. Appl. Genet. 2014, 127, 2465–2477. [Google Scholar] [CrossRef]
- Moullet, O.; Fossati, D.; Mascher, F.; Guadagnolo, R.; Schori, A. Use of marker-assisted selection (MAS) for pyramiding leaf rust resistance genes (Lr9, Lr24, Lr22a) in wheat. In Proceedings of the Tagung der Vereinigung der Pfl anzenzüchter und Saatgutkaufl eute Österreichs, Irdning, Austria, 24–26 November 2009; pp. 143–146. [Google Scholar]
- Randhawa, H.S.; Mutti, J.S.; Kidwell, K.; Morris, C.F.; Chen, X.; Gill, K.S. Rapid and Targeted Introgression of Genes into Popular Wheat Cultivars Using Marker-Assisted Background Selection. PLoS ONE 2009, 4, e5752. [Google Scholar] [CrossRef] [Green Version]
- Johnson, R. Breeding Strategies for Resistance to the Rusts of Wheat; DF CIMMYt: Mexico City, Mexico, 1988; pp. 63–75. [Google Scholar]
- Qi, A.-Y.; Zhang, P.-P.; Zhou, Y.; Yao, Z.-J.; Li, Z.-F.; Liu, D.-Q. Mapping of QTL conferring leaf rust resistance in Chinese wheat lines W014204 and Fuyu 3 at adult plant stage. J. Integr. Agric. 2016, 15, 18–28. [Google Scholar] [CrossRef]
- Darino, M.A.; Diéguez, M.J.; Singh, D.; Ingala, L.R.; Pergolesi, M.F.; Park, R.F.; McIntosh, R.A.; Sacco, F. Detection and location of Lr11 and other leaf rust resistance genes in the durably resistant wheat cultivar Buck Poncho. Euphytica 2015, 206, 135–147. [Google Scholar] [CrossRef]
- Lowe, I.; Cantu, D.; Dubcovsky, J. Durable resistance to the wheat rusts: Integrating systems biology and traditional phenotype-based research methods to guide the deployment of resistance genes. Euphytica 2011, 179, 69–79. [Google Scholar] [CrossRef] [Green Version]
- Cloutier, S.; McCallum, B.D.; Loutre, C.; Banks, T.W.; Wicker, T.; Feuillet, C.; Keller, B.; Jordan, M.C. Leaf rust resistance gene Lr1, isolated from bread wheat (Triticum aestivum L.) is a member of the large psr567 gene family. Plant Mol. Biol. 2007, 65, 93–106. [Google Scholar] [CrossRef] [PubMed]
- Feuillet, C.; Travella, S.; Stein, N.; Albar, L.; Nublat, A.; Keller, B. Map-based isolation of the leaf rust disease resistance gene Lr10 from the hexaploid wheat (Triticum aestivum L.) genome. Proc. Natl. Acad. Sci. USA 2003, 100, 15253–15258. [Google Scholar] [CrossRef] [Green Version]
- Huang, L.; Brooks, S.A.; Li, W.; Fellers, J.P.; Trick, H.; Gill, B.S. Map-Based Cloning of Leaf Rust Resistance Gene Lr21 From the Large and Polyploid Genome of Bread Wheat. Genetics 2003, 164, 655–664. [Google Scholar] [CrossRef] [PubMed]
- Krattinger, S.G.; Lagudah, E.S.; Spielmeyer, W.; Singh, R.P.; Huerta-Espino, J.; McFadden, H.; Bossolini, E.; Selter, L.L.; Keller, B. A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 2009, 623, 1360–1363. [Google Scholar] [CrossRef] [Green Version]
- Stein, N.; Feuillet, C.; Wicker, T.; Schlagenhauf, E.; Keller, B. Subgenome chromosome walking in wheat: A 450-kb physical contig in Triticum monococcum l. spans the Lr10 resistance locus in hexaploid wheat (Triticum aestivum L.). Proc. Natl. Acad. Sci. USA 2000, 97, 13436–13441. [Google Scholar] [CrossRef] [Green Version]
