STOP Strategy to Inhibit P. falciparum and S. aureus Growth: Molecular Mechanism Studies on Purposely Designed Hybrids
Abstract
1. Introduction
2. Results
2.1. In Vitro Antimicrobial Activity
2.2. In Vitro Antimalarial Activity
2.3. Cytotoxicity and Haemolytic Activity
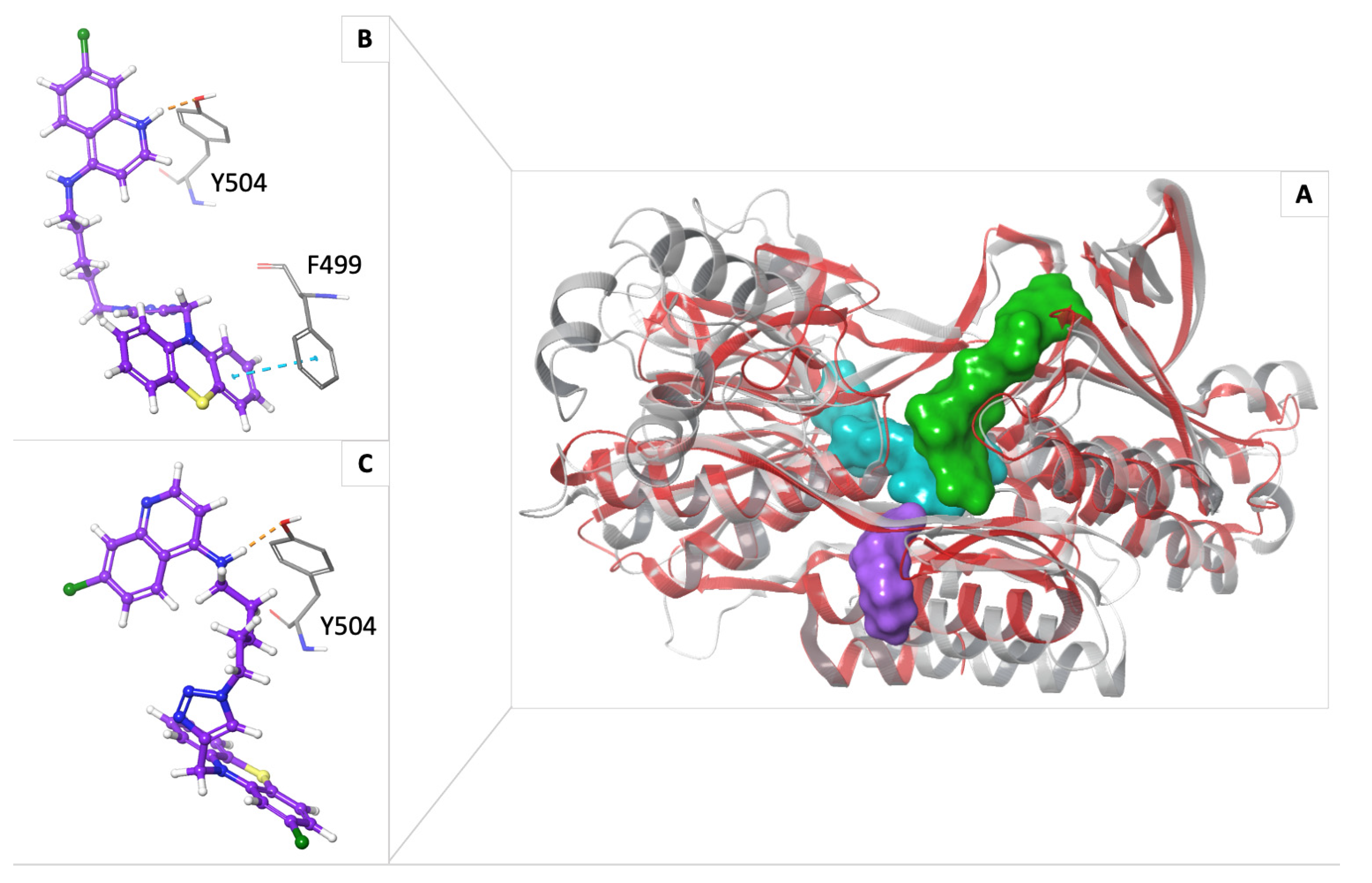
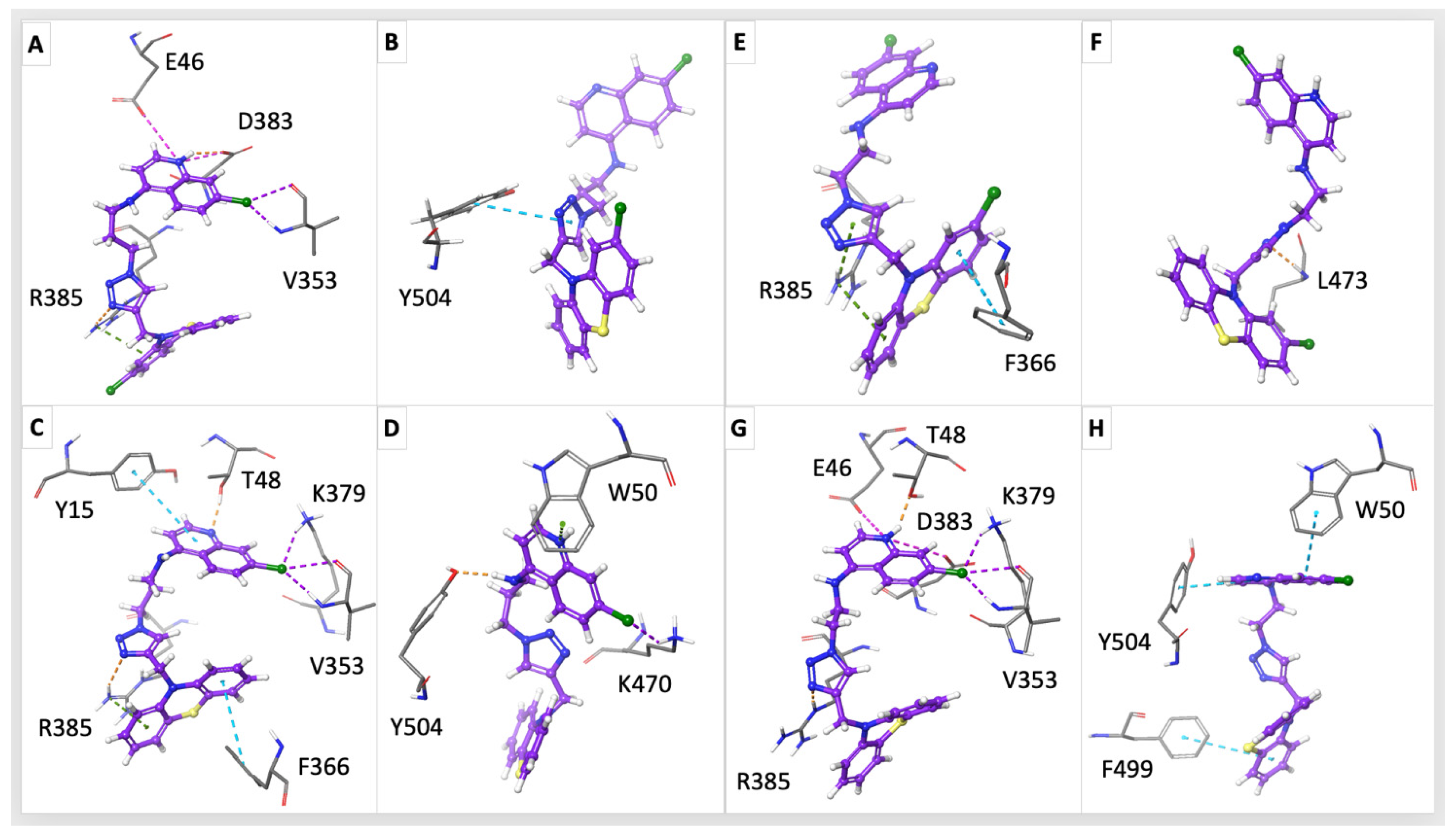
2.4. In Silico Study on NDH-2
3. Discussion
4. Materials and Methods
4.1. Chemistry
4.1.1. General Methods
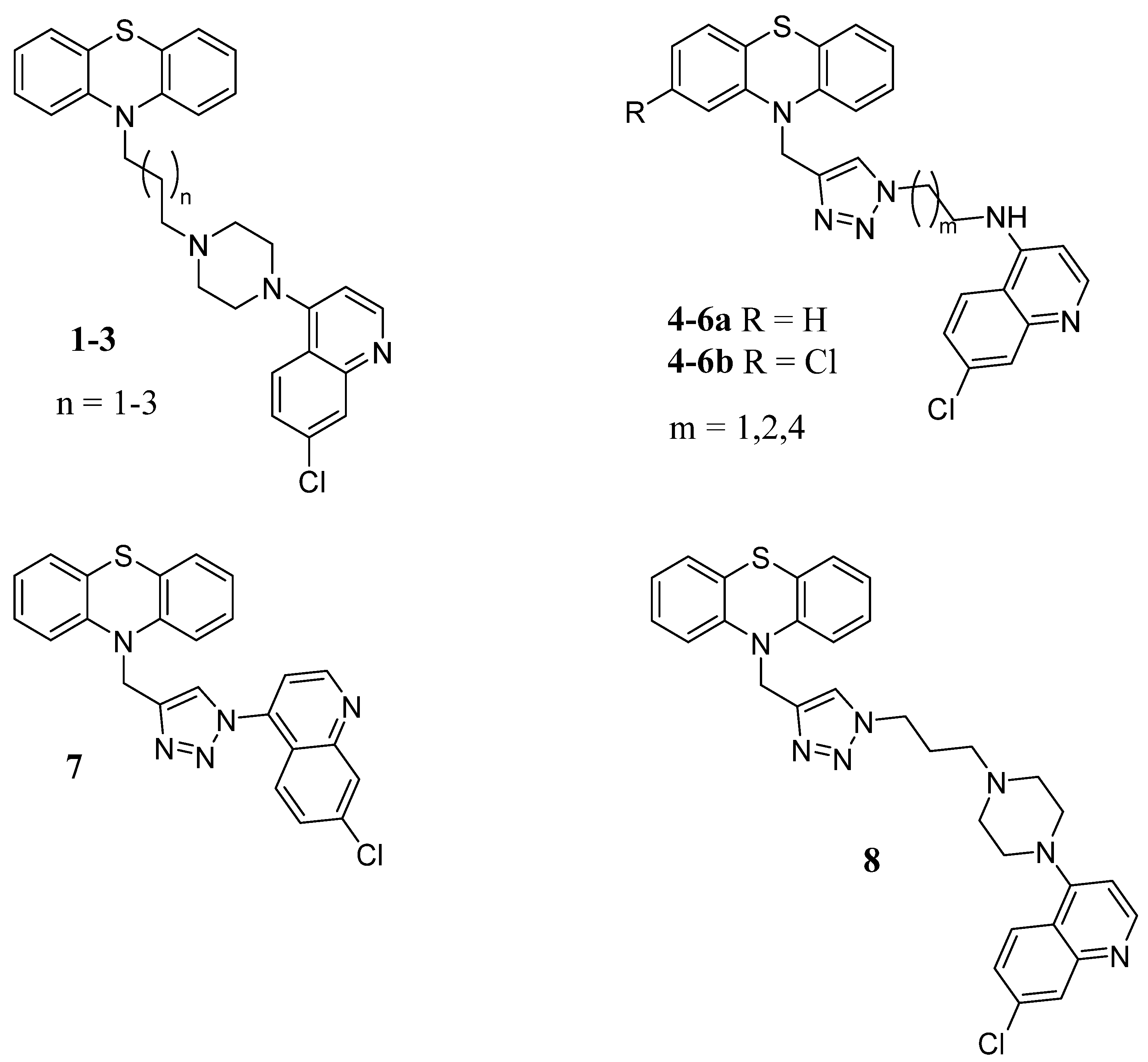
