Characterization of LysBC17, a Lytic Endopeptidase from Bacillus cereus
Abstract
1. Introduction
2. Results and Discussion
2.1. Bioinformatics
2.2. SDS-PAGE and Zymogram Analysis of LysBC17
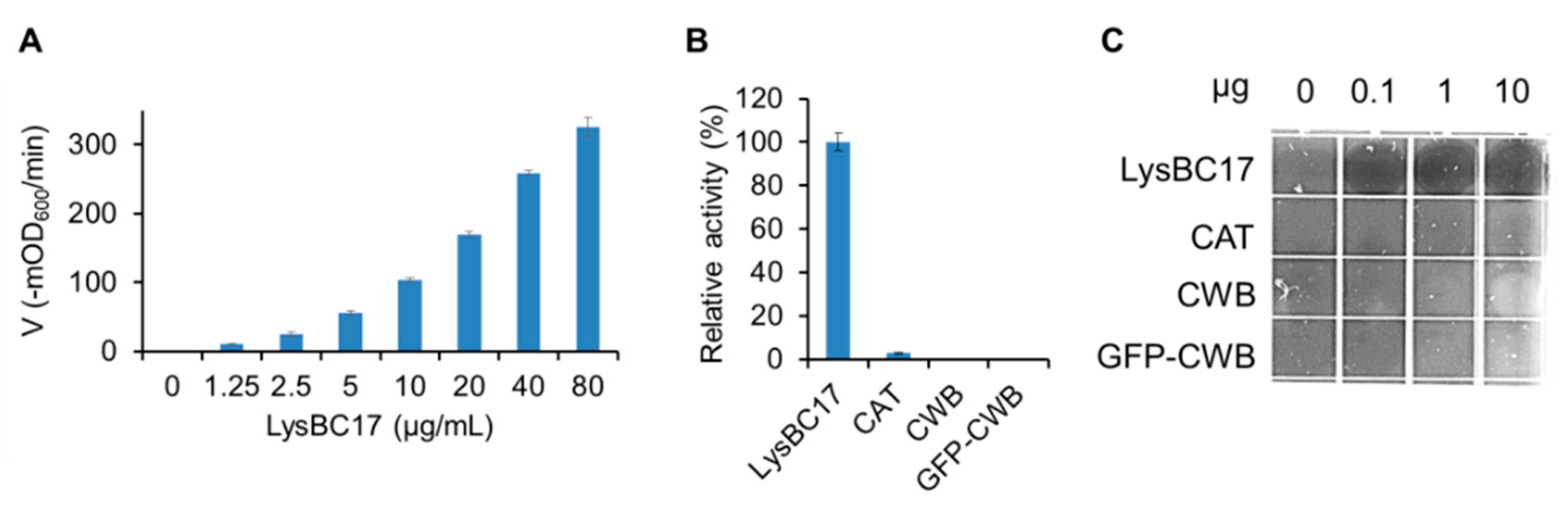
2.3. Characterization of LysBC17 by TRA
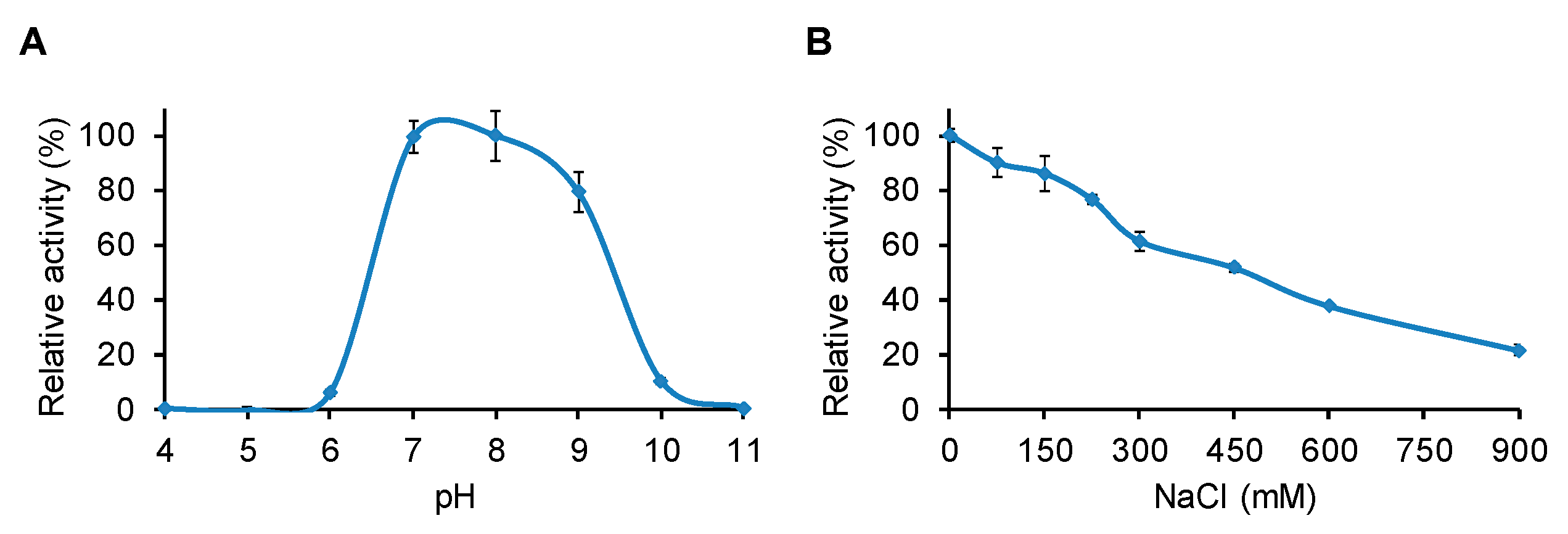
2.4. Determination of pH and NaCl Optima for LysBC17
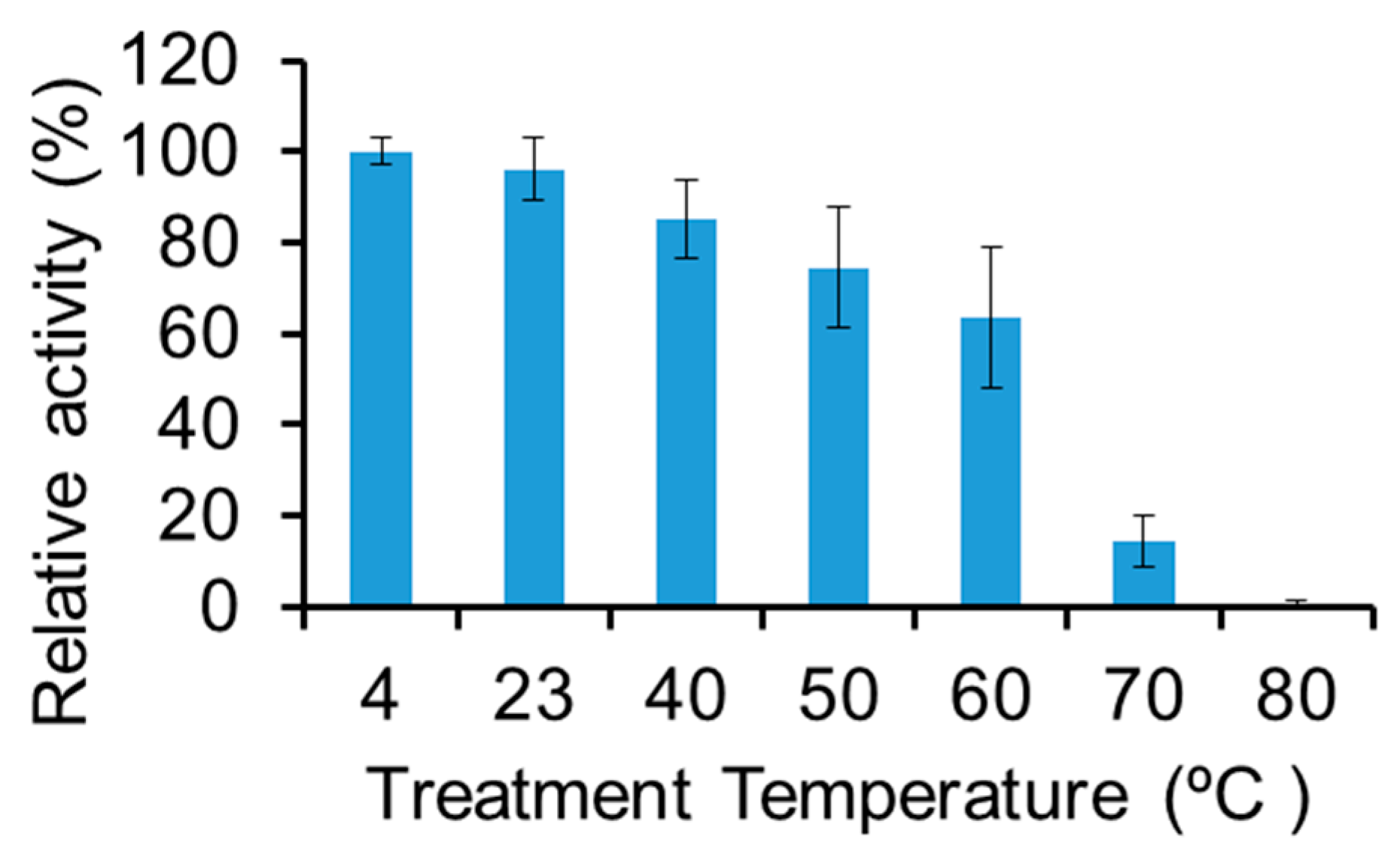
2.5. Thermostability of LysBC17
2.6. Screening the LysBC17 Lytic Host Range
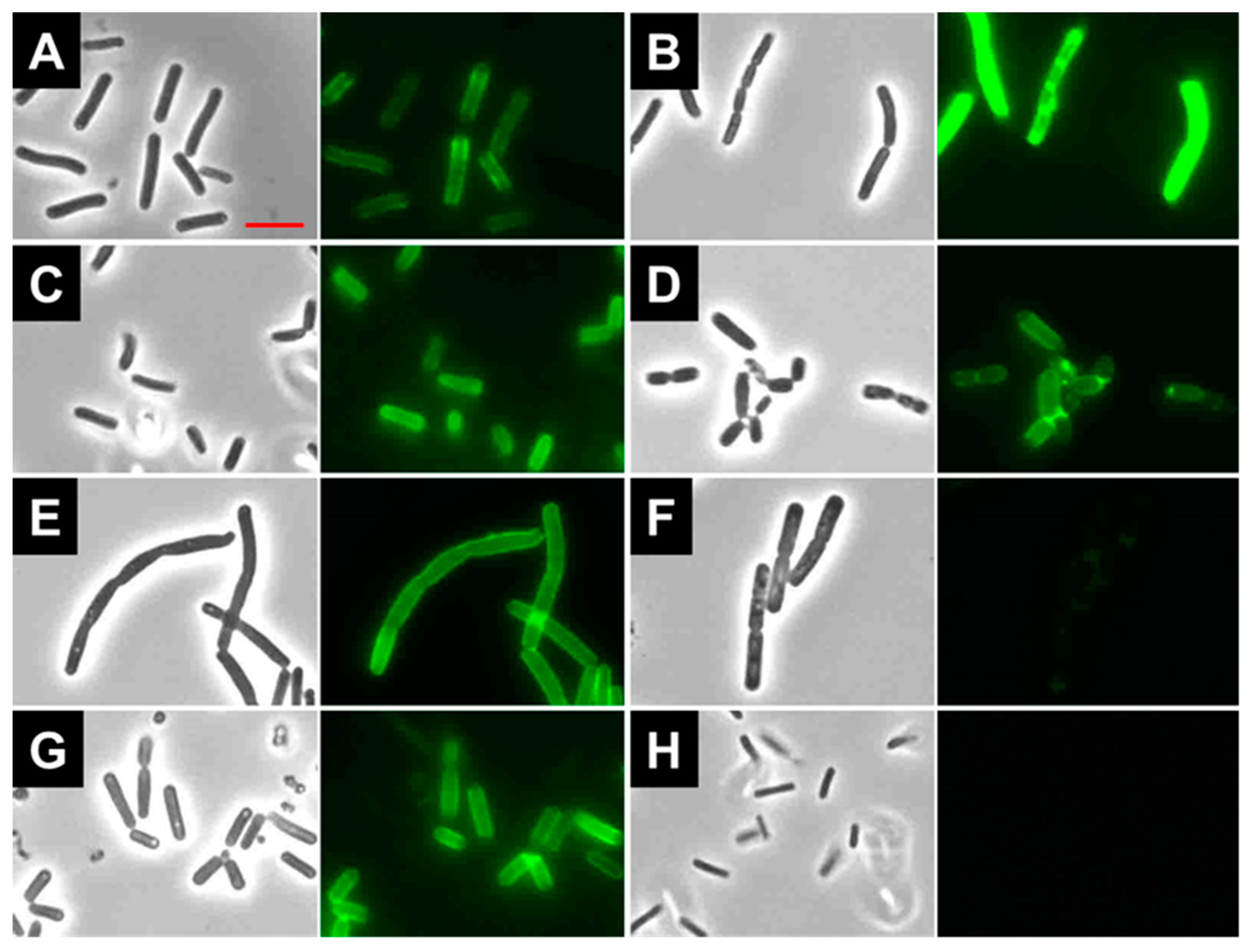
2.7. Binding of LysBC17 CWB
3. Materials and Methods
3.1. Bioinformatics
3.2. Bacteria
3.3. Cloning of LysBC17 and Derivatives
3.4. Recombinant Protein Expression and Purification
3.5. SDS-PAGE and Zymograms
3.6. Plate lysis Assays
