Abstract
Corynebacterium urealyticum is a non-diphtherial urease-producing clinically relevant corynebacterial, most frequently involved in urinary tract infections. Most of the C. urealyticum clinical isolates are frequently resistant to several antibiotics. We investigated the susceptibility of 40 C. urealyticum isolated in our institution during the period 2005–2017 to eight compounds representative of the main clinically relevant classes of antimicrobial agents. Antimicrobial susceptibility was determined by the Epsilometer test. Resistance genes were searched by PCR. All strains were susceptible to vancomycin whereas linezolid and rifampicin also showed good activity (MICs90 = 1 and 0.4 mg/L, respectively). Almost all isolates (39/40, 97.5%) were multidrug resistant. The highest resistance rate was observed for ampicillin (100%), followed by erythromycin (95%) and levofloxacin (95%). Ampicillin resistance was associated with the presence of the blaA gene, encoding a class A β-lactamase. The two rifampicin-resistant strains showed point mutations driving amino acid replacements in conserved residues of RNA polymerase subunit β (RpoB). Tetracycline resistance was due to an efflux-mediated mechanism. Thirty-nine PFGE patterns were identified among the 40 C. urealyticum, indicating that they were not clonally related, but producing sporadic infections. These findings raise the need of maintaining surveillance strategies among this multidrug resistant pathogen.
1. Introduction
The genus Corynebacterium include Gram positive aerobic bacteria, which are widely distributed in the microbiota of humans and animals. Medically relevant Corynebacterium species consist of Corynebacterium diphtheriae, the pathogenic bacterium that causes diphtheria, and the non-diphtherial corynebacterial (as Corynebacterium urealyticum, Corynebacterium striatum, and Corynebacterium jeikeium, among others), which are part of the skin and mucous membranes flora [1]. They are usually not pathogenic but can occasionally opportunistically capitalize on atypical access to tissues or weakened host defenses. A key role of Corynebacterium species as attenuator of Staphylococcus aureus virulence in the nose microbiota has been recently suggested [2]. Corynebacterium urealyticum is a slow growing, asaccharolytic, and lipophilic microorganism, whose name refers its ability to split urea [3]. It possesses a strong urease activity that leads to the formation of struvite stones following ammonium magnesium phosphate precipitation due to the increase of urine pH. C. urealyticum behaves as an opportunistic human pathogen, causing acute and chronic urinary tract infections (UTIs), eventually leading to bacteraemia [3]. It has also been isolated from the skin of healthy elderly individuals, mainly females [4]. Some of the risk factors pre-disposing to an infection by C. urealyticum are prolonged use of urinary catheters, hospitalization for long periods, previous treatment with broad-spectrum antibiotics or immunosuppressants, and history of previous UTIs [5].
The majority of C. urealyticum currently isolated from clinical samples are multidrug resistant, thus potentially limiting effective empirical treatment [1,6]. Development of resistance has been observed during treatment with different antimicrobial classes: β-lactams, gentamicin, fluoroquinolones, macrolides, rifampicin, and tetracycline [7]. However, the resistance mechanisms for most of these compounds have not been described.
Previous [8,9] and recent studies [10] about antimicrobial activity against C. urealyticum and other Corynebacterium species have proven that vancomycin and linezolid were uniformly active against these bacteria, whereas most of them displayed high level resistance against quinolones, β-lactams, and macrolides.
The aim of this study was to evaluate the prevalence of multidrug resistant strains as well as determine the resistance mechanisms of C. urealyticum isolates from a Spanish hospital (Santander) during the period 2005–2017. The genomes of five of these isolates have been sequenced and the main antimicrobial resistance determinants identified.
2. Results
2.1. Susceptibility of C. urealyticum to Antimicrobial Agents
The MIC50 and MIC90 distributions, as well as the percentage of resistance to the different antibiotics for the 40 C. urealyticum included in this study, are shown in Table 1.

Table 1.
Susceptibility of 40 Corynebacterium urealyticum clinical isolates against eight antimicrobial agents. Isolates were classified as resistant or susceptible according to criteria defined by EUCAST (2020). MIC, minimum inhibitory concentration; MIC50, MIC that inhibits 50% of the isolates; MIC90, MIC that inhibits 90% of the isolates. S, susceptible; R, resistant. * Staphylococcus spp. breakpoint.
Ampicillin (100%), erythromycin (95%), and levofloxacin (95%) showed the highest number of resistant isolates, with a monomodal distribution of their MICs. Interestingly, for each antimicrobial agent, the two susceptible strains were different (VH4696 and VH4851 susceptible to erythromycin, VH6223 and VH4248 susceptible to levofloxacin). All the levofloxacin and erythromycin-resistant isolates had high level resistance (MIC > 32 mg/L and MIC > 256 mg/L, respectively). Concerning ampicillin-resistant isolates, 39 isolates showed a MIC > 256 mg/L whereas one isolate showed a MIC = 24 mg/L (the urine isolate VH2234).
Gentamicin and tetracycline also showed high number of resistant C. urealyticum strains, with 82.5% and 50% of isolates, respectively. In both cases, a bimodal MICs distribution was observed. The MICs values for gentamicin were in the range 1.5–256 mg/L, and between 3 and 256 mg/L for tetracycline, with relatively low MIC90 values, 9 and 4 mg/L, respectively.
Rifampicin and linezolid were the compounds with the lowest percentage of resistance, with only two and one resistant strains, respectively. Rifampicin-resistant isolates showed a high level resistance (MICs > 32 mg/L), whereas the MIC of the linezolid resistant isolate was 3 mg/L. Vancomycin was the only compound uniformly active against all tested isolates.
