Functional Analysis of the Acinetobacter baumannii XerC and XerD Site-Specific Recombinases: Potential Role in Dissemination of Resistance Genes
Abstract
1. Introduction
2. Results
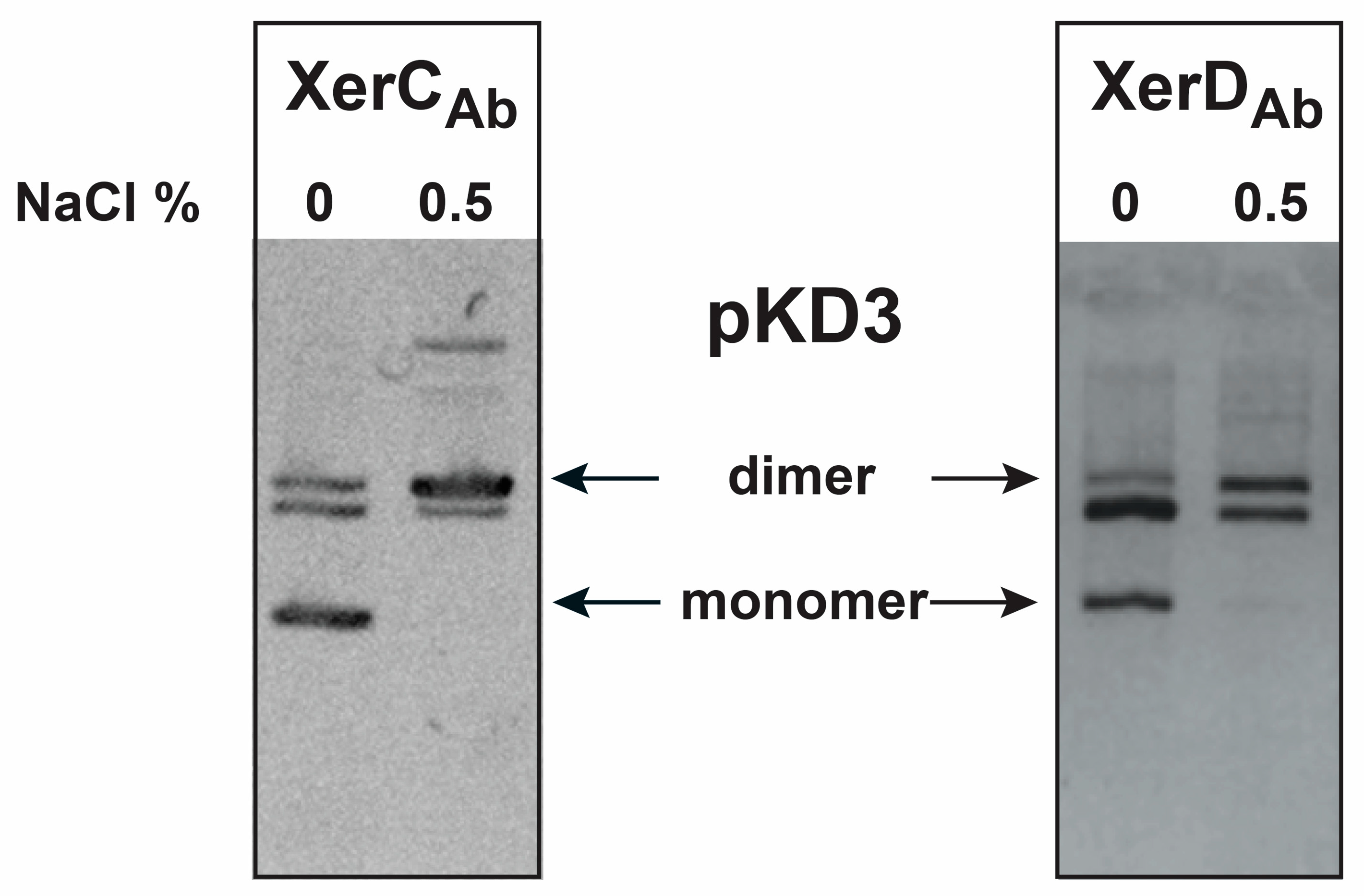
2.1. Cloning and Complementation Analysis of A. baumannii Xer Recombinases
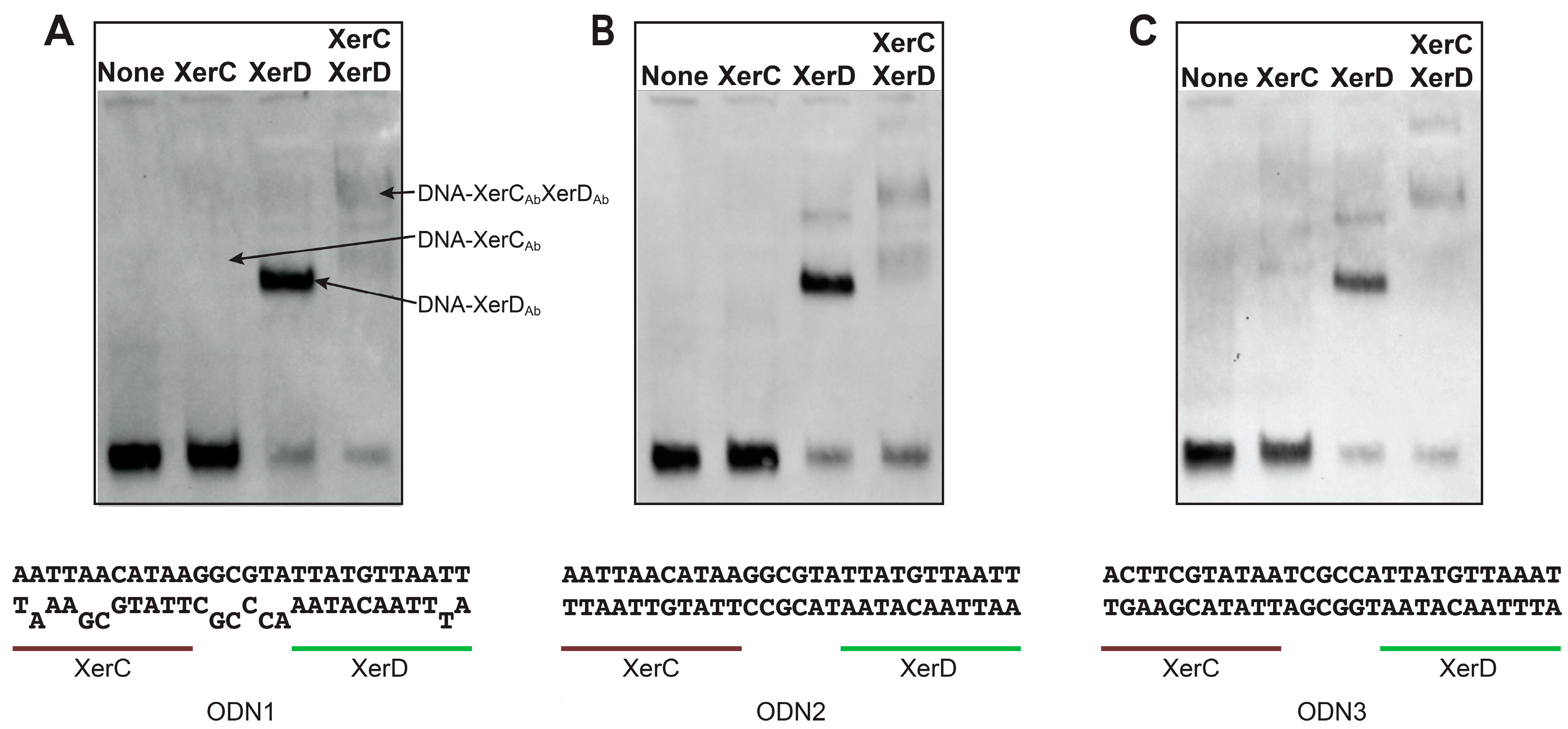
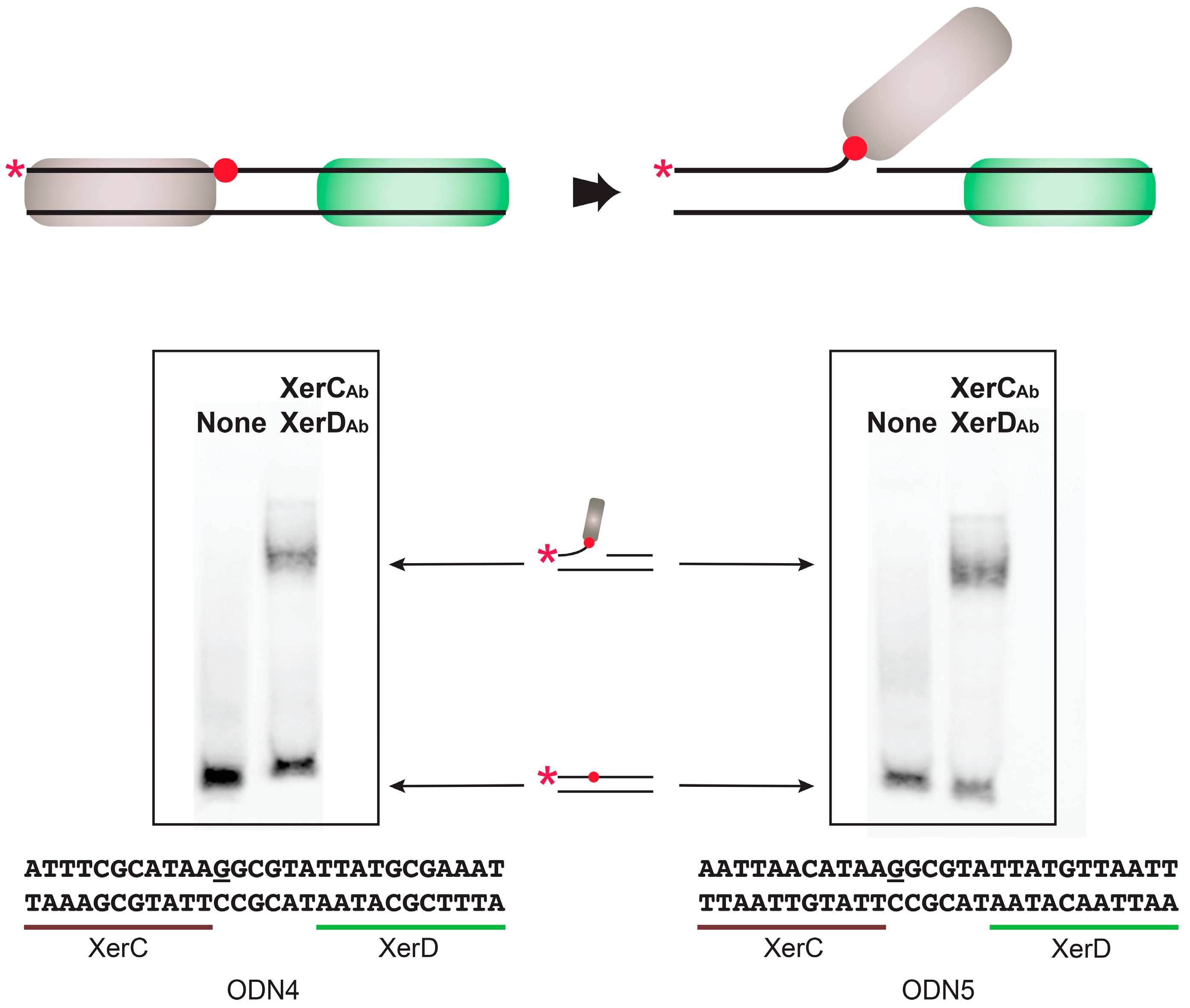
2.2. Binding of A. baumannii Xer Recombinases to XerC/D Binding Sites
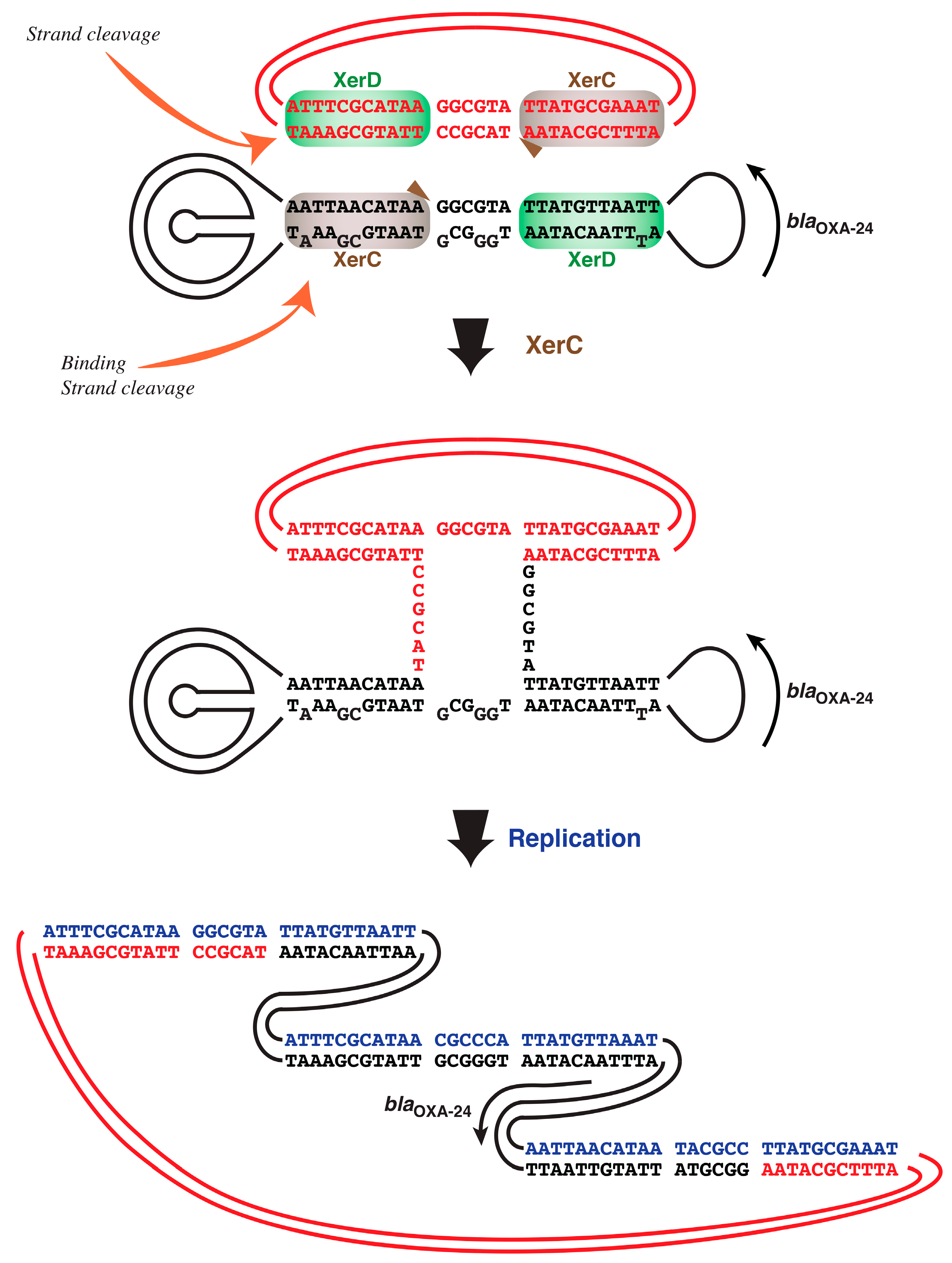
2.3. A. baumannii Xer Recombinases-Mediated Strand Exchange
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Plasmids
4.2. General DNA Procedures
4.3. Protein Purification
4.4. In Vivo Resolution Assay
4.5. DNA-Binding Assay
4.6. In Vitro Xer-Mediated DNA Cleavage
| Bacterial Strain or Plasmid | Relevant Characteristics, Genotype, or Phenotype a | Source or Reference |
|---|---|---|
| E. coli strains | ||
| DS941 | AB1157 recF143 lacIq lacZΔM15 | [65] |
| DS981 | DS941 xerC (Kanr) | [56] |
| DS9028 | DS941 xerD (Tmpr) | [57] |
| DS981XerCAb | DS981 (pMSR1) (Kanr Tetr) | This work |
| DS9028XerDAb | DS9028 (pMSR2) (Tmpr Tetr) | This work |
| DS9040 | DS941 xerC xerD (Kanr Genr) | [34] |
| JC8679 | DS945 recBC sbcA (hyperrecombinogenic) | [39] |
| A. baumannii strain | ||
| A118 | Human clinical isolate | [58] |
| Plasmids | ||
| pMSR1 | xerCAb cloned into the pACYC184 EcoRI site (Tetr) | This work |
| pMSR2 | xerDAb cloned into the pACYC184 EcoRI site (Tetr) | This work |
| pBAD102xerCAb | xerCAb cloned into pBAD102 (Ampr) | This work |
| pBAD102xerDAb | xerDAb cloned into pBAD102 (Ampr) | This work |
| pKD3 | EcoRI-SacI fragment containing the pJHCMW1 mwr site with substitution C to T at the ArgR binding site cloned in pUC18 (Ampr) | [8] |
| pUC18 | Cloning vector (Ampr) | [60] |
| pCR2.1 | Cloning vector (Ampr, Kanr) | ThermoFisher |
| pACYC184 | Cloning vector, p15A replicon (Chlr Tetr) | [61] |
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Aussel, L.; Barre, F.X.; Aroyo, M.; Stasiak, A.; Stasiak, A.Z.; Sherratt, D. FtsK Is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell 2002, 108, 195–205. [Google Scholar] [CrossRef]
- Sherratt, D.J.; Soballe, B.; Barre, F.X.; Filipe, S.; Lau, I.; Massey, T.; Yates, J. Recombination and chromosome segregation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2004, 359, 61–69. [Google Scholar] [CrossRef]