3.7. Turbidity Reduction Assays
3.8. Binding Assays
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Arnesen, L.P.S.; Fagerlund, A.; Granum, P.E. From soil to gut: Bacillus cereus and its food poisoning toxins. Fems. Microbiol. Rev. 2008, 32, 579–606. [Google Scholar] [CrossRef] [PubMed]
- Jensen, G.B.; Hansen, B.M.; Eilenberg, J.; Mahillon, J. The hidden lifestyles of Bacillus cereus and relatives. Environ. Microbiol. 2003, 5, 631–640. [Google Scholar] [CrossRef] [PubMed]
- Bottone, E.J. Bacillus cereus, a volatile human pathogen. Clin. Microbiol. Rev. 2010, 23, 382–398. [Google Scholar] [CrossRef] [PubMed]
- Hede, K. Antibiotic resistance: An infectious arms race. Nature 2014, 509, S2–S3. [Google Scholar] [CrossRef] [PubMed]
- Ventola, C.L. The antibiotic resistance crisis: Part 2: Management strategies and new agents. P. T. 2015, 40, 344–352. [Google Scholar]
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. P. T. 2015, 40, 277–283. [Google Scholar] [PubMed]
- Dibner, J.J.; Richards, J.D. Antibiotic growth promoters in agriculture: History and mode of action. Poult. Sci. 2005, 84, 634–643. [Google Scholar] [CrossRef]
- Castanon, J.I.R. History of the use of antibiotic as growth promoters in European poultry feeds. Poultry Sci. 2007, 86, 2466–2471. [Google Scholar] [CrossRef]
- Millet, S.; Maertens, L. The European ban on antibiotic growth promoters in animal feed: From challenges to opportunities. Vet. J. 2011, 187, 143–144. [Google Scholar] [CrossRef]
- FDA. New Animal Drugs and New Animal Drug Combination Products Administered in or on Medicated Feed or Drinking Water of Food-Producing Animals: Recommendations for Drug Sponsors for Voluntarily Aligning Product Use Conditions with GFI #209; U.S. Food & Drug Administration: Rockville, MD, USA, 2013; Volume GFI. [Google Scholar]
- FDA. FDA Announces Implementation of GFI #213, Outlines Continuing Efforts to Address Antimicrobial Resistance. Available online: https://www.fda.gov/AnimalVeterinary/NewsEvents/CVMUpdates/ucm535154.htm (accessed on 29 October 2018).
- McKenna, M. After Years of Debate, the FDA Finally Curtails Antibiotic Use in Livestock. Available online: https://www.newsweek.com/after-years-debate-fda-curtails-antibiotic-use-livestock-542428 (accessed on 13 January 2017).