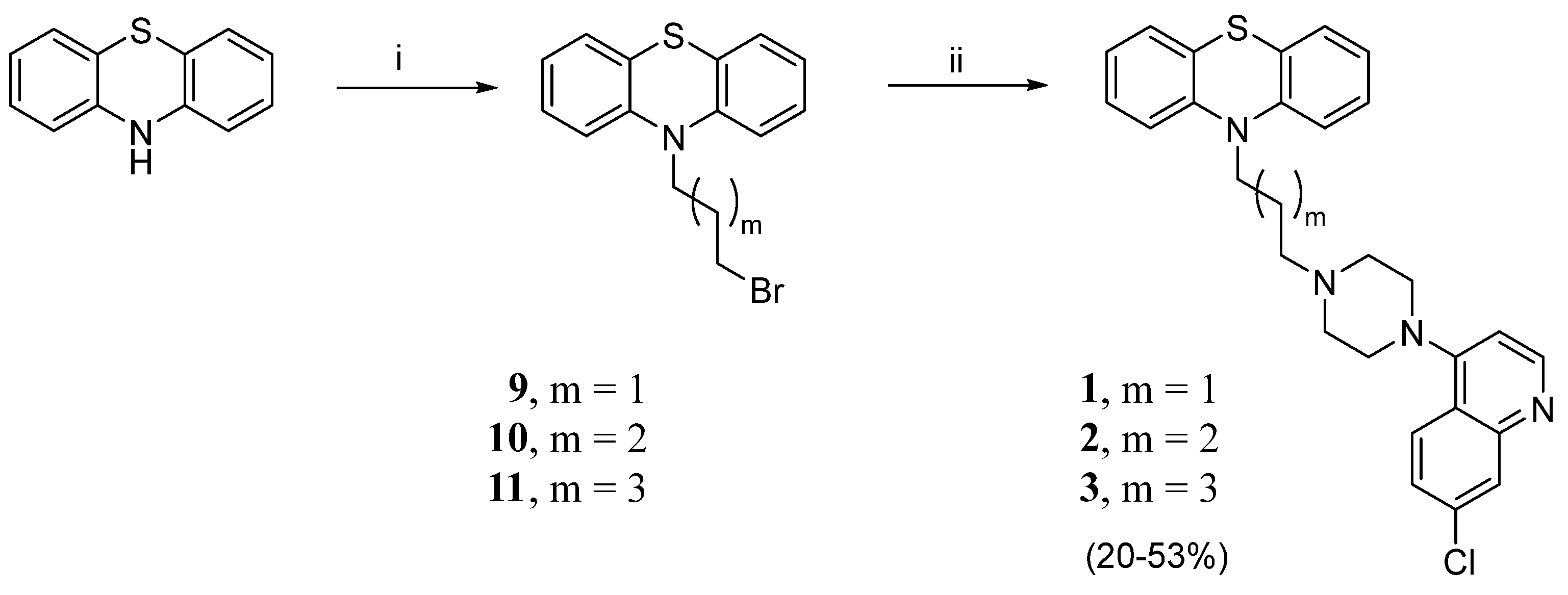
4.1.2. General Method for the Preparation of N-Bromoalkyl-Phenothiazine Derivatives 9–11
- 10-(3-chloropropyl)-10H-phenothiazine (9). Using the previous procedure and starting from 1.0 g of phenothiazine (0.005 mol) and 1-bromo-3-chloropropane, 0.85 g of 9 was obtained (PE/EtOAc 4.9:0.1). Yield: 61%, oil. 1H NMR δ 2.23–2.28 (m, 2H, -CH2-), 3.68 (t, J = 6.4 Hz, 2H, N-CH2-), 4.10 (t, J = 6.4 Hz, 2H, -CH2-Cl), 6.91–6.97 (m, 4H, Ar), 7.16–7.20 (m, 4H, Ar).