- Zawadzki, P.; May, P.F.; Baker, R.A.; Pinkney, J.N.; Kapanidis, A.N.; Sherratt, D.J.; Arciszewska, L.K. Conformational transitions during FtsK translocase activation of individual XerCD-dif recombination complexes. Proc. Natl. Acad. Sci. USA 2013, 110, 17302–17307. [Google Scholar] [CrossRef]
- Summers, D.K.; Beton, C.W.; Withers, H.L. Multicopy plasmid instability: The dimer catastrophe hypothesis. Mol. Microbiol. 1993, 8, 1031–1038. [Google Scholar] [CrossRef]
- Colloms, S.D.; Alen, C.; Sherratt, D.J. The ArcA/ArcB two-component regulatory system of Escherichia coli is essential for Xer site-specific recombination at psi. Mol. Microbiol. 1998, 28, 521–530. [Google Scholar] [CrossRef] [PubMed]
- Colloms, S.D. The topology of plasmid-monomerizing Xer site-specific recombination. Biochem. Soc. Trans. 2013, 41, 589–594. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bui, D.; Ramiscal, J.; Trigueros, S.; Newmark, J.S.; Do, A.; Sherratt, D.J.; Tolmasky, M.E. Differences in resolution of mwr-containing plasmid dimers mediated by the Klebsiella pneumoniae and Escherichia coli XerC recombinases: Potential implications in dissemination of antibiotic resistance genes. J. Bacteriol. 2006, 188, 2812–2820. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pham, H.; Dery, K.J.; Sherratt, D.J.; Tolmasky, M.E. Osmoregulation of dimer resolution at the plasmid pJHCMW1 mwr locus by Escherichia coli XerCD recombination. J. Bacteriol. 2002, 184, 1607–1616. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tolmasky, M.E.; Colloms, S.; Blakely, G.; Sherratt, D.J. Stability by multimer resolution of pJHCMW1 is due to the Tn1331 resolvase and not to the Escherichia coli Xer system. Microbiology 2000, 146, 581–589. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Trigueros, S.; Tran, T.; Sorto, N.; Newmark, J.; Colloms, S.D.; Sherratt, D.J.; Tolmasky, M.E. mwr Xer site-specific recombination is hypersensitive to DNA supercoiling. Nucleic Acids Res. 2009, 37, 3580–3587. [Google Scholar] [CrossRef]
- Das, B.; Martinez, E.; Midonet, C.; Barre, F.X. Integrative mobile elements exploiting Xer recombination. Trends Microbiol. 2013, 21, 23–30. [Google Scholar] [CrossRef]
- Midonet, C.; Barre, F.X. Xer site-specific recombination: Promoting vertical and horizontal transmission of genetic information. Microbiol Spectr 2014, 2. MDNA3-0056-2014. [Google Scholar]
- Val, M.E.; Bouvier, M.; Campos, J.; Sherratt, D.; Cornet, F.; Mazel, D.; Barre, F.X. The single-stranded genome of phage CTX is the form used for integration into the genome of Vibrio cholerae. Mol. Cell 2005, 19, 559–566. [Google Scholar] [CrossRef]