On the whole, multidrug resistance, defined as nonsusceptibility to at least one agent in three or more antimicrobial categories [11], was observed in 39 out of 40 isolates.
All the strains were resistant to at least two antimicrobial compounds. They presented nine different resistance profiles, the resistance combination for levofloxacin (LVX), ampicillin (AMP), gentamicin (GEN) and erythromycin (ERY) being the most prevalent (LVX-AMP-GEN-ERY = 14 isolates), as well as in combination with tetracycline (TET) resistance (LVX-AMP-GEN-ERY-TET = 13 isolates). The relationship between antibiotic resistance profiles and sample origin could not be established.
Strain VH4549, isolated from a urine sample, was resistant against six of the eight tested compounds (it showed LVX-AMP-GEN-ERY-TET-RIF profile), being the isolate with less therapeutic options. On the other hand, the strain VH6223, from placenta, was the most susceptible isolate, showing resistance only against ampicillin and erythromycin.
The susceptibility of the five C. urealyticum whose genomes were sequenced against the eight compounds tested is shown in Table 2.

Table 2.
MIC values of five C. urealyticum whose genomes were sequenced. MICs are expressed in mg/L.
2.2. Detection of Resistance Genes by PCR and Genome Sequencing
The 38 C. urealyticum resistant to erythromycin carried the ermX gene. The PCR amplification product of strain VH2234 was sequenced and compared with the ermX gene of C. urealyticum DSM 7109 [4], showing a 95% identity. Whole genome sequencing of five erythromycin-resistant C. urealyticum confirmed the presence of the ermX gene.
Thirty-eight out of 40 C. urealyticum were resistant to levofloxacin. The 38 resistant strains showed a MIC of levofloxacin >32 mg/L. The sequences of the QRDR region of the gyrA gene of 28 isolates categorized as resistant and one isolate categorized as susceptible were compared to the sequence of this region in the gyrA gene of C. urealyticum DSM 7109 (quinolone-susceptible). Twenty-two levofloxacin-resistant isolates showed the double mutation Ser-90→Val and Asp-94→Tyr, whereas in three resistant strains Asp-94 was replaced by Ala. Three levofloxacin-resistant strains were single Ser-90→Val mutants. The levofloxacin susceptible strain VH4248 did not show mutations at residues 90 and 94, as the reference strain DSM 7109. One strain resistant to levofloxacin showed no mutation at the QRDR region of gyrA, suggesting a different resistance mechanism.
Two of our isolates (VH3073 and VH4549), as well as C. urealyticum DSM 7109, were resistant to rifampicin (MIC > 32 mg/L). Rifampicin resistance is nearly always due to a genetic change in the β subunit of RNA polymerase (RpoB). Alignment of the RpoB sequences of these three rifampicin-resistant strains with the corresponding proteins of three rifampicin-susceptible C. urealyticum revealed non-conservative changes in Ser-444 (VH4549 and DSM 7109) or Gln-511 (VH3073), which can be related with the rifampicin-resistant phenotype (Figure 1).

Figure 1.
Alignment of RNA polymerase β subunit (RpoB) (residues 421–540) of six Corynebacterium urealyticum strains. Strains VH4549, DSM 7109, and VH3073 are resistant to rifampicin whereas VH4248, DSM 7111, and VH5913 are susceptible.
The 40 C. urealyticum strains were ampicillin-resistant. The high ampicillin resistance phenotype (MIC90 > 256 mg/L) was associated with the presence of the blaA gene. Ampicillin-resistant C. urealyticum rendered the expected 0.8 Kb PCR product when amplified with blaA specific primers. Conversely, C. urealyticum 18408721, a clinical isolate susceptible to ampicillin, was negative by the blaA-based PCR. The C. urealyticum blaA gene encodes a serine hydrolase belonging to the class A β-lactamase protein family. In order to know the genomic context of the resistance genes and inquiry about their transfer mechanisms, we sequenced the genomes of five C. urealyticum. Genome analysis revealed that the region containing the blaA gene is highly conserved among the five C. urealyticum and the previously sequenced strains C. urealyticum DSM 7109 [4] and DSM 7111 [12] (Figure 2A). This region spans 20 Kbp of the assembled genomes, including a tnp gene (transposase), lysR (transcriptional regulator), and a Penicillin-Binding-Protein (PBP) type 1 gene. A similar genomic organization can be found in C. striatum KC-Na-01 (Figure 2B). We compared the amino acid sequence encoded by the C. urealyticum blaA gene with its counterparts in other species and we found that it is highly conserved in C. striatum strain KC-Na-01 (NCBI’s protein accession #WP_049063072), in C. jeikeium K411 (#WP_034987125), in C. amycolatum SK46 (#WP_076773763.1), as well as in C. resistens DSM 45100 (#WP_042378726.1) [13]. There is a particular region in the BlaA sequence (between amino acid positions 157–164) that concentrates most of the variability when comparing all species.
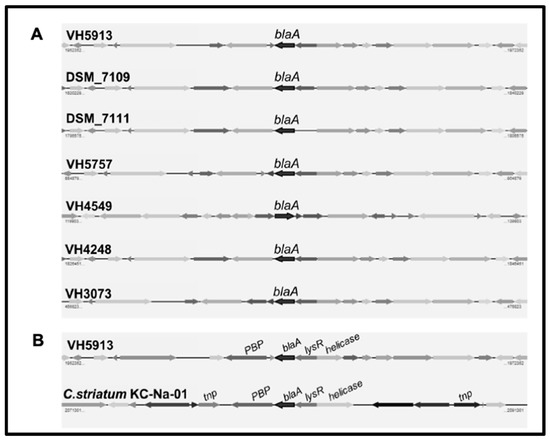
Figure 2.