- 10-(4-bromobutyl)-10H-phenothiazine (10). Using the previous procedure and starting from 1.0 g of phenothiazine (0.005 mol) and 1,4-dibromobutane, 0.80 g of 10 was obtained (PE/EtOAc 4.75:0.25). Yield: 48%, oil. 1H NMR δ 2.08–2.10 (m, 4H, -CH2-CH2-), 3.51 (t, J = 6.0 Hz, 2H, N-CH2-), 4.02 (t, J = 6.0 Hz, 2H, -CH2-Br), 6.97–7.06 (m, 4H, Ar), 7.26–7.29 (m, 4H, Ar).
- 10-(5-bromopentyl)-10H-phenothiazine (11). Using the previous procedure and starting from 1.0 g of phenothiazine (0.005 mol) and 1,5 dibromopentane, 0.90 g of 11 was obtained (PE/EtOAc 4.9:0.1). Yield: 52%, oil. 1H NMR δ 1.56–1.60 (m, 2H, -CH2-), 1.80–1.88 (m, 4H, -CH2-CH2-), 3.36 (t, J = 6.4 Hz, 2H, N-CH2-), 3.86 (t, J = 6.4 Hz, 2H, -CH2-Br), 6.83–6.92 (m, 4H, Ar), 7.11–7.15 (m, 4H, Ar).
- 10-(3-(4-(7-chloroquinolin-4-yl)piperazin-1-yl)propyl)-10H-phenothiazine (1). In a Schlenk tube, 9 (0.35 g, 1.3 mmol, 1 eq), 7-chloro-4-(piperazin-1-yl)quinoline (0.47 g, 1.9 mmol, 1.5 eq), and triethylamine (TEA) (0.19 g, 0.26 mL, 1.5 eq) were dissolved in acetonitrile. The reaction mixture was heated at 70 °C for 24 h. The solvent was removed under reduced pressure, and the crude product was purified by flash column chromatography (PE/EtOAc 4.75:0.25) to yield 0.12 g of 1 (yield 20%), m.p. 72–73 °C. 1H NMR δ 2.18–2.21 (m, 2H, -CH2-), 2.85–2.91 (m, 6H, N-CH2- + piperazine), 3.37 (s, 4H, piperazine), 4.05 (t, J = 6.0 Hz, 2H, -CH2-N-PTZ), 6.84 (d, J = 6.0 Hz, 1H, Ar), 6.92–6.97 (m, 4H, Ar), 7.17–7.20 (m, 4H, Ar), 7.42 (d, J = 12.0 Hz, 1H, Ar), 7.84 (d, J = 12.0 Hz, 1H, Ar), 8.09 (s, 1H, Ar), 8.71 (d, J = 6.0 Hz, 1H, Ar). 13C NMR: δ 45.1, 53.1, 55.6, 109.1, 115.8, 121.9, 122.8, 125.2, 125.5, 126.4, 127.4, 127.7, 128.9, 135.2, 145.3. MS (ES) m/z: 488.1 (M + H).
- 10-(4-(4-(7-chloroquinolin-4-yl)piperazin-1-yl)butyl)-10H-phenothiazine (2). 0.36 g of 10 (1.1 mmol, 1.0 eq), 0.27 g of 7-chloro-4-(piperazin-1-yl)quinoline (1.1 mmol, 1.0 eq), and 0.11 g (0.16 mL, 1.0 eq) of TEA were dissolved in toluene. The reaction mixture was heated under reflux for 8 h, cooled at r.t., washed with H2O, dried over anhydrous Na2SO4, and the solvent evaporated to dryness. The crude was purified by flash column chromatography (PE/EtOAc (NH4OH 5%) 8:2) to yield 0.15 g of 2 (yield 27%), m.p. 78–80 °C. 1H NMR: δ