Shape-Memory Nanofiber Meshes with Programmable Cell Orientation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Polymerization and Characterization
2.3. Electrospinning Method and Characterization of Nanofibers
2.4. Shape Memory Behavior
2.5. Cell Culture on Nanofiber Mesh
3. Results
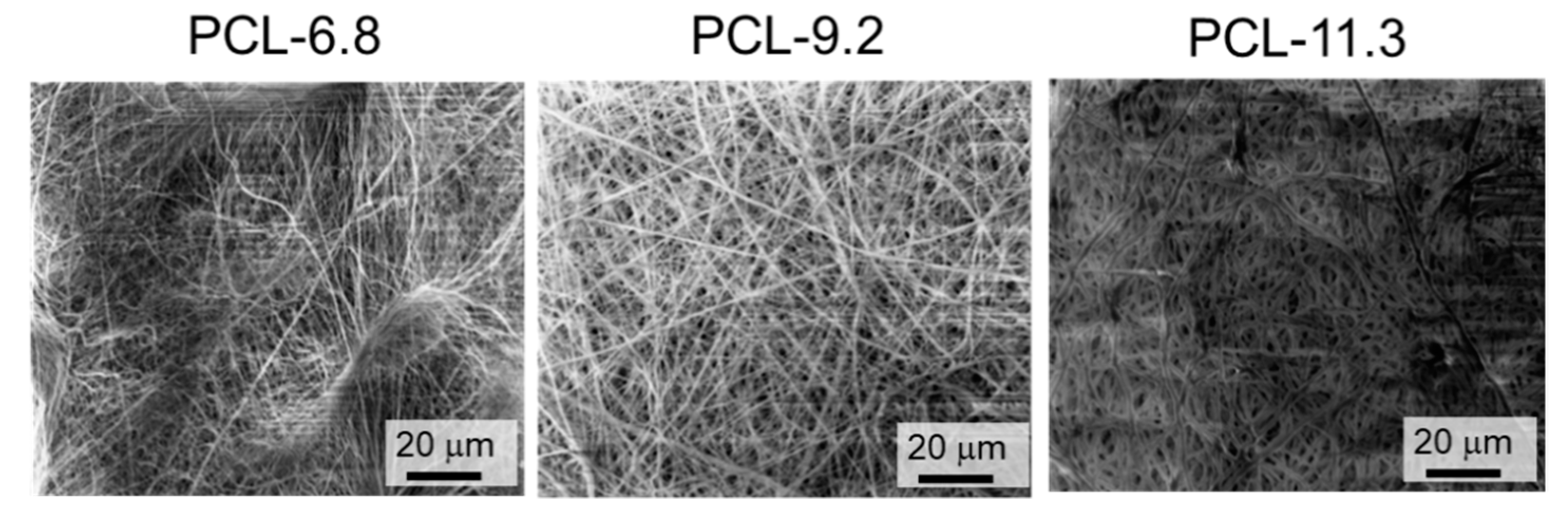
3.1. Fabrication of Poly(ε-Caprolactone) PCL-Based Polyurethane Nanofiber Meshes
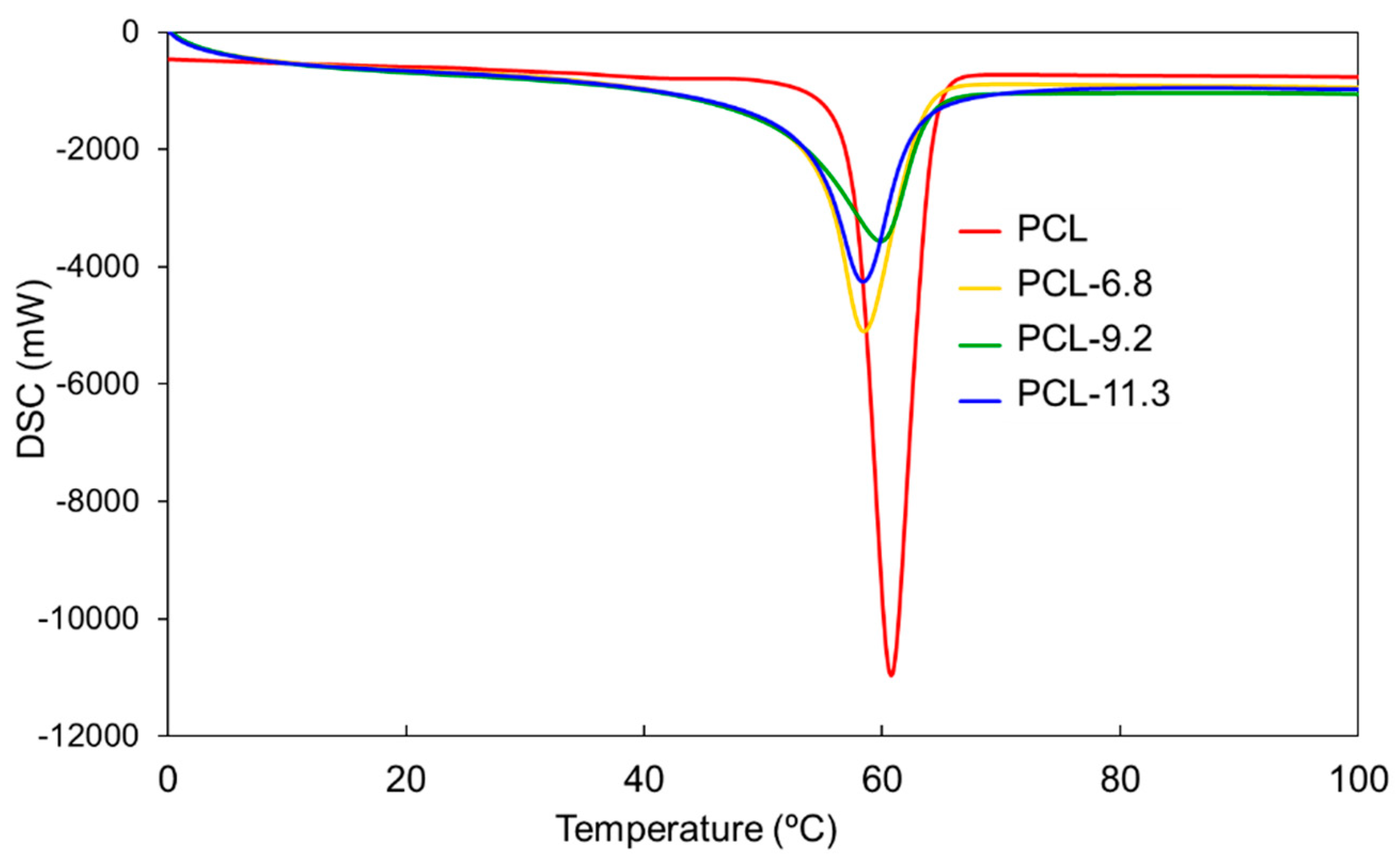
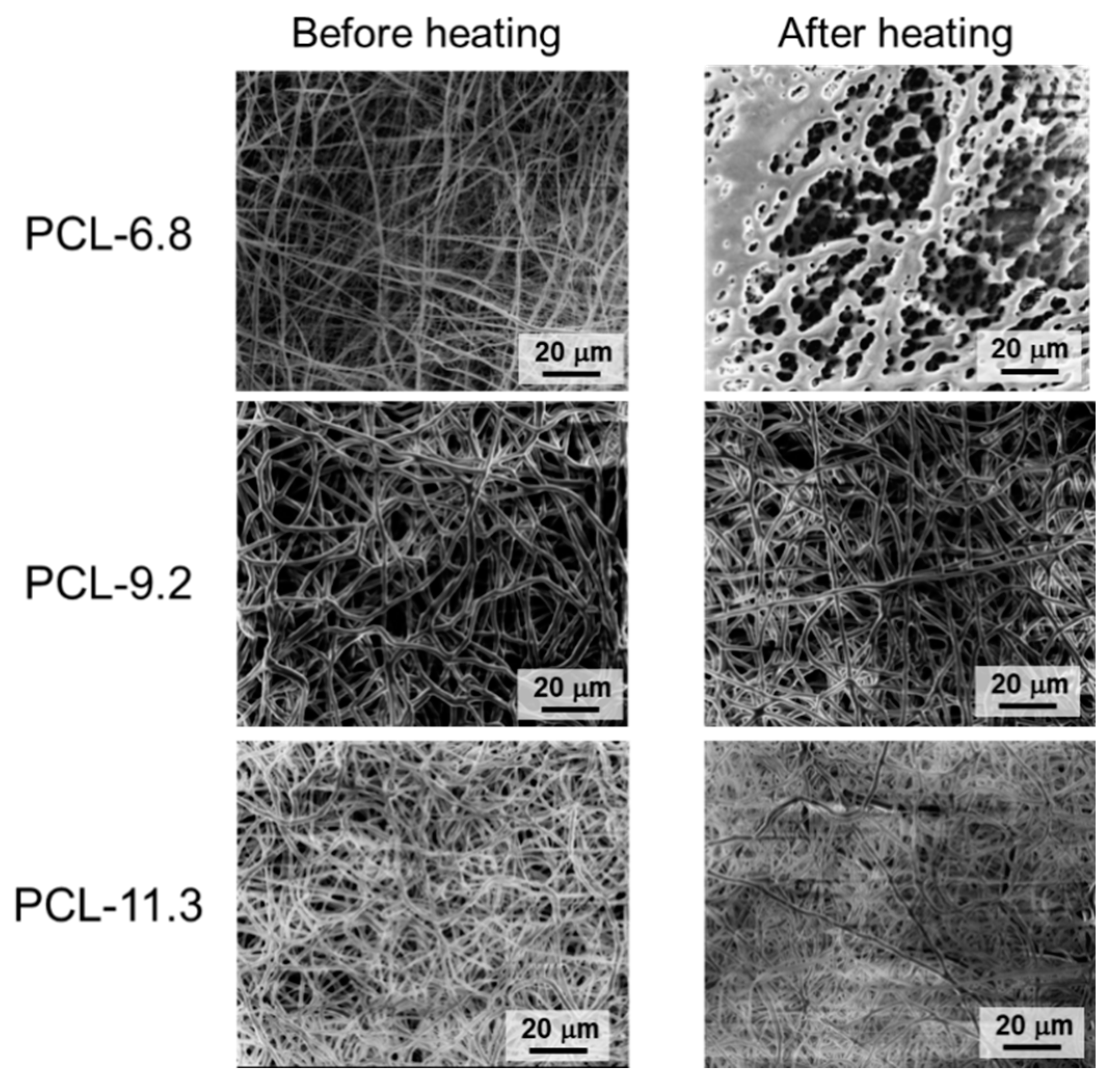
3.2. Thermal Properties
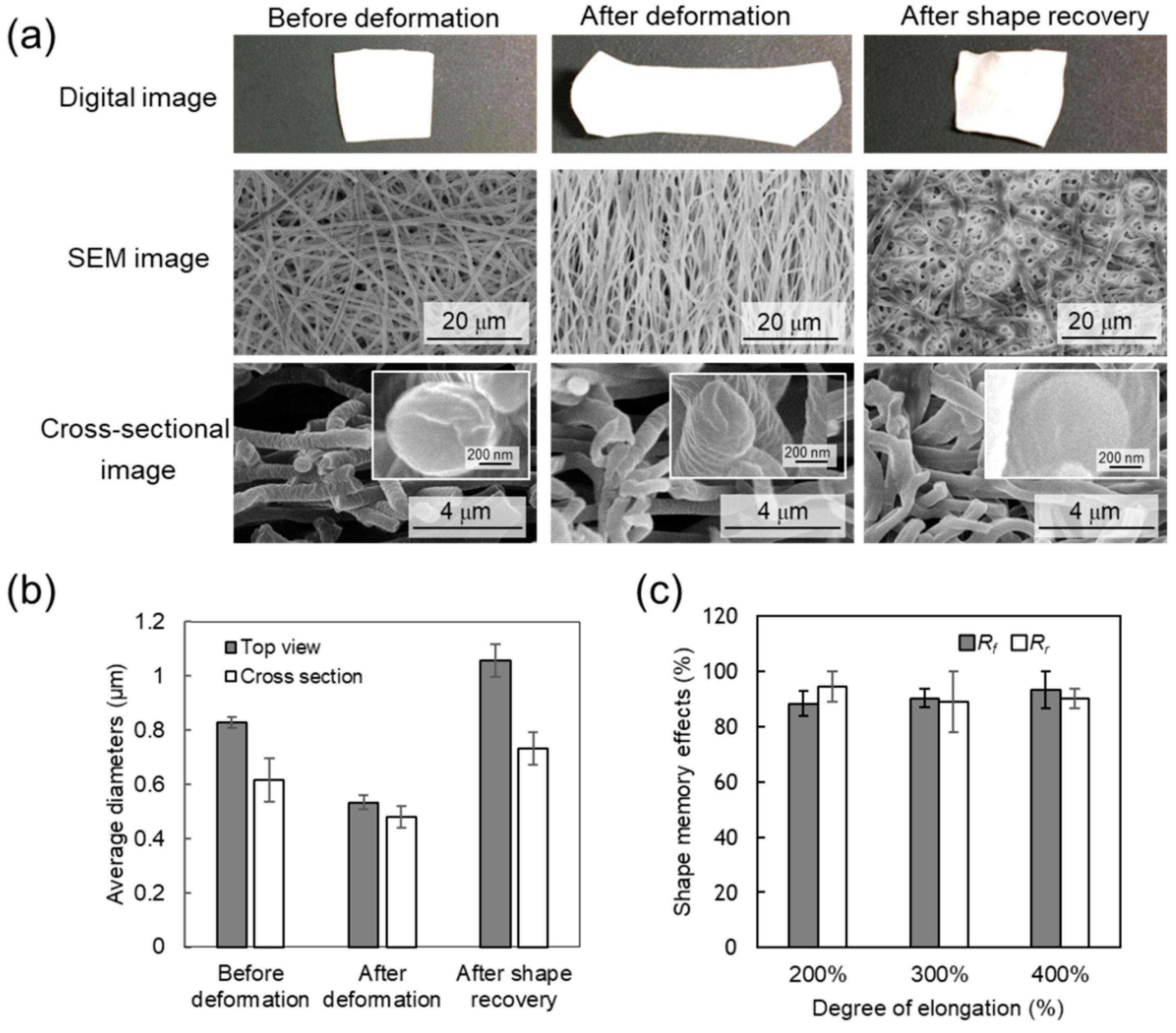
3.3. Shape-Memory Properties
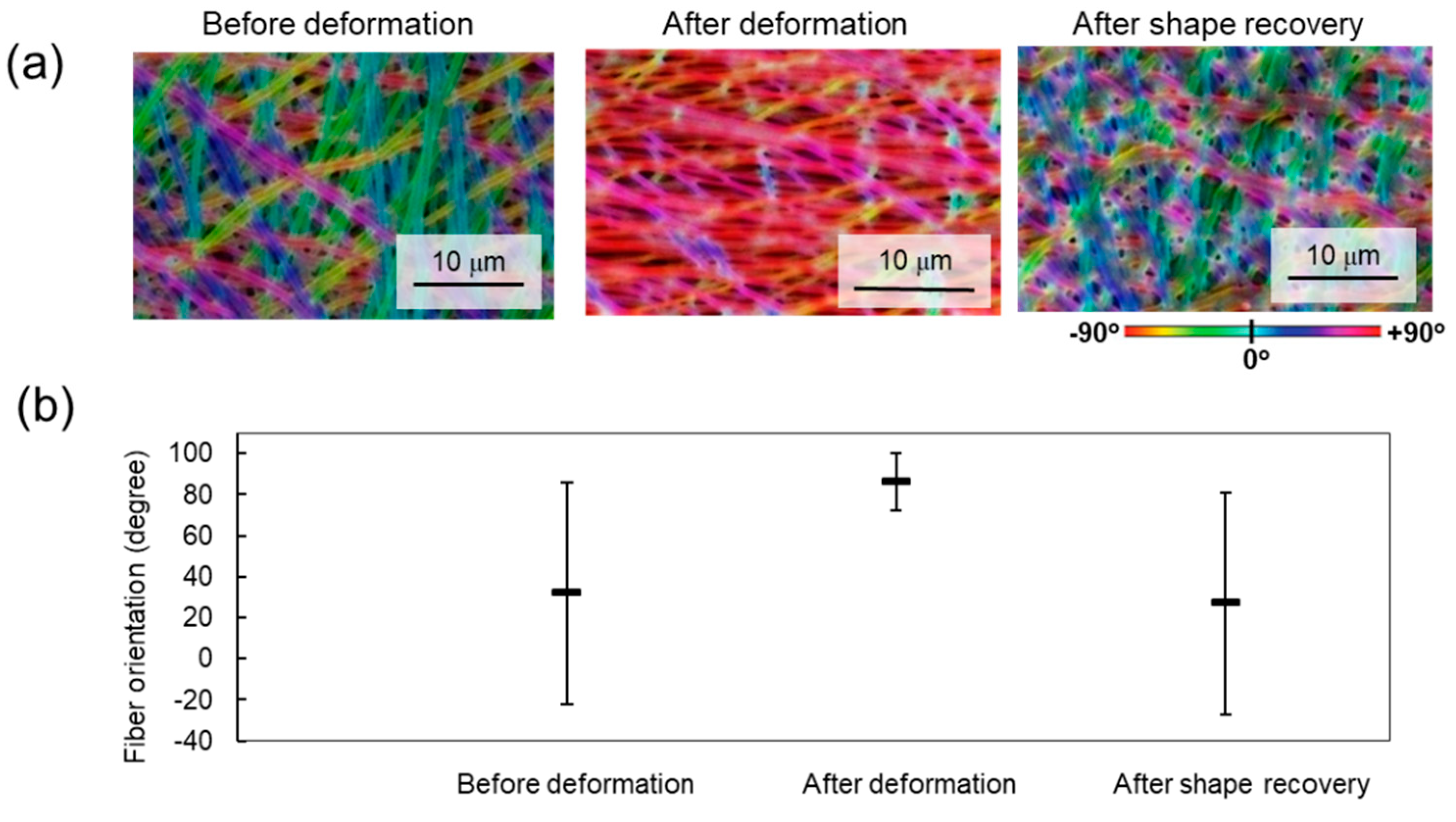
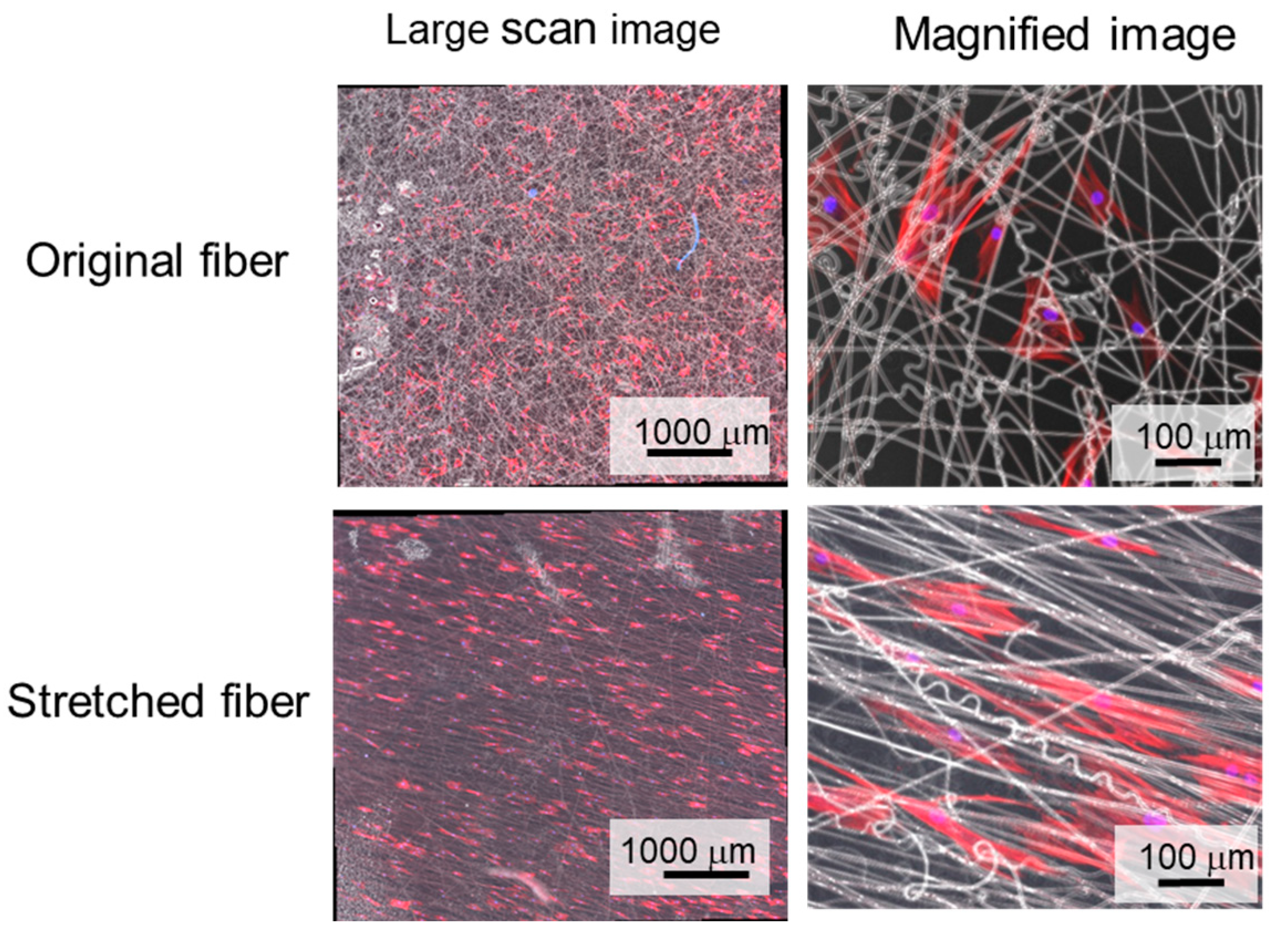
3.4. Cellular Alignment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gall, K.; Dunn, M.L.; Liu, Y.; Finch, D.; Lake, M.; Munshi, N.A. Shape memory polymer nanocomposites. Acta Biomater. 2002, 50, 5115–5126. [Google Scholar] [CrossRef]
- Lendlein, A.; Langer, R. Biodegradable, elastic shape-memory polymers for potential biomedical applications. Science 2002, 296, 1673–1676. [Google Scholar] [CrossRef] [PubMed]
- Nagahama, K.; Ueda, Y.; Ouchi, T.; Ohya, Y. Biodegradable shape-memory polymers exhibiting shpar thermal transitions and controlled drug release. Biomacromolecules 2009, 10, 1789–1794. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Zhu, Y.; Huang, H.; Lu, J. Recent advances in shape–memory polymers: Structure, mechanism, functionality, modeling and applications. Prog. Mater. Sci. 2012, 37, 1720–1763. [Google Scholar] [CrossRef]
- Liu, Y.; Du, H.; Liu, L.; Leng, J. Shape memory polymers and their composites in aerospace applications: A review. Smart Mater. Struct. 2014, 23, 023001. [Google Scholar] [CrossRef]