A: Genomic context of the blaA gene in seven C. urealyticum. B: Comparison among the blaA-containing regions of C. urealyticum VH5913 and Corynebacterium striatum KC-Na-01.
Thirty-three of the C. urealyticum showed low level gentamicin resistance. The presence of the gene aac(3)-XI encoding for an aminoglycoside 3-N acetyltransferase was evaluated by PCR using primers based on the C. striatum aac(3)-XI gene [14], giving negative results in all strains. However, analysis of the five C. urealyticum sequenced genomes revealed the presence of a 447-bp open reading frame showing 79% identity with the C. striatum aac(3)-XI gene in four strains but not in strain VH4248 (Figure 3). Homology search in databases revealed that the C. urealyticum aac(3)-XI orthologous encodes an aminoglycoside 3-N acetyltransferase also present in C. coyleae (#WP_092102070.1) (80% identity), C. fournierii (#WP_085957501) (75% identity), and several Corynebacterium spp. The aac(3)-XI gene is flanked by arfB, encoding the peptidyl-tRNA hydrolase ArfB, at the upstream region, and the luxR-family two-component transcriptional response regulator, at the downstream region (Figure 3). On the other hand, a search for additional aminoglycoside resistance genes revealed the presence of the gene aph(3′)-Ic, encoding resistance to kanamycin and other aminoglycosides rarely used in clinical practice, and the pair of genes aph(3″)-Ib and aph(6)-Id, conferring streptomycin resistance, in four strains, but not in strain VH4248, which does not present any of these genes.
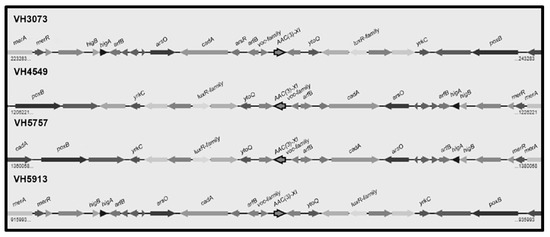
Figure 3.
Genomic map of the region including the aac(3)-XI gene and its neighbors in four C. urealyticum clinical isolates: merA (mercuric ion reductase); merR (transcriptional regulator, MerR family); higB (toxin HigB); higA (antitoxin HigA); arfB (peptidyl tRNA hydrolase ArfB); arsO (flavin-dependent monooxygenase ArsO, associated with arsenic resistance); cadA (copper-translocating P-type ATPase); arsR (transcriptional regulator, ArsR family); voc family (VOC family protein); ytoQ (uncharacterized protein YtoQ); luxR-family (two-component transcriptional response regulator, LuxR family); yrkC (uncharacterized protein YrkC); poxB (pyruvate dehydrogenase).
Twenty of our C. urealyticum were resistant to tetracycline. Tetracycline-resistant strains showed a bimodal MICs distribution: strains VH638 and VH2234 showed high resistance level (MICs ≥ 256 and 32 mg/L, respectively), whereas 18 strains showed low resistance level (MICs range = 3–6 mg/L). The C. urealyticum reference strain DSM 7109 is tetracycline-resistant (MIC = 32 mg/L) and this resistance is associated to the tetAB genes [4]. The tetAB genes were neither detected by genome analysis of the strains VH4549, VH5757, and VH5913, nor by PCR analysis of the remaining 17 tetracycline-resistant strains, which suggests the existence of alternative resistance mechanisms. When the tetracycline MICs of seven of our tetracycline-resistant strains were measured in the presence of the efflux-pump inhibitor Phe-Arg-β-naphthylamide (PAβN), a dramatic increase of tetracycline susceptibility was observed (Table 3). However, the tetracycline MIC of strain DSM 7109 remained unchanged.

Table 3.
MIC values (mg/L) of seven C. urealyticum and the reference strain DSM 7109 determined in absence and presence of PAβN.
2.3. Molecular Epidemiology of the C. urealyticum Isolates
The PFGE method displayed a high typeability and discriminatory power. XbaI digestion of the 40 C. urealyticum isolates revealed 39 distinct PFGE patterns which were labelled from 1 to 39 (Figure 4). The reference strain DSM 7109 PFGE pattern was also included in the dendrogram. Only two strains were assigned into the same PFGE pattern (VH4696 and VH4851), designated as pattern 18. These two strains were isolated from urine samples taken from the same patient at an interval of eight days, and showed the same antibiotic resistance profile (LVX-AMP-GEN-TET). The relationship between PFGE patterns and antibiotic resistance profiles or sample origin could not be established.
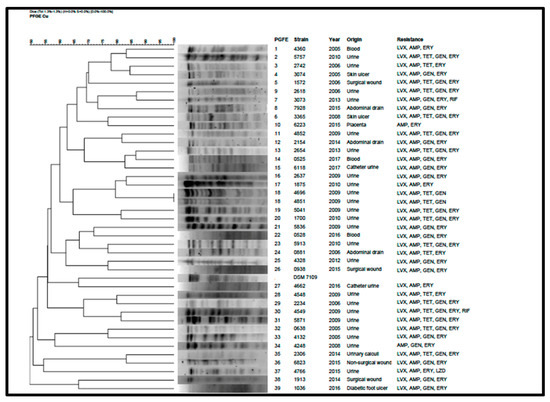
Figure 4.