- Campos, J.; Martinez, E.; Suzarte, E.; Rodriguez, B.L.; Marrero, K.; Silva, Y.; Ledon, T.; del Sol, R.; Fando, R. VGJ phi, a novel filamentous phage of Vibrio cholerae, integrates into the same chromosomal site as CTX phi. J. Bacteriol. 2003, 185, 5685–5696. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hassan, F.; Kamruzzaman, M.; Mekalanos, J.J.; Faruque, S.M. Satellite phage TLCphi enables toxigenic conversion by CTX phage through dif site alteration. Nature 2010, 467, 982–985. [Google Scholar] [CrossRef] [PubMed]
- Zakharova, M.V.; Beletskaya, I.V.; Denjmukhametov, M.M.; Yurkova, T.V.; Semenova, L.M.; Shlyapnikov, M.G.; Solonin, A.S. Characterization of pECL18 and pKPN2: A proposed pathway for the evolution of two plasmids that carry identical genes for a Type II restriction-modification system. Mol. Genet. Genomics 2002, 267, 171–178. [Google Scholar] [CrossRef]
- Tran, T.; Andres, P.; Petroni, A.; Soler-Bistue, A.; Albornoz, E.; Zorreguieta, A.; Reyes-Lamothe, R.; Sherratt, D.J.; Corso, A.; Tolmasky, M.E. Small plasmids harboring qnrB19: A model for plasmid evolution mediated by site-specific recombination at oriT and Xer sites. Antimicrob. Agents Chemother. 2012, 56, 1821–1827. [Google Scholar] [CrossRef]
- Ramirez, M.S.; Traglia, G.M.; Lin, D.L.; Tran, T.; Tolmasky, M.E. Plasmid-mediated antibiotic resistance and virulence in gram-negatives: The Klebsiella pneumoniae paradigm. Microbiol. Spectr. 2014, 2. PLAS-0016-2013. [Google Scholar] [CrossRef]
- Blackwell, G.A.; Hall, R.M. The tet39 determinant and the msrE-mphE genes in Acinetobacter plasmids are each part of discrete modules flanked by inversely oriented pdif (XerC-XerD) sites. Antimicrob. Agents Chemother. 2017, 61, e00717–e00780. [Google Scholar] [CrossRef]
- Poirel, L.; Nordmann, P. Genetic structures at the origin of acquisition and expression of the carbapenem-hydrolyzing oxacillinase gene blaOXA-58 in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2006, 50, 1442–1448. [Google Scholar] [CrossRef]
- Merino, M.; Acosta, J.; Poza, M.; Sanz, F.; Beceiro, A.; Chaves, F.; Bou, G. OXA-24 carbapenemase gene flanked by XerC/XerD-like recombination sites in different plasmids from different Acinetobacter species isolated during a nosocomial outbreak. Antimicrob. Agents Chemother. 2010, 54, 2427–2727. [Google Scholar] [CrossRef] [PubMed]