- Wang, J.; Quach, A.; Brasch, M.E.; Turner, C.E.; Henderson, J.H. On-command on/off switching of progenitor cell and cancer cell polarized motility and aligned morphology via a cytocompatible shape memory polymer scaffold. Biomaterials 2017, 140, 150–161. [Google Scholar] [CrossRef] [PubMed]
- Tseng, L.F.; Mather, P.T.; Henderson, J.H. Shape-memory-actuated change in scaffold fiber alignment directs stem cell morphology. Acta Biomater. 2013, 9, 8790–8801. [Google Scholar] [CrossRef] [PubMed]
- Bothe, M.; Emmerling, F.; Pretsch, T. Poly(ester urethane) with Varying Polyester Chain Length: Polymorphism and Shape-Memory Behavior. Macromol. Chem. Phys. 2013, 214, 2683–2693. [Google Scholar] [CrossRef]
- Hsu, S.-H.; Hung, K.-C.; Lin, Y.-Y.; Su, C.-H.; Yeh, H.-Y.; Jeng, U.S.; Lu, C.-Y.; Dai, S.A.; Fu, W.-E.; Lin, J.-C. Water-based synthesis and processing of novel biodegradable elastomers for medical applications. J. Mater. Chem. B 2014, 2, 5083–5092. [Google Scholar] [CrossRef]
- Chien, Y.C.; Chuang, W.T.; Jeng, U.S.; Hsu, S.H. Preparation, Characterization, and Mechanism for Biodegradable and Biocompatible Polyurethane Shape Memory Elastomers. ACS Appl. Mater. Interfaces 2017, 9, 5419–5429. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, T.; Hayashi, N.; Hayashi, S. Structure and Properties of Shape-Memory Polyurethane Block Copolymers. J. Appl. Polym. Sci. 1996, 60, 1061–1069. [Google Scholar] [CrossRef]
- Leng, J.; Lan, X.; Liu, Y.; Du, S. Shape-memory polymers and their composites: Stimulus methods and applications. Prog. Mater. Sci. 2011, 56, 1077–1135. [Google Scholar] [CrossRef]