PFGE patterns, dendrogram, sample origin, and resistance profiles of the 40 C. urealyticum and the reference strain DSM 7109. LVX: levofloxacin; AMP: ampicillin; TET: tetracycline; GEN: gentamicin; ERY: erythromycin; RIF: rifampicin; LZD: linezolid.
The high number of pulsotypes obtained in our C. urealyticum isolates highlighted the elevated genetic diversity in this specie. Among 40 strains, 39 were unrelated, producing sporadic infections.
3. Discussion
Management of C. urealyticum infections usually relies on glycopeptides (vancomycin or teicoplanin), to which this microorganism is uniformly susceptible [15]. All our C. urealyticum were susceptible to vancomycin. In two previous studies, we have reported full activity of vancomycin [16] and teicoplanin [17] against C. urealyticum. In a more recent report including 52 C. urealyticum clinical strains, vancomycin was also active against all the isolates [10]. In vitro studies indicated that linezolid can also be effective [5], and subsequent studies confirmed that linezolid was fully active against C. urealyticum [8,9,10,18]. The data presented in this work basically agree with linezolid susceptibility data from the mentioned studies, highlighting its option as a therapeutic alternative for vancomycin. However, one of our isolates showed low level resistance to this compound (MIC = 3 mg/L), which raises the need of maintaining surveillance strategies among this multidrug resistant pathogen as well as defining its resistance profile before treatment.
Isolates resistant against erythromycin, levofloxacin, and ampicillin showed a monomodal distribution of their MICs, which suggests the existence of a unique or major mechanism of resistance for each antimicrobial. Erythromycin was inactive against the majority of our C. urealyticum, in accordance with previously reported data [19]. All the erythromycin-resistant C. urealyticum carried the ermX gene. It is well established that the ermX gene encodes an N-6-methyltransferase that modifies an adenine of the 23S rRNA, conferring resistance against erythromycin [4]. Whole genome sequencing of five C. urealyticum confirmed the presence of the ermX gene.
Fluoroquinolones have been extensively used in the empirical treatment of urinary tract infections. Upon antibiotic administration, these drugs tend to accumulate in the organs of the body leading to the selection of spontaneous mutants in large bacterial populations, including those that colonize the skin and mucous membranes such as corynebacteria. In fluoroquinolone-resistant Corynebacterium spp. mutations are circumscribed to the gyrA gene (QRDR region), since these bacteria lack the parC gene. Thirty-eight of our 40 C. urealyticum (95%) showed high level resistance to levofloxacin. López-Medrano et al. reported that 79% of their isolates were resistant to ciprofloxacin [18]. Sequencing of the QRDR region of the gyrA gene of 29 levofloxacin-resistant C. urealyticum revealed that resistance is associated with single or double amino acid substitutions in residues Ser-90 and Asp-94 (C. urealyticum numbering). Ramos et al. have recently reported three C. urealyticum strains showing high level quinolone resistance associated to double amino-acid substitutions in Ser-90 and Asp-94 [20]. However, in three of our strains, one amino acid replacement (Asp-94 by Tyr) was enough to display high resistance level to levofloxacin. In one C. urealyticum, no link between mutations in this region and levofloxacin-resistant phenotype could be established, suggesting the existence of additional resistance mechanisms.
Rifampicin has been used as complementary agent for the management of C. striatum infections [21,22]. Rifampicin showed good activity against our C. urealyticum, since only two out of 40 isolates were resistant. Of note, the C. urealyticum reference strain (DSM 7109) is also rifampicin-resistant. A recent study including 52 C. urealyticum in Canada showed the same MIC50 and MIC90 values for rifampicin (≤0.05 mg/L) [10]. Resistance to this compound typically results from the substitution of some highly conserved residues in the RNA polymerase β subunit [23]. In Mycobacterium tuberculosis, more than 96% of rifampicin-resistant strains have mutations within the 81-bp rifampicin resistance-determining region (RRDR) of the rpoB gene (codons 507–533) [24]. We compared RpoB sequences of the three rifampicin-resistant C. urealyticum with that of susceptible strains. Considering the presumptive location of the RpoB active site and discarding the influence of conservative replacements in rifampicin susceptibility, we propose that the high rifampicin MICs can be explained by amino acid replacements in RpoB of the two rifampicin-resistant strains (Ser-444→Phe in VH4549 and Gln-511→Lys in VH3073) (Figure 1). We have also identified the substitution Ser-444→Asn in DSM 7109.
All our C. urealyticum displayed high level resistance to ampicillin (MIC90 > 256 mg/L). Hydrolysis of β-lactam antibiotics by β-lactamases is the most common mechanism of β-lactam resistance in clinically relevant bacteria. The ampicillin-resistant C. urealyticum were positive for the blaA-based PCR whereas a susceptible strain was negative. Whole genome analysis of five C. urealyticum isolates confirmed the presence of the blaA gene, flanked by transposase encoding genes (Figure 2A). The blaA gene encodes a serine hydrolase belonging to the class A β-lactamase protein family, which is highly conserved in several Corynebacterium species. In seven C. urealyticum strains and in C. striatum KC-Na-01 the blaA gene is in close vicinity to a transposase encoding genes (Figure 2B), suggesting that it has been horizontally propagated.