- Grosso, F.; Quinteira, S.; Poirel, L.; Novais, A.; Peixe, L. Role of common blaOXA-24/OXA-40-carrying platforms and plasmids in the spread of OXA-24/OXA-40 among Acinetobacter species clinical isolates. Antimicrob. Agents Chemother. 2012, 56, 3969–3972. [Google Scholar] [CrossRef] [PubMed]
- D’Andrea, M.; Giani, T.; D’Arezzo, S.; Capone, A.; Petrosillo, N.; Visca, P.; Luzzaro, F.; Rossolini, G.M. Characterization of pABVA01, a plasmid encoding the OXA-24 carbapenemase from italian isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 2009, 53, 3528–3533. [Google Scholar] [CrossRef] [PubMed]
- Povilonis, J.; Seputiene, V.; Krasauskas, R.; Juskaite, R.; Miskinyte, M.; Suziedelis, K.; Suziedeliene, E. Spread of carbapenem-resistant Acinetobacter baumannii carrying a plasmid with two genes encoding OXA-72 carbapenemase in Lithuanian hospitals. J. Antimicrob. Chemother. 2013, 68, 1000–1006. [Google Scholar] [CrossRef] [PubMed]
- Tian, G.B.; Adams-Haduch, J.M.; Bogdanovich, T.; Pasculle, A.W.; Quinn, J.P.; Wang, H.N.; Doi, Y. Identification of diverse OXA-40 group carbapenemases, including a novel variant, OXA-160, from Acinetobacter baumannii in Pennsylvania. Antimicrob. Agents Chemother. 2011, 55, 429–432. [Google Scholar] [CrossRef] [PubMed]
- Girlich, D.; Bonnin, R.A.; Bogaerts, P.; De Laveleye, M.; Huang, D.T.; Dortet, L.; Glaser, P.; Glupczynski, Y.; Naas, T. Chromosomal amplification of the blaOXA-58 carbapenemase gene in a Proteus mirabilis clinical isolate. Antimicrob. Agents Chemother. 2017, 61, e01616–e01697. [Google Scholar] [CrossRef]
- Boucher, H.W.; Talbot, G.H.; Benjamin, D.K.; Bradley, J.; Guidos, R.J.; Jones, R.N.; Murray, B.E.; Bonomo, R.A.; Gilbert, D. 10 × ‘20 Progress—Development of new drugs active against gram-negative bacilli: An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2013, 56, 1685–1694. [Google Scholar] [CrossRef]
- Ramirez, M.S.; Bonomo, R.A.; Tolmasky, M.E. Carbapenemases: Transforming Acinetobacter baumannii into a yet more dangerous menace. Biomolecules 2020, 10, 720. [Google Scholar] [CrossRef]
- Mindlin, S.; Petrenko, A.; Petrova, M. Chromium resistance genetic element flanked by XerC/XerD recombination sites and its distribution in environmental and clinical Acinetobacter strains. FEMS Microbiol. Lett. 2018, 365, fny047. [Google Scholar] [CrossRef]
- Mindlin, S.; Beletsky, A.; Mardanov, A.; Petrova, M. Adaptive dif modules in permafrost strains of Acinetobacter iwoffii and their distribution and abundance among present day Acinetobacter strains. Front. Microbiol. 2019, 10, 632. [Google Scholar] [CrossRef]