- Ebara, M.; Uto, K.; Idota, N.; Hoffman, J.M.; Aoyagi, T. Shape-memory surface with dynamically tunable nano-geometry activated by body heat. Adv. Mater. 2012, 24, 273–278. [Google Scholar] [CrossRef] [PubMed]
- Ebara, M.; Akimoto, M.; Uto, K.; Shiba, K.; Yoshikawa, G.; Aoyagi, T. Focus on the interlude between topographic transition and cell response on shape-memory surfaces. Polymer 2014, 55, 5961–5968. [Google Scholar] [CrossRef]
- Ebara, M.; Uto, K.; Idota, N.; Hoffman, J.M.; Aoyagi, T. The taming of the cell: Shape-memory nanopatterns direct cell orientation. Int. J. Nanomed. 2014, 9, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Uto, K.; Ebara, M. Magnetic-Responsive Microparticles that Switch Shape at 37 °C. Appl. Sci. 2017, 7, 1203. [Google Scholar] [CrossRef]
- Uto, K.; Mano, S.S.; Aoyagi, T.; Ebara, M. Substrate Fluidity Regulates Cell Adhesion and Morphology on Poly(ε-caprolactone)-Based Materials. ACS Biomater. Sci. Eng. 2016, 2, 446–453. [Google Scholar] [CrossRef]
- Uto, K.; Aoyagi, T.; DeForest, C.A.; Hoffman, A.S.; Ebara, M. A Combinational Effect of “Bulk” and “Surface” Shape-Memory Transitions on the Regulation of Cell Alignment. Adv. Healthc. Mater. 2017, 6, 1601439. [Google Scholar] [CrossRef] [PubMed]
- Naheed, S.; Zuber, M.; Barikani, M. Synthesis and thermo-mechanical investigation of macrodiol-based shape memory polyurethane elastomers. Int. J. Mater. Res. 2017, 108, 515–522. [Google Scholar] [CrossRef]
- Cha, D.; Kim, H.Y.; Lee, K.H.; Jung, Y.C.; Cho, J.W.; Chun, B.C. Electrospun nonwovens of shape-memory polyurethane block copolymers. J. Appl. Polym. Sci. 2005, 96, 460–465. [Google Scholar] [CrossRef]
- Kotek, R. Recent advances in polymer fibers. Polym. Rev. 2008, 48, 221–229. [Google Scholar] [CrossRef]