Aminoglycosides are complementary antibiotics for the treatment of infections caused by Corynebacterium spp. However, it has been reported that C. urealyticum is mostly resistant to aminoglycosides [25]. The MICs of C. urealyticum DSM 7109 for kanamycin and streptomycin are >256 and >128 mg/L, respectively [4]. Thirty-three out of our 40 C. urealyticum were resistant to gentamicin, with MIC50 and MIC90 values of 4 and 9 mg/L, respectively, indicating a high prevalence but a low level of resistance (range tested 0.016–256 mg/L). The gene aac(3)-XI, encoding an aminoglycoside 3-N acetyltransferase, which confers resistance to gentamicin and other aminoglycosides, has been recently identified in C. striatum [14], whose presence was correlated to low level of resistance to gentamicin [26]. Search by PCR with primers based on C. striatum aac(3)-XI sequence gave negative results in our 40 C. urealyticum, since these primers did not match with the C. urealyticum aac(3)-XI gene. However, by means of whole genome sequencing, we detected this gene as part of a highly conserved region in strains VH3073, VH4549, VH5757, and VH5913, but not in strain VH4248. We hypothesize that low level gentamicin resistance in these four strains is related with the presence of the aac(3)-XI gene, whereas in strain VH4248 is due to another mechanism. This analysis also revealed the presence in these four strains of a region including the gene aph(3′)-Ic (related to kanamycin resistance) and the pair of genes aph(3″)-Ib and aph(6)-Id (related to streptomycin resistance), which is also present in C. urealyticum DSM 7109 [4] as well as in other Corynebacterium spp.
Fifty percent of our C. urealyticum were resistant to tetracycline. Tetracycline-resistant strains showed a bimodal MICs distribution, which suggests the existence of different resistance mechanisms. In C. striatum, tetracycline resistance is mediated by an ATP gradient efflux mechanism encoded by the pair of genes tetA-tetB [27], which is also found in C. urealyticum DSM 7109 [4]. However, in our C. urealyticum, the tetA-tetB genes were not detected. The remarkable decrease of tetracycline MICs of seven of our C. urealyticum observed in presence of PAβN, a broad-spectrum efflux pump inhibitor, indicates that tetracycline resistance is mediated by an efflux mechanism.
PFGE is considered as the “gold standard” technique to assess epidemiological relationships for most clinically-relevant bacteria [28]. Our results showed almost the same number of isolates as PFGE patterns, with only two strains sharing the same pulsotype. This high diversity among C. urealyticum isolates revealed that they are not related but causing sporadic infections. While whole genome sequencing provides more detailed and accurate information, its use is not still viable in the daily clinical routine. Thus, PFGE remains as an important tool at epidemiological level. However, increasing the number of sequenced strains will provide more information, such as antimicrobial resistance and virulence, in comparison to PFGE, which will improve, in turn, our knowledge about the resistance and virulence mechanisms of this pathogen.
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
Forty C. urealyticum isolated from clinical samples at Clinical Microbiology Laboratory, Hospital Universitario Marqués de Valdecilla (HUMV), Santander (Spain), during the period 2005–2017, were used in this study. C. urealyticum DSM 7109 was also included as the reference strain. The origin of the samples was diverse: urine (25), abdominal drainage (3), surgical wound (3), blood (3), skin ulcer (2), urinary stone (1), non-surgical wound (1), diabetic foot ulcer (1), and placenta (1). They were initially identified by the API Coryne system (bioMérieux, Marcy l’Etoile, France) and confirmed by MALDI-TOF mass spectrometry using the Vitek MS (bioMérieux) platform, according to manufacturer’s instructions. C. urealyticum 18408721, isolated at Hospital Universitario Central de Asturias (HUCA), Oviedo (Spain), was used as an ampicillin-susceptible control strain. All strains were grown in blood agar (BA) plates at 37 °C for 72 h and kept frozen at −80 °C in Brain Heart Infusion (BHI) broth with 20% glycerol until use.
4.2. Antimicrobial Susceptibility Assays
To study the activity of eight antimicrobial compounds (ampicillin, erythromycin, gentamicin, levofloxacin, linezolid, rifampicin, tetracycline, and vancomycin) against the 40 C. urealyticum, minimal inhibitory concentrations (MICs) were determined using Etest® strips (bioMérieux) on Mueller-Hinton (MH) agar plates supplemented with horse blood and β-NAD (Oxoid, Madrid, Spain). Briefly, agar plates were inoculated with a 100 μL aliquot of a bacterial suspension at OD600 = 0.1, and incubated for 48 h. Tetracycline MICs were also determined in presence of PAβN (50 mg/L).
Clinical categories were established according to the breakpoints for the microdilution susceptibility assay defined by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (http://www.eucast.org) guidelines [29]. EUCAST defined Corynebacterium spp. specific breakpoints for vancomycin, linezolid, tetracycline, and rifampicin; for ampicillin, gentamicin, and levofloxacin we considered EUCAST PK-PD breakpoints. For erythromycin, Staphylococcus spp. cut-offs defined by EUCAST were used.
4.3. Search of Resistance Genes by PCR
The 40 C. urealyticum were screened for the presence of resistance genes commonly found in Corynebacterium spp. and other Gram positive multidrug resistant bacteria. Specific primers are listed in Table 4. The gyrA and rpoB genes were amplified and sequenced for mapping mutations associated with levofloxacin and rifampicin resistance, respectively. C. urealyticum DNA for PCR reactions was prepared using InstaGeneTM Matrix (Bio-Rad, Madrid, Spain) following manufacturer’s instructions.

Table 4.
Primers used in the detection and sequencing of resistance genes. Tª indicates annealing temperatures. Size refers amplicon sizes in base pairs.
4.4. PCR Products Sequencing
PCR products were purified with silica gel columns using NucleoSpin® Gel and PCR clean-up kit (Macherey-Nagel, Düren, Germany). Purified DNA was sequenced by Macrogen (Madrid, Spain) with the primers outlined in Table 4. Mutations in gyrA and rpoB genes and amino acid changes in their corresponding proteins were identified by pairwise and multiple alignment of sequences between resistant and susceptible isolates using MEGA7 [33] and Clustal W [34] programs.