- Girlich, D.; Damaceno, Q.S.; Oliveira, A.C.; Nordmann, P. OXA-253, a variant of the carbapenem-hydrolyzing class D beta-lactamase OXA-143 in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2014, 58, 2976–2978. [Google Scholar] [CrossRef] [PubMed]
- Bonnin, R.A.; Girlich, D.; Jousset, A.B.; Gauthier, L.; Cuzon, G.; Bogaerts, P.; Haenni, M.; Madec, J.Y.; Couve-Deacon, E.; Barraud, O.; et al. A single Proteus mirabilis lineage from human and animal sources: A hidden reservoir of OXA-23 or OXA-58 carbapenemases in Enterobacterales. Sci. Rep. 2020, 10, 9160. [Google Scholar] [CrossRef] [PubMed]
- Cameranesi, M.M.; Moran-Barrio, J.; Limansky, A.S.; Repizo, G.D.; Viale, A.M. Site-Specific recombination at XerC/D sites mediates the formation and resolution of plasmid co-integrates carrying a blaOXA-58- and TnaphA6-Resistance module in Acinetobacter baumannii. Front. Microbiol. 2018, 9, 66. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, H.; Butler-Cole, B.; Burgin, A.; Baker, R.; Sherratt, D.J.; Arciszewska, L.K. Functional analysis of the C-terminal domains of the site-specific recombinases XerC and XerD. J. Mol. Biol. 2003, 330, 15–27. [Google Scholar] [CrossRef]
- Subramanya, H.S.; Arciszewska, L.K.; Baker, R.A.; Bird, L.E.; Sherratt, D.J.; Wigley, D.B. Crystal structure of the site-specific recombinase, XerD. EMBO J. 1997, 16, 5178–5187. [Google Scholar] [CrossRef] [PubMed]
- Hallet, B.; Arciszewska, L.K.; Sherratt, D. Reciprocal control of catalysis by the tyrosine recombinases XerC and XerD: An enzymatic switch in site-specific recombination. Mol. Cell 1999, 4, 949–959. [Google Scholar] [CrossRef]
- Spiers, A.J.; Sherratt, D.J. C-terminal interactions between the XerC and XerD site-specific recombinases. Mol. Microbiol. 1999, 32, 1031–1042. [Google Scholar] [CrossRef]
- Ferreira, H.; Sherratt, D.; Arciszewska, L. Switching catalytic activity in the XerCD site-specific recombination machine. J. Mol. Biol. 2001, 312, 45–57. [Google Scholar] [CrossRef]
- Summers, D.K.; Sherratt, D.J. Multimerization of high copy number plasmids causes instability: CoIE1 encodes a determinant essential for plasmid monomerization and stability. Cell 1984, 36, 1097–1103. [Google Scholar] [CrossRef]
- Sarno, R.; McGillivary, G.; Sherratt, D.J.; Actis, L.A.; Tolmasky, M.E. Complete nucleotide sequence of Klebsiella pneumoniae multiresistance plasmid pJHCMW1. Antimicrob. Agents Chemother. 2002, 46, 3422–3427. [Google Scholar] [CrossRef]