- Zhuo, H.; Hu, J.; Chen, S. Electrospun polyurethane nanofibres having shape memory effect. Mater. Lett. 2008, 62, 2074–2076. [Google Scholar] [CrossRef]
- Chung, S.E.; Park, C.H.; Yu, W.-R.; Kang, T.J. Thermoresponsive shape memory characteristics of polyurethane electrospun web. J. Appl. Polym. Sci. 2011, 120, 492–500. [Google Scholar] [CrossRef]
- Okada, T.; Niiyama, E.; Uto, K.; Aoyagi, T.; Ebara, M. Inactivated Sendai virus (HVJ-E) immobilized electrospun nanofiber for cancer therapy. Materials 2016, 9, 12. [Google Scholar] [CrossRef] [PubMed]
- Garrett, R.; Niiyama, E.; Kotsuchibashi, Y.; Uto, K.; Ebara, M. Biodegradable nanofiber for delivery of immunomodulating agent in the treatment of basal cell carcinoma. Fibers 2015, 3, 478–490. [Google Scholar] [CrossRef]
- Brannmark, C.; Paul, A.; Ribeiro, D.; Magnusson, B.; Brolen, G.; Enejder, A.; Forslow, A. Increased adipogenesis of human adipose-derived stem cells on polycaprolactone fiber matrices. PLoS ONE 2014, 9, e113620. [Google Scholar] [CrossRef] [PubMed]
- Hyysalo, A.; Ristola, M.; Joki, T.; Honkanen, M.; Vippola, M.; Narkilahti, S. Aligned Poly(epsilon-caprolactone) Nanofibers Guide the Orientation and Migration of Human Pluripotent Stem Cell-Derived Neurons, Astrocytes, and Oligodendrocyte Precursor Cells In Vitro. Macromol. Biosci. 2017, 17, 1600517. [Google Scholar] [CrossRef] [PubMed]
- Stitzel, J.; Liu, J.; Lee, S.J.; Komura, M.; Berry, J.; Soker, S.; Lim, G.; Van Dyke, M.; Czerw, R.; Yoo, J.J.; et al. Controlled fabrication of a biological vascular substitute. Biomaterials 2006, 27, 1088–1094. [Google Scholar] [CrossRef] [PubMed]