- Ling, H.-Q.; Zhu, Y.; Keller, B. High-resolution mapping of the leaf rust disease resistance gene Lr1 in wheat and characterization of BAC clones from the Lr1 locus. Theor. Appl. Genet. 2003, 106, 875–882. [Google Scholar] [CrossRef]
- Clark, R.M.; Tavaré, S.; Doebley, J. Estimating a Nucleotide Substitution Rate for Maize from Polymorphism at a Major Domestication Locus. Mol. Biol. Evol. 2005, 22, 2304–2312. [Google Scholar] [CrossRef]
- Rabinowicz, P.D.; Citek, R.; Budiman, M.A.; Nunberg, A.; Bedell, J.A.; Lakey, N.; O’Shaughnessy, A.L.; Nascimento, L.U.; McCombie, W.R.; Martienssen, R.A. Differential methylation of genes and repeats in land plants. Genome Res. 2005, 15, 1431–1440. [Google Scholar] [CrossRef] [Green Version]
- Mago, R.; Tabe, L.; Vautrin, S.; Simková, H.; Kubaláková, M.; Upadhyaya, N.; Appels, R. Major haplotype divergence including multiple germin-like protein genes, at the wheat Sr2 adult plant stem rust resistance locus. BMC Plant Biol. 2014, 14, 379. [Google Scholar] [CrossRef] [Green Version]
- Mago, R.; Tabe, L.; McIntosh, R.A.; Pretorius, Z.; Kota, R.; Paux, E.; Wicker, T.; Breen, J.; Lagudah, E.S.; Ellis, J.G.; et al. A multiple resistance locus on chromosome arm 3BS in wheat confers resistance to stem rust (Sr2), leaf rust (Lr27) and powdery mildew. Theor. Appl. Genet. 2011, 123, 615–623. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.P.; McIntosh, R.A. Complementary genes for reaction to Puccinia recondita tritici in Triticum aestivum. I. Genetic and linkage studies. Can. J. Genet. Cytol. 1984, 26, 723–735. [Google Scholar] [CrossRef]
- Singh, D.; Park, R.F.; McIntosh, R.A. Genetic relationship between the adult plant resistance gene Lr12 and the complementary gene Lr31 for seedling resistance to leaf rust in common wheat. Plant Pathol. 1999, 48, 567–573. [Google Scholar] [CrossRef]
- Isidore, E.; Scherrer, B.; Chalhoub, B.; Feuillet, C.; Keller, B. Ancient haplotypes resulting from extensive molecular rearrangements in the wheat A genome have been maintained in species of three different ploidy levels. Genome Res. 2005, 15, 526–536. [Google Scholar] [CrossRef] [Green Version]
- Marande, W.; Vautrin, S.; Bellec, A.; Prat, E.; Beydon, G.; Pergolesi, M.F.; Diéguez, M.J.; Sacco, F.; Berges, H. Non-gridded BAC libraries to iso-late genomic regions involved in different cellular processes of interest in wheat varieties. In Proceedings of the 22nd International Triticeae Mapping Initiative Workshop, Fargo, ND, USA, 25–29 June 2012. [Google Scholar]
- Steuernagel, B.; Periyannan, S.; Hernández-Pinzón, I.; Witek, K.; Rouse, M.N.; Yu, G.; Hatta, A.; Ayliffe, M.; Bariana, H.; Jones, J.; et al. Rapid cloning of disease-resistance genes in plants using mutagenesis and sequence capture. Nat. Biotechnol. 2016, 34, 652–655. [Google Scholar] [CrossRef]