- Arciszewska, L.K.; Baker, R.A.; Hallet, B.; Sherratt, D.J. Coordinated control of XerC and XerD catalytic activities during Holliday junction resolution. J. Mol. Biol. 2000, 299, 391–403. [Google Scholar] [CrossRef]
- Colloms, S.D.; McCulloch, R.; Grant, K.; Neilson, L.; Sherratt, D.J. Xer-mediated site-specific recombination in vitro. EMBO J. 1996, 15, 1172–1181. [Google Scholar] [CrossRef] [PubMed]
- Riley, M.; Abe, T.; Arnaud, M.B.; Berlyn, M.K.; Blattner, F.R.; Chaudhuri, R.R.; Glasner, J.D.; Horiuchi, T.; Keseler, I.M.; Kosuge, T.; et al. Escherichia coli K-12: A cooperatively developed annotation snapshot—2005. Nucleic Acids Res. 2006, 34, 1–9. [Google Scholar] [CrossRef]
- Martinez, J.; Fernandez, J.S.; Liu, C.; Hoard, A.; Mendoza, A.; Nakanouchi, J.; Rodman, N.; Courville, R.; Tuttobene, M.R.; Lopez, C.; et al. Human pleural fluid triggers global changes in the transcriptional landscape of Acinetobacter baumannii as an adaptive response to stress. Sci. Rep. 2019, 9, 17251. [Google Scholar] [CrossRef]
- Tran, T.; Sherratt, D.J.; Tolmasky, M.E. fpr, a deficient Xer recombination site from a Salmonella plasmid, fails to confer stability by dimer resolution: Comparative studies with the pJHCMW1 mwr site. J. Bacteriol. 2010, 192, 883–887. [Google Scholar] [CrossRef]
- Acosta, J.; Merino, M.; Viedma, E.; Poza, M.; Sanz, F.; Otero, J.R.; Chaves, F.; Bou, G. Multidrug-resistant Acinetobacter baumannii Harboring OXA-24 carbapenemase, Spain. Emerg. Infect. Dis. 2011, 17, 1064–1067. [Google Scholar] [CrossRef] [PubMed]
- Larouche, A.; Roy, P.H. Effect of attC structure on cassette excision by integron integrases. Mob DNA 2011, 2, 3. [Google Scholar] [CrossRef] [PubMed]
- Escudero, J.A.; Loot, C.; Nivina, A.; Mazel, D. The Integron: Adaptation on demand. Microbiol. Spectr. 2015, 3. MDNA3-0019-2014. [Google Scholar]
- Vallenet, D.; Nordmann, P.; Barbe, V.; Poirel, L.; Mangenot, S.; Bataille, E.; Dossat, C.; Gas, S.; Kreimeyer, A.; Lenoble, P.; et al. Comparative analysis of Acinetobacters: Three genomes for three lifestyles. PLoS ONE 2008, 3, e1805. [Google Scholar] [CrossRef]
- Blakely, G.; Sherratt, D. Determinants of selectivity in Xer site-specific recombination. Genes Dev. 1996, 10, 762–773. [Google Scholar] [CrossRef][Green Version]
- Grainge, I.; Bregu, M.; Vazquez, M.; Sivanathan, V.; Ip, S.C.; Sherratt, D.J. Unlinking chromosome catenanes in vivo by site-specific recombination. EMBO J. 2007, 26, 4228–4238. [Google Scholar] [CrossRef] [PubMed]