- Stankus, J.J.; Guan, J.; Fujimoto, K.; Wagner, W.R. Microintegrating smooth muscle cells into a biodegradable, elastomeric fiber matrix. Biomaterials 2006, 27, 735–744. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Inai, R.; Kotaki, M.; Ramakrishna, S. Electrospun nanofiber fabrication as synthetic extracelular matrix and its potential for vascular tissue engineering. Tissue Eng. 2004, 10, 1160–1168. [Google Scholar] [CrossRef] [PubMed]
- Riboldi, S.A.; Sampaolesi, M.; Neuenschwander, P.; Cossu, G.; Mantero, S. Electrospun degradable polyesterurethane membranes: Potential scaffolds for skeletal muscle tissue engineering. Biomaterials 2005, 26, 4606–4615. [Google Scholar] [CrossRef] [PubMed]
- Shields, K.J.; Beckman, M.J.; Bowlin, G.L.; Wayne, J.S. Mechanical properties and cellular proliferation of electrospun collagen type. Tissue Eng. 2004, 10, 1510–1517. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Murugan, R.; Wang, S.; Ramakrishna, S. Electrospinning of nano/micro scale poly(L-lactic acid) aligned fibers and their potential in neural tissue engineering. Biomaterials 2005, 26, 2603–2610. [Google Scholar] [CrossRef] [PubMed]
- Jin, H. Human bone marrow stromal cell responses on electrospun silk fibroin mats. Biomaterials 2004, 25, 1039–1047. [Google Scholar] [CrossRef]
- Li, W.J.; Tuli, R.; Huang, X.; Laquerriere, P.; Tuan, R.S. Multilineage differentiation of human mesenchymal stem cells in a three-dimensional nanofibrous scaffold. Biomaterials 2005, 26, 5158–5166. [Google Scholar] [CrossRef] [PubMed]
- Dalby, M.J.; McCloy, D.; Robertson, M.; Agheli, H.; Sutherland, D.; Affrossman, S.; Oreffo, R.O. Osteoprogenitor response to semi-ordered and random nanotopographies. Biomaterials 2006, 27, 2980–2987. [Google Scholar] [CrossRef] [PubMed]