4.5. Genome Sequencing and Analysis
For whole genome sequencing, genomic DNAs of strains VH3073, VH4248, VH4549, VH5757, and VH5913 (selected on the basis of their resistance profile), were extracted using the NucleoSpin® Microbial DNA kit (Macherey-Nagel). Library preparation followed the NEBNext Fast DNA Fragmentation and Library Preparation Kit (New England Biolabs, Beverly, MA, USA) protocol and sequencing was performed in an Illumina HiSeq 2500 machine, at above 1000× coverage for all strains. De novo genome assembly was done with the SPAdes assembler and built-in on the PATRIC assembly server [35]. Structural and functional annotations were performed in RAST server (https://rast.nmpdr.org) [36]. Prediction of antimicrobial resistance profiles and comparative genomic analyses were performed using the bioinformatic platforms PATRIC (https://www.patricbrc.org/) [35] and EDGAR [37]. Multiple alignments were performed using T-coffee server in its variant M-coffee [38].
4.6. Pulsed-Field Gel Electrophoresis (PFGE)
PFGE was performed with a CHEF-DRIII system (Bio-Rad). Bacteria were grown in BHI broth with shaking at 37 °C for 48−72 h. Cultures were adjusted to OD600 = 2.0, cells from 250 μL were pelleted and resuspended in 300 μL of TE buffer (10 mM Tris, 1 mM EDTA) containing 2 mg/mL lysozyme. This suspension was incubated at 37 °C for 1 h, inverting the tubes every 10 min. An equal volume of 2% LM agarose (Pronadisa, Madrid, Spain) in TE buffer containing 1% SDS and 0.2 mg/mL proteinase K was added, and plugs were cast with a standard casting tray. After the plugs solidified, they were incubated overnight at 55 °C with shaking in 4 mL of TE buffer containing 1% sarcosyl and 0.15 mg/mL proteinase K. The plugs were washed six times with pre-warmed TE buffer and then digested with 30 U of XbaI at 37 °C overnight. Electrophoresis was performed in a 1.2% agarose gel at 6 V/cm and at 14 °C with 0.5× TBE buffer (0.5 mM Tris, 45 mM boric acid, 0.5 mM EDTA). Pulse times ramped from 0.1 to 5 s for 24 h. Low range PFGE marker (New England Biolabs) was used as the molecular size marker.
Cluster analysis was performed with Fingerprinting II v4.5 software (Bio-Rad) by using the Dice similarity coefficient and the Unweighted Pair Group Method with Arithmetic means (UPGMA), with 1.3% of optimization and tolerance. Isolates were classified as indistinguishable if they showed 100% similarity, as closely related subtypes if they showed 95–99% similarity, and as different strains if they showed <95% similarity.
4.7. Data Availability
The whole genomes of the five C. urealyticum have been deposited in the Integrated Microbial Genomes Database under the following accession numbers: VH3073 (2833973259); VH4248 (2833948465); VH4549 (2833950470); VH5757 (2833952634); VH5913 (2830819286). Four of them have also been deposited in GenBank under the following accession numbers: VH3073: GCA_008244525.1; VH4248: GCA_008180085.1; VH4549: GCA_008180045.1; VH5913: GCA_008180065.1.
5. Conclusions
This study illustrates the high prevalence of multidrug resistance among the C. urealyticum isolated in a Spanish hospital, in particular resistances to ampicillin, erythromycin, and levofloxacin. Our isolates were still susceptible to low concentrations of vancomycin and linezolid. In any case, appropriate antimicrobial therapy must be given in accordance with the results of the antimicrobial susceptibility test for each infection. One C. urealyticum showed resistance to linezolid. This finding raises the need of maintaining surveillance strategies among this multidrug resistant pathogen.
Author Contributions
Conceptualization, L.M.-M. and J.N.; methodology, I.C.-G., M.F.-M., A.R.-F. and L.G.C.P.; software, I.C.-G., D.J.P.R., E.R.G.R.A., L.G.C.P. and J.N.; validation, L.G.C.P., J.C., L.M.-M. and J.N.; formal analysis, I.C.-G., J.R.-V. and J.N.; investigation, I.C.-G., and M.F.-M.; resources, J.C., L.M.-M. and J.N.; data curation, I.C.-G., J.R.-V. and J.N.; writing—original draft preparation, J.N.; writing—review and editing, I.C.-G., L.M.-M. and J.N.; supervision, L.M.-M. and J.N.; project administration, J.N.; funding acquisition, J.C., L.M.-M. and J.N. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Plan Nacional de I+D+i 2013-2016 and Instituto de Salud Carlos III, Subdirección General de Redes y Centros de Investigación Cooperativa, Ministerio de Ciencia, Innovación y Universidades, Spanish Network for Research in Infectious Diseases (REIPI) D16/0016/0007 and RD16/0016/0008), and co-financed by European Development Regional Fund “A way to achieve Europe”, Operative program Intelligent Growth 2014-2020.