- Shimokawa, K.; Ishihara, K.; Grainge, I.; Sherratt, D.J.; Vazquez, M. FtsK-dependent XerCD-dif recombination unlinks replication catenanes in a stepwise manner. Proc. Natl. Acad. Sci. USA 2013, 110, 20906–20911. [Google Scholar] [CrossRef]
- Isler, B.; Doi, Y.; Bonomo, R.A.; Paterson, D.L. New treatment options against carbapenem-resistant Acinetobacter baumannii infections. Antimicrob. Agents Chemother. 2019, 63, e01110–e01118. [Google Scholar] [PubMed]
- Hartstein, A.I.; Rashad, A.L.; Liebler, J.M.; Actis, L.A.; Freeman, J.; Rourke, J.W., Jr.; Stibolt, T.B.; Tolmasky, M.E.; Ellis, G.R.; Crosa, J.H. Multiple intensive care unit outbreak of Acinetobacter calcoaceticus subspecies anitratus respiratory infection and colonization associated with contaminated, reusable ventilator circuits and resuscitation bags. Am. J. Med. 1988, 85, 624–631. [Google Scholar] [CrossRef]
- Summers, D.K.; Sherratt, D.J. Resolution of ColE1 dimers requires a DNA sequence implicated in the three-dimensional organization of the cer site. EMBO J. 1988, 7, 851–858. [Google Scholar] [CrossRef] [PubMed]
- Cornet, F.; Mortier, I.; Patte, J.; Louarn, J.M. Plasmid pSC101 harbors a recombination site, psi, which is able to resolve plasmid multimers and to substitute for the analogous chromosomal Escherichia coli site dif. J. Bacteriol. 1994, 176, 3188–3195. [Google Scholar] [CrossRef]
- Spiers, A.J.; Sherratt, D.J. Relating primary structure to function in the Escherichia coli XerD site-specific recombinase. Mol. Microbiol. 1997, 24, 1071–1082. [Google Scholar] [CrossRef]
- Ramirez, M.S.; Don, M.; Merkier, A.K.; Bistue, A.J.; Zorreguieta, A.; Centron, D.; Tolmasky, M.E. Naturally competent Acinetobacter baumannii clinical isolate as a convenient model for genetic studies. J. Clin. Microbiol. 2010, 48, 1488–1490. [Google Scholar] [CrossRef]
- Traglia, G.M.; Chua, K.; Centron, D.; Tolmasky, M.E.; Ramirez, M.S. Whole-genome sequence analysis of the naturally competent Acinetobacter baumannii clinical isolate A118. Genome Biol. Evol. 2014, 6, 2235–2239. [Google Scholar] [CrossRef]
- Yanisch-Perron, C.; Vieira, J.; Messing, J. Improved M13 phage cloning vectors and host strains: Nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 1985, 33, 103–119. [Google Scholar] [CrossRef]