- Silva, G.A.; Czeisler, C.; Niece, K.L.; Beniash, E.; Harrington, D.A.; Kessler, J.A.; Stupp, S.L. Selective Differetiation of Neural progenitor cells by high-epitope density nanofibers. Science 2004, 303, 1352–1355. [Google Scholar] [CrossRef] [PubMed]
- Curtis, A.; Wilkinson, C. Nantotechniquies and approaches in biotechnology. Trends Biotechnol. 2001, 19, 97–101. [Google Scholar] [CrossRef]
- Mengsteab, P.Y.; Uto, K.; Smith, A.S.; Frankel, S.; Fisher, E.; Nawas, Z.; Macadangdang, J.; Ebara, M.; Kim, D.H. Spatiotemporal control of cardiac anisotropy using dynamic nanotopographic cues. Biomaterials 2016, 86, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, H.; Ishiguro, T.; Konosu, Y.; Minagawa, M.; Tanioka, A.; Richau, K.; Kratz, K.; Lendlein, A. Shape-memory properties of electrospun non-woven fabrics prepared from degradable polyesterurethanes containing poly(ω-pentadecalactone) hard segments. Eur. Polym. J. 2012, 48, 1866–1874. [Google Scholar] [CrossRef]
- Fejos, M.; Molnar, K.; Karger-Kocsis, J. Epoxy/Polycaprolactone Systems with Triple-Shape Memory Effect: Electrospun Nanoweb with and without Graphene versus Co-Continuous Morphology. Materials 2013, 6, 4489–4504. [Google Scholar] [CrossRef] [PubMed]
- Ji, F.; Zhu, Y.; Hu, J.; Liu, Y.; Yeung, L.; Ye, G. Smart polymer fibers with shape memory effect. Smart Mater Struct. 2006, 15, 1547–1554. [Google Scholar] [CrossRef]
- Barmouz, M.; Behravesh, A.H. Shape memory behaviors in cylindrical shell PLA/TPU-cellulose nanofiber bio-nanocomposites: Analytical and experimental assessment. Compos. Part A 2017, 101, 160–172. [Google Scholar] [CrossRef]