- Sánchez-Martín, J.; Steuernagel, B.; Ghosh, S.; Herren, G.; Hurni, S.; Adamski, N.; Vrána, J.; Kubaláková, M.; Krattinger, S.G.; Wicker, T.; et al. Rapid gene isolation in barley and wheat by mutant chromosome sequencing. Genome Biol. 2016, 17, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thind, A.K.; Wicker, T.; Simkova, H.; Fossati, D.; Moullet, O.; Brabant, C.; Vrana, J.; Dolezel, J.; Krattinger, S.G. Rapid cloning of genes in hexaploid wheat using cultivar-specific long-range chromosome assembly. Nat. Biotechnol. 2017, 35, 793–796. [Google Scholar] [CrossRef] [PubMed]
- Miedaner, T.; Juroszek, P. Climate change will influence disease resistance breeding in wheat in Northwestern Europe. Theor. Appl. Genet. 2021, 134, 1–15. [Google Scholar] [CrossRef]
- Mago, R.; Simkova, H.; Brown-Guedira, G.; Dreisigacker, S.; Breen, J.; Jin, Y.; Singh, R.; Appels, R.; Lagudah, E.S.; Ellis, J.; et al. An accurate DNA marker assay for stem rust resistance gene Sr2 in wheat. Theor. Appl. Genet. 2011, 122, 735–744. [Google Scholar] [CrossRef] [PubMed]
| Cross | Rust Race | Genes Detected | Resistance Type | Nomenclature * |
|---|---|---|---|---|
| R46 × KDE | Ca04KDE | LrG6 | ASR | MCT |
| R46 × R.Siriri | Ma07Bg9 | LrG6 | ASR | MFT |
| R46 × Onix | Ma07Bg9 | LrG6 | ASR | MFT |
| BM × Baguette9 | Ma07Bg9 | Lr16 | ASR | MFT |
| BM × ACA801 | Ma07Bg9 | Lr16 | ASR | MFT |
| BM × Buck Biguá | Ma07Bg9 | Lr16 | ASR | MFT |
| BM × BioINTA1001 | Ma07Bg9 | Lr16 | ASR | MFT |
| BM × P | Ma04BuckGuapo | Lr16 | ASR | MBT |
| BM × P | Rq05Cronox | Lr16 + Lr17 | ASR | MDR |
| BM × P | 66 | Lr3 + Lr16 + Lr17 | ASR | JBB |
| BM × P | Ca02Lr17 | LrBMP1 | APR | MHP |
| SV × G6 | Ca02G1R | LrSV2 | APR | MCG |
| SV × P | Ca02G1R | LrcSV2 | APR | MCG |
| BM × ACA801 | BM × Baguette9 | BM × BioINTA1001 | BM × Buck Biguá |
|---|---|---|---|
| barc124 1 | barc18 | gwm148 | barc124 |
| gwm148 | gwm374 | gwm374 | gwm257 |
| gwm257 1 | gwm403 | wmc597 | gwm374 |
| gwm374 | wmc597 | wmc764 2 | gwm630 |
| gwm403 | wmc764 2 | wmc764 2 | |
| gwm630 | |||
| gwm636 1 | |||
| wmc597 wmc764 2 |
| Gene | Resistance Type | Associated Marker |
|---|---|---|
| LrSV1 | APR | gwm261/P31/M42 |
| LrSV2 | APR | gwm533/P31/M37 |
| LrcSV2 | APR | gwm149 |
| Lr16 | ASR | wmc764 |
| Lr26 | ASR | SCAR SCSS30.2 |
| Place/Year | Mean Number of Pustules/cm2 LrBMP1 Presence vs. Absence | t-Tests p Value |
|---|---|---|
| Ca04 | 34.8 vs. 69.7 | p > 0.001 |
| Ca05 | 41.5 vs. 70.6 | p > 0.001 |
| Ca06 | 29.8 vs. 61.4 | p > 0.001 |
| Ca07 | 33.7 vs. 67.3 | p > 0.001 |
| Ma06 | 38.1 vs. 57.4 | p > 0.001 |
| Ma07 | 32.2 vs. 62.7 | p > 0.001 |
| Ma09 | 30.6 vs. 60.2 | p > 0.001 |
| Rq06 | 45.4 vs. 66 | p > 0.001 |
| Ca04 + Ca05 + Ca06 + Ca07 | 35 vs. 67 | p > 0.001 |
| Ma06 + Ma07 + Ma09 | 33.6 vs. 60.1 | p > 0.001 |
| Ca04 + Ca05 + Ca06 + Ca07+ Ma06 + Ma07 + Ma09 + Rq06 | 36.1 vs. 64.1 | p > 0.001 |