- Chang, A.C.; Cohen, S.N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 1978, 134, 1141–1156. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.N.; Chang, A.C.; Hsu, L. Nonchromosomal antibiotic resistance in bacteria: Genetic transformation of Escherichia coli by R-factor DNA. Proc. Natl. Acad. Sci. USA 1972, 69, 2110–2114. [Google Scholar] [CrossRef] [PubMed]
- Blakely, G.; May, G.; McCulloch, R.; Arciszewska, L.K.; Burke, M.; Lovett, S.T.; Sherratt, D.J. Two related recombinases are required for site-specific recombination at dif and cer in E. coli K12. Cell 1993, 75, 351–361. [Google Scholar] [CrossRef]
- Nunes-Duby, S.E.; Radman-Livaja, M.; Kuimelis, R.G.; Pearline, R.V.; McLaughlin, L.W.; Landy, A. Gamma integrase complementation at the level of DNA binding and complex formation. J. Bacteriol. 2002, 184, 1385–1394. [Google Scholar] [CrossRef]
- Summers, D. Timing, self-control and a sense of direction are the secrets of multicopy plasmid stability. Mol. Microbiol. 1998, 29, 1137–1145. [Google Scholar] [CrossRef] [PubMed]

| Name | Sequence |
|---|---|
| ODN1 | A(A/A)TT(A/G)(A/C)CATAAG(G/G)(C/C)G(T/C)(A/A)TTATGTTAATT |
| ODN2 | AATTAACATAAGGCGTATTATGTTAATT |
| ODN3 | ACTTCGTATAATCGCCATTATGTTAAAT |
| ODN4 | ATTTCGCATAAGGCGTATTATGCGAAAT |
| ODN5 | AATTAACATAAGGCGTATTATGTTAATT |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, D.L.; Traglia, G.M.; Baker, R.; Sherratt, D.J.; Ramirez, M.S.; Tolmasky, M.E. Functional Analysis of the Acinetobacter baumannii XerC and XerD Site-Specific Recombinases: Potential Role in Dissemination of Resistance Genes. Antibiotics 2020, 9, 405. https://doi.org/10.3390/antibiotics9070405
Lin DL, Traglia GM, Baker R, Sherratt DJ, Ramirez MS, Tolmasky ME. Functional Analysis of the Acinetobacter baumannii XerC and XerD Site-Specific Recombinases: Potential Role in Dissemination of Resistance Genes. Antibiotics. 2020; 9(7):405. https://doi.org/10.3390/antibiotics9070405
Chicago/Turabian StyleLin, David L., German M. Traglia, Rachel Baker, David J. Sherratt, Maria Soledad Ramirez, and Marcelo E. Tolmasky. 2020. "Functional Analysis of the Acinetobacter baumannii XerC and XerD Site-Specific Recombinases: Potential Role in Dissemination of Resistance Genes" Antibiotics 9, no. 7: 405. https://doi.org/10.3390/antibiotics9070405
APA StyleLin, D. L., Traglia, G. M., Baker, R., Sherratt, D. J., Ramirez, M. S., & Tolmasky, M. E. (2020). Functional Analysis of the Acinetobacter baumannii XerC and XerD Site-Specific Recombinases: Potential Role in Dissemination of Resistance Genes. Antibiotics, 9(7), 405. https://doi.org/10.3390/antibiotics9070405