- Kawaguchi, K.; Iijima, M.; Miyakawa, H.; Ohta, M.; Muguruma, T.; Endo, K.; Nakazawa, F.; Mizoguchi, I. Effects of chitosan fiber addition on the properties of polyurethane with thermo-responsive shape memory. J. Biomed. Mater. Res. B Appl. Biomater. 2017, 105, 1151–1156. [Google Scholar] [CrossRef] [PubMed]
- Aslan, S.; Kaplan, S. Thermomechanical and Shape Memory Performances of Thermo-sensitive Polyurethane Fibers. Fibers Polym. 2018, 19, 272–280. [Google Scholar] [CrossRef]
- Karger-Kocsis, J.; Keki, S. Biodegradable polyester-based shape memory polymers: Concepts of (supra)molecular architecturing. Express Polym. Lett. 2014, 8, 397–412. [Google Scholar] [CrossRef]
- Lim, D.I.; Park, H.S.; Park, J.H.; Knowles, J.C.; Gong, M.S. Application of high-strength biodegradable polyurethanes containing different ratios of biobased isomannide and poly (-caprolactone) diol. J. Bioact. Compat. Polym. 2013, 28, 274–288. [Google Scholar] [CrossRef] [PubMed]
- Maeda, T.; Kim, Y.J.; Aoyagi, T.; Ebara, M. The design of temperature-responsive nanofiber meshes for cell storage applications. Fibers 2017, 5, 13. [Google Scholar] [CrossRef]
- Zhu, Y.; Hu, J.; Yeung, L.-Y.; Liu, Y.; Ji, F.; Yeung, K.-W. Development of shape memory polyurethane fiber with complete shape recoverability. Smart Mater. Struct. 2006, 15, 1385–1394. [Google Scholar] [CrossRef]
- Ping, P.; Wang, W.; Chen, X.; Jing, X. Poly(e-caprolactone) polyurethane and its shape-memory property. Biomacromolecules 2004, 6, 587–592. [Google Scholar] [CrossRef] [PubMed]
- Guan, J.-L. Role of focal adhesion kinese integrin Signaling. Int. J. Biochem. Cell. Biol. 1997, 29, 1085–1096. [Google Scholar] [CrossRef]