- Nelson, D.C.; Schmelcher, M.; Rodriguez-Rubio, L.; Klumpp, J.; Pritchard, D.G.; Dong, S.; Donovan, D.M. Endolysins as antimicrobials. Adv. Virus Res. 2012, 83, 299–365. [Google Scholar]
- Schmelcher, M.; Donovan, D.M.; Loessner, M.J. Bacteriophage endolysins as novel antimicrobials. Future Microbiol. 2012, 7, 1147–1171. [Google Scholar] [CrossRef] [PubMed]
- Geng, P.L.; Hu, Y.M.; Zhou, G.P.; Yuan, Z.M.; Hu, X.M. Characterization of three autolysins with activity against cereulide-producing Bacillus isolates in food matrices. Int. J. Food. Microbiol. 2017, 241, 291–297. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Rubio, L.; Gutierrez, D.; Donovan, D.M.; Martinez, B.; Rodriguez, A.; Garcia, P. Phage lytic proteins: Biotechnological applications beyond clinical antimicrobials. Crit. Rev. Biotechnol. 2015, 36, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Casjens, S. Prophages and bacterial genomics: What have we learned so far? Mol. Microbiol. 2003, 49, 277–300. [Google Scholar] [CrossRef]
- Schmitz, J.E.; Ossiprandi, M.C.; Rumah, K.R.; Fischetti, V.A. Lytic enzyme discovery through multigenomic sequence analysis in Clostridium perfringens. Appl. Microbiol. Biot. 2011, 89, 1783–1795. [Google Scholar] [CrossRef]
- Swift, S.M.; Waters, J.J.; Rowley, D.T.; Oakley, B.B.; Donovanl, D.M. Characterization of two glycosyl hydrolases, putative prophage endolysins, that target Clostridium perfringens. Fems. Microbiol. Lett. 2018, 365. [Google Scholar] [CrossRef]
- Ivanova, N.; Sorokin, A.; Anderson, I.; Galleron, N.; Candelon, B.; Kapatral, V.; Bhattacharyya, A.; Reznik, G.; Mikhailova, N.; Lapidus, A.; et al. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 2003, 423, 87–91. [Google Scholar] [CrossRef]
- Lu, J.Z.; Fujiwara, T.; Komatsuzawa, H.; Sugai, M.; Sakon, J. Cell wall-targeting domain of glycylglycine endopeptidase distinguishes among peptidoglycan cross-bridges. J. Biol. Chem. 2006, 281, 549–558. [Google Scholar] [CrossRef]
- Etobayeva, I.; Linden, S.B.; Alem, F.; Harb, L.; Rizkalla, L.; Mosier, P.D.; Johnson, A.A.; Temple, L.; Hakami, R.M.; Nelson, D.C. Discovery and biochemical characterization of PlyP56, PlyN74, and PlyTB40-Bacillus specific endolysins. Viruses-Basel 2018, 10. [Google Scholar] [CrossRef]
- Peng, Q.; Yuan, Y.H. Characterization of a novel phage infecting the pathogenic multidrug-resistant Bacillus cereus and functional analysis of its endolysin. Appl. Microbiol. Biot. 2018, 102, 7901–7912. [Google Scholar] [CrossRef]
- Son, B.; Yun, J.; Lim, J.A.; Shin, H.; Heu, S.; Ryu, S. Characterization of LysB4, an endolysin from the Bacillus cereus-infecting bacteriophage B4. BMC Microbiol. 2012, 12. [Google Scholar] [CrossRef] [PubMed]
- Jun, S.Y.; Jang, I.J.; Yoon, S.; Jang, K.; Yu, K.S.; Cho, J.Y.; Seong, M.W.; Jung, G.M.; Yoon, S.J.; Kang, S.H. Pharmacokinetics and tolerance of the phage endolysin-based candidate drug SAL200 after a single intravenous administration among healthy volunteers. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef]
- Cutlip, S.E.; Hott, J.M.; Buchanan, N.P.; Rack, A.L.; Latshaw, J.D.; Moritz, J.S. The effect of steam-conditioning practices on pellet quality and growing broiler nutritional value. J. Appl. Poultry. Res. 2008, 17, 249–261. [Google Scholar] [CrossRef]
- Svihus, B.; Zimonja, O. Chemical alterations with nutritional consequences due to pelleting animal feeds: A review. Anim. Prod. Sci. 2011, 51, 590–596. [Google Scholar] [CrossRef]