Acknowledgments
We thank Javier Fernández for the gift of the strain C. urealyticum 18408721 and María Luisa Junco for technical assistance.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bernard, K. The genus Corynebacterium and other medically relevant coryneform-like bacteria. J. Clin. Microbiol. 2012, 50, 3152–3158. [Google Scholar] [CrossRef]
- Ramsey, M.M.; Freire, M.O.; Gabrilska, R.A.; Rumbaugh, K.P.; Lemon, K.P. Staphylococcus aureus Shifts toward Commensalism in Response to Corynebacterium Species. Front. Microbiol. 2016, 7, 1230. [Google Scholar] [CrossRef] [PubMed]
- Soriano, F.; Ponte, C.; Santamaria, M.; Castilla, C.; Fernández Roblas, R. In vitro and in vivo study of stone formation by Corynebacterium group D2 (Corynebacterium urealyticum). J. Clin. Microbiol. 1986, 23, 691–694. [Google Scholar] [CrossRef]
- Tauch, A.; Trost, E.; Tilker, A.; Ludewig, U.; Schneiker, S.; Goesmann, A.; Arnold, W.; Bekel, T.; Brinkrolf, K.; Brune, I.; et al. The lifestyle of Corynebacterium urealyticum derived from its complete genome sequence established by pyrosequencing. J. Biotechnol. 2008, 136, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Pagnoux, C.; Bérezné, A.; Damade, R.; Paillot, J.; Aouizerate, J.; Le Guern, V.; Salmon, D.; Guillevin, D. Encrusting Cystitis Due to Corynebacterium urealyticum in a Patient with ANCA-Associated Vasculitis: Case Report and Review of the Literature. Semin. Arthritis. Rheum. 2011, 41, 297–300. [Google Scholar] [CrossRef] [PubMed]
- Funke, G.; Von Graevenitz, A.; Clarridge, J.E.; Bernard, K.A.; Funke, G. Clinical Microbiology of Coryneform Bacteria. Clin. Microbiol. Rev. 1997, 10, 125–159. [Google Scholar] [CrossRef]
- Fish, D.N.; Piscitelli, S.C.; Danzinger, L.H. Development of Resistance During Antimicrobial Therapy. Pharmacotherapy 1995, 15, 279–291. [Google Scholar] [CrossRef]
- Gómez-Garcés, J.L.; Alos, J.I.; Tamayo, J. In vitro activity of linezolid and 12 other antimicrobials against coryneform bacteria. Int. J. Antimicrob. Agents 2007, 29, 688–692. [Google Scholar] [CrossRef]
- Fernández-Roblas, R.; Adames, H.; Martín-de-Hijas, N.Z.; García Almeida, D.; Gadea, I.; Esteban, J. In vitro activity of tigecycline and 10 other antimicrobials against clinical isolates of the genus Corynebacterium. Int. J. Antimicrob. Agents 2009, 33, 453–455. [Google Scholar] [CrossRef]
- Neemuchwala, A.; Soares, D.; Ravirajan, V.; Marchand-Austin, A.; Kus, J.V.; Patel, S.N. In Vitro Antibiotic Susceptibility Pattern of Non-diphtheriae Corynebacterium Isolates in Ontario, Canada, from 2011 to 2016. Antimicrob. Agents Chemother. 2018, 62, e01776-17. [Google Scholar] [CrossRef]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Guimaraes, L.C.; Soares, S.C.; Albersmeier, A.; Blom, J.; Jaenicke, S.; Azevedo, V.; Soriano, F.; Tauch, A.; Trost, E. Complete Genome Sequence of Corynebacterium urealyticum Strain DSM 7111, Isolated from a 9-Year-Old Patient with Alkaline-Encrusted Cystitis. Genome Announc. 2013, 1, e00264-13. [Google Scholar] [CrossRef] [PubMed]
- Schröder, J.; Maus, I.; Meyer, K.; Wördemann, S.; Blom, J.; Jaenicke, S.; Schneider, J.; Trost, E.; Tauch, A. Complete genome sequence, lifestyle, and multi-drug resistance of the human pathogen Corynebacterium resistens DSM 45100 isolated from blood samples of a leukemia patient. BMC Genom. 2012, 13, 141. [Google Scholar] [CrossRef] [PubMed]
- Galimand, M.; Fishovitz, J.; Lambert, T.; Barbe, V.; Zajicek, J.; Mobashery, S.; Courvalin, P. AAC(3)-XI, a new aminoglycoside 3-N-acetyltransferase from Corynebacterium striatum. Antimicrob. Agents Chemother. 2015, 59, 5647–5653. [Google Scholar] [CrossRef] [PubMed]
- Soriano, F.; Tauch, A. Microbiological and clinical features of Corynebacterium urealyticum: Urinary tract stones and genomics as the Rosetta Stone. Clin. Microbiol. Infect. 2008, 14, 632–643. [Google Scholar] [CrossRef] [PubMed]
- Salas, C.; Calvo, J.; Martinez-Martinez, L. Activity of Tigecycline against Coryneform Bacteria of Clinical Interest and Listeria monocytogenes. Antimicrob. Agents Chemother. 2008, 52, 1503–1505. [Google Scholar] [CrossRef][Green Version]
- Navas, J.; Salas, C.; Calvo, J.; Martinez-Martinez, L. Activity of daptomycin and three comparator agents against non-diphtheriae Corynebacterium isolates of clinical interest. J. Antimicrob. Chemother. 2012, 67, 776–778. [Google Scholar] [CrossRef]
- Lopez-Medrano, F.; Garcia-Bravo, M.; Morales, J.M.; Andrés, A.; San Juan, R.; Lizasoain, M.; Aguado, J.M. Urinary tract infection due to Corynebacterium urealyticum in kidney transplant recipients: An underdiagnosed etiology for obstructive uropathy and graft dysfunction-results of a prospective cohort study. Clin. Infect. Dis. 2008, 46, 825–830. [Google Scholar] [CrossRef]