| Cross | BC1 | BC2 | Selfed BC2 | Progeny of Selfed BC2 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | Lr161 | Lr162 + BM-2BS 3 | Selected (%RG 4) | Total | Lr161 | Lr162 + BM-2BS 3 | Selected (%RG 4) | Total | Lr161 | Lr162 + BM-2BS 3 | Final Field Selection 5 | %RG 6 | |
| BM × ACA801 | 210 | 94 | 22 | 2 (71–77) | 303 | 139 | 22 | 7 (80–86) | 1115 | 299 | 19 | 3 | >80 |
| BM × Baguette9 | 138 | 75 | 22 | 5 (78–86) | 337 | 177 | 19 | 8 (85–94) | 1997 | 529 | 21 | 1 | >85 |
| BM × BioINTA1001 | 308 | NT | 22 | 6 (78–85) | 474 | NT | 22 | 10 (83–90) | 1734 | 428 | 11 | 4 | >83 |
| BM × Buck Biguá | 283 | 139 | 19 | 5 (72–80) | 409 | 207 | 22 | 7 (81–88) | 2214 | 509 | 11 | 3 | >81 |
| Cross | BC1 | BC2 | Selfed BC2 | Progeny of Selfed BC2 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | LrG6 1 | LrSV1 + LrSV2 2 | Selected (%RG 3) | Total | LrG6 1 | LrSV1 + LrSV2 2 | Selected (%RG 3) | Total | LrSV1, LrSV22, LrG61 and LrcSV2 4 | Final Field Selection 5 | %RG 6 | |
| R46 × Onix | 198 | 94 | 19 | 4 (79–87) | 225 | 124 | 22 | 9 (85–96) | 1458 | 19 | 2 | >85 |
| R46 × R.Siriri | 215 | 106 | 21 | 2 (77–84) | 143 | 75 | 16 | 8 (84–97) | 997 | 8 | 2 | >84 |
| Cross | BC1 | Selfed BC1 | BC2 of Selfed BC1 | BC2F3 | BC2F4 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | LrG6 1 | LrSV1 + LrSV2 2 | Selected (%RG 3) | Total | LrG6 1 | LrSV1 + LrSV2 2 | Selected (%RG 3) | Total | LrG61 | LrSV1 + LrSV2 4 | Selected (%RG 3) | Total | LrSV1 + LrSV2 4, LrcSV2 5, Lr26 6 | Final Field Selection 7 | %RG 8 | |
| R46 × KDE | 115 | 59 | 15 | 4 (37–52) | 156 | 110 | 28 | 2 (65–71) | 66 | 66 | 21 | 4 (93–96) | 83 | 8 | 5 | >93 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Diéguez, M.J.; López, M.; Altieri, E.; Pergolesi, M.F.; Dabove, M.A.; Cuyeu, A.R.; Justus, N.; Kandus, M.; Ingala, L.; Sacco, F. Characterization and Use in Wheat Breeding of Leaf Rust Resistance Genes from Durable Varieties. Biology 2021, 10, 1168. https://doi.org/10.3390/biology10111168
Diéguez MJ, López M, Altieri E, Pergolesi MF, Dabove MA, Cuyeu AR, Justus N, Kandus M, Ingala L, Sacco F. Characterization and Use in Wheat Breeding of Leaf Rust Resistance Genes from Durable Varieties. Biology. 2021; 10(11):1168. https://doi.org/10.3390/biology10111168
Chicago/Turabian StyleDiéguez, María José, Micaela López, Emiliano Altieri, María Fernanda Pergolesi, Marisol Alicia Dabove, Alba Romina Cuyeu, Nadia Justus, Mariana Kandus, Lorena Ingala, and Francisco Sacco. 2021. "Characterization and Use in Wheat Breeding of Leaf Rust Resistance Genes from Durable Varieties" Biology 10, no. 11: 1168. https://doi.org/10.3390/biology10111168
APA StyleDiéguez, M. J., López, M., Altieri, E., Pergolesi, M. F., Dabove, M. A., Cuyeu, A. R., Justus, N., Kandus, M., Ingala, L., & Sacco, F. (2021). Characterization and Use in Wheat Breeding of Leaf Rust Resistance Genes from Durable Varieties. Biology, 10(11), 1168. https://doi.org/10.3390/biology10111168