- Wijma, H.J.; Floor, R.J.; Janssen, D.B. Structure- and sequence-analysis inspired engineering of proteins for enhanced thermostability. Curr. Opin. Struc. Biol. 2013, 23, 588–594. [Google Scholar] [CrossRef]
- Busto, M.D.; Apenten, R.K.O.; Robinson, D.S.; Wu, Z.; Casey, R.; Hughes, R.K. Kinetics of thermal inactivation of pea seed lipoxygenases and the effect of additives on their thermostability. Food Chem. 1999, 65, 323–329. [Google Scholar] [CrossRef]
- Gekko, K.; Timasheff, S.N. Thermodynamic and kinetic examination of protein stabilization by glycerol. Biochemistry 1981, 20, 4677–4686. [Google Scholar] [CrossRef] [PubMed]
- Coordinators, N.R. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2018, 46, D8–D13. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef] [PubMed]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.A.J.; Wu, M.H.; Yan, C.H.; Chau, B.K.H.; So, H.; Ng, A.; Chan, A.; Cheah, K.S.E.; Briscoe, J.; Cheung, M. Phosphorylation of Sox9 is required for neural crest delamination and is regulated downstream of BMP and canonical Wnt signaling. Proc. Natl. Acad. Sci. USA 2013, 110, 2882–2887. [Google Scholar] [CrossRef] [PubMed]
- Becker, S.C.; Dong, S.; Baker, J.R.; Foster-Frey, J.; Pritchard, D.G.; Donovan, D.M. LysK CHAP endopeptidase domain is required for lysis of live staphylococcal cells. FEMS Microbiol. Lett. 2009, 294, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Donovan, D.M.; Foster-Frey, J. LambdaSa2 prophage endolysin requires Cpl-7-binding domains and amidase-5 domain for antimicrobial lysis of streptococci. FEMS Microbiol. Lett. 2008, 287, 22–33. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Donovan, D.M.; Lardeo, M.; Foster-Frey, J. Lysis of staphylococcal mastitis pathogens by bacteriophage phi11 endolysin. FEMS Microbiol. Lett. 2006, 265, 133–139. [Google Scholar] [CrossRef] [PubMed]

| Bacteria | LysBC17 Activity a |
|---|---|
| Bacillus cereus Bc17 | +++ |
| Bacillus cereus ATCC 4342 | +++ |
| Bacillus cereus ATCC 11778 | +++ |
| Bacillus cereus ATCC 13061 | +++ |
| Bacillus cereus ATCC 14579 | +++ |
| Bacillus anthracis Ames 35 | ++ |
| Bacillus anthracis UM23 | + |
| Bacillus thuringiensis ATCC 13061 | ++ |
| Bacillus pumilus BJ0050 | +++ |
| Bacillus pumilus BJ0055 | +/- |
| Clostridium perfringens Cp35 | - |
| Staphylococcus aureus Newman | - |
| Streptococcus uberis | - |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Swift, S.M.; Etobayeva, I.V.; Reid, K.P.; Waters, J.J.; Oakley, B.B.; Donovan, D.M.; Nelson, D.C. Characterization of LysBC17, a Lytic Endopeptidase from Bacillus cereus. Antibiotics 2019, 8, 155. https://doi.org/10.3390/antibiotics8030155
Swift SM, Etobayeva IV, Reid KP, Waters JJ, Oakley BB, Donovan DM, Nelson DC. Characterization of LysBC17, a Lytic Endopeptidase from Bacillus cereus. Antibiotics. 2019; 8(3):155. https://doi.org/10.3390/antibiotics8030155
Chicago/Turabian StyleSwift, Steven M., Irina V. Etobayeva, Kevin P. Reid, Jerel J. Waters, Brian B. Oakley, David M. Donovan, and Daniel C. Nelson. 2019. "Characterization of LysBC17, a Lytic Endopeptidase from Bacillus cereus" Antibiotics 8, no. 3: 155. https://doi.org/10.3390/antibiotics8030155
APA StyleSwift, S. M., Etobayeva, I. V., Reid, K. P., Waters, J. J., Oakley, B. B., Donovan, D. M., & Nelson, D. C. (2019). Characterization of LysBC17, a Lytic Endopeptidase from Bacillus cereus. Antibiotics, 8(3), 155. https://doi.org/10.3390/antibiotics8030155