- Ortiz-Pérez, A.; Martín-De-Hijas, N.Z.; Esteban, J.; Fernández-Natal, M.I.; García-Cía, J.I.; Fernández-Roblas, R. High Frequency of Macrolide Resistance Mechanisms in Clinical Isolates of Corynebacterium Species. Microb. Drug Resist. 2010, 16, 273–277. [Google Scholar] [CrossRef]
- Ramos, J.N.; Valadao, T.B.; Baio, P.V.P.; Mattos-Guaraldi, A.L.; Vieira, V.V. Novel mutations in the QRDR region gyrA gene in multidrug-resistance Corynebacterium spp. isolates from intravenous sites. Antonie Van Leeuwenhoek 2020, 113, 589–592. [Google Scholar] [CrossRef]
- Noussair, L.; Salomon, E.; El Sayed, F.; Duran, C.; Bouchand, F.; Roux, A.L.; Gaillard, J.L.; Bauer, T.; Rottman, M.; Dinh, A. Monomicrobial bone and joint infection due to Corynebacterium striatum: Literature review and amoxicillin-rifampin combination as treatment perspective. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 1269–1278. [Google Scholar] [CrossRef]
- Shah, M.; Murillo, J.L. Successful treatment of Corynebacterium striatum endocarditis with daptomycin plus rifampin. Ann. Pharmacother. 2005, 39, 1741–1744. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Koga, H.; Ohno, H.; Ogawa, K.; Fukuda, M.; Hirakata, Y.; Maesaki, S.; Tomono, K.; Tashiro, T.; Kohno, S. Relationship between antimycobacterial activities of rifampicin, rifabutin and KRM-1648 and rpoB mutations of Mycobacterium tuberculosis. J. Antimicrob. Chemother. 1998, 42, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Horng, Y.T.; Jeng, W.Y.; Chen, Y.Y.; Liu, C.H.; Dou, H.Y.; Lee, J.J.; Chang, K.C.; Chien, C.C.; Soo, P.C. Molecular analysis of codon 548 in the rpoB gene involved in Mycobacterium tuberculosis resistance to rifampin. Antimicrob. Agents Chemother. 2015, 59, 1542–1548. [Google Scholar] [CrossRef] [PubMed]
- Salem, N.; Salem, L.; Saber, S.; Ismail, G.; Bluth, M. Corynebacterium urealyticum: A comprehensive review of an understated organism. Infect. Drug Resist. 2015, 8, 129–145. [Google Scholar] [CrossRef]
- Navas, J.; Fernández-Martínez, M.; Salas, C.; Cano, M.E.; Martinez-Martinez, L. Susceptibility to Aminoglycosides and Distribution of aph and aac(3)-XI Genes among Corynebacterium striatum Clinical Isolates. PLoS ONE 2016, 11, e0167856. [Google Scholar] [CrossRef]
- Tauch, A.; Krieft, S.; Pühler, A.; Kalinowski, J. The tetAB genes of the Corynebacterium striatum R-plasmid pTP10 encode an ABC transporter and confer tetracycline, oxytetracycline and oxacillin resistance in Corynebacterium glutamicum. FEMS Microbiol. Lett. 1999, 173, 203–209. [Google Scholar] [CrossRef][Green Version]
- Goering, R.V. Pulsed field gel electrophoresis: A review of application and interpretation in the molecular epidemiology of infectious disease. Infect. Genet. Evol. 2010, 10, 866–875. [Google Scholar] [CrossRef]
- European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters; Version 10.0; EUCAST: Växjö, Sweden, 2020; Available online: http://www.eucast.org (accessed on 25 January 2020).
- Sutcliffe, J.; Grebe, T.; Tait-Kamradt, A.; Wondrack, L. Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 1996, 40, 2562–2566. [Google Scholar] [CrossRef]
- Alibi, S.; Ferjani, A.; Boukadida, J.; Cano, M.E.; Fernandez-Martinez, M.; Martinez-Martinez, L.; Navas, J. Occurrence of Corynebacterium striatum as an emerging antibiotic-resistant nosocomial pathogen in a Tunisian hospital. Sci. Rep. 2017, 7, 9704. [Google Scholar] [CrossRef]
- Sierra, J.M.; Martínez-Martínez, L.; Vázquez, F.; Giralt, E.; Vila, J. Relationship between mutations in the gyrA gene and quinolone resistance in clinical isolates of Corynebacterium striatum and Corynebacterium amycolatum. Antimicrob. Agents Chemother. 2005, 49, 1714–1719. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; Mcgettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, P.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Wattam, A.R.; Davis, J.J.; Assaf, R.; Boisvert, S.; Brettin, T.; Bun, C.; Conrad, A.; Dietrich, E.M.; Disz, T.; Gabbard, J.L.; et al. Improvements to PATRIC, the all-bacterial Bioinformatics Database and Analysis Resource Center. Nucleic. Acids. Res. 2017, 45, D535–D542. [Google Scholar] [CrossRef]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic. Acids. Res. 2014, 42, 206–214. [Google Scholar] [CrossRef]
- Blom, J.; Kreis, J.; Spanig, S.; Juhre, T.; Bertelli, C.; Ernst, C.; Goesmann, A. EDGAR 2.0: An enhanced software platform for comparative gene content analyses. Nucleic. Acids. Res. 2016, 44, W22–W28. [Google Scholar] [CrossRef]
- Moretti, S.; Armougom, F.; Wallace, I.M.; Higgins, D.G.; Jongeneel, C.V.; Notredame, C. The M-Coffee web server: A meta-method for computing multiple sequence alignments by combining alternative alignment methods. Nucleic. Acids. Res. 2007, 35, W645–W648. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).