- Li, F.; Hou, J.; Zhu, W.; Zhang, X.; Xu, M.; Luo, X.; Ma, D.; Kim, B.K. Crystallinity and morphology of Segmented Polyurethanes with Different Soft-Segment Length. J. Appl. Polym. Sci. 1996, 62, 631–638. [Google Scholar] [CrossRef]

| Samples | Composition (Molar Ratio) | Segment Ratio 1) (Molar Ratio) | Segment Ratio (w/w%) | Feed | Molecular Weight 2) | PDI 2) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PCL:HDI:BD | Soft | Hard | Soft | Hard | PCL (mg) | HDI (µL) | BD (µL) | Mw | Mn | (Mw/Mn) | |
| PCL | - | - | - | - | - | - | - | - | 59,700 | 46,300 | 1.29 |
| PCL-6.8 | 1:3:2 | 1 | 5 | 93.2 | 6.8 | 800 | 44 | 16 | 78,900 | 54,000 | 1.46 |
| PCL-9.2 | 1:4:3 | 1 | 7 | 90.8 | 9.2 | 800 | 58 | 24 | 73,200 | 53,000 | 1.38 |
| PCL-11.3 | 1:5:4 | 1 | 9 | 88.7 | 11.3 | 800 | 73 | 32 | 93,500 | 68,000 | 1.37 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niiyama, E.; Tanabe, K.; Uto, K.; Kikuchi, A.; Ebara, M. Shape-Memory Nanofiber Meshes with Programmable Cell Orientation. Fibers 2019, 7, 20. https://doi.org/10.3390/fib7030020
Niiyama E, Tanabe K, Uto K, Kikuchi A, Ebara M. Shape-Memory Nanofiber Meshes with Programmable Cell Orientation. Fibers. 2019; 7(3):20. https://doi.org/10.3390/fib7030020
Chicago/Turabian StyleNiiyama, Eri, Kanta Tanabe, Koichiro Uto, Akihiko Kikuchi, and Mitsuhiro Ebara. 2019. "Shape-Memory Nanofiber Meshes with Programmable Cell Orientation" Fibers 7, no. 3: 20. https://doi.org/10.3390/fib7030020
APA StyleNiiyama, E., Tanabe, K., Uto, K., Kikuchi, A., & Ebara, M. (2019). Shape-Memory Nanofiber Meshes with Programmable Cell Orientation. Fibers, 7(3), 20. https://doi.org/10.3390/fib7030020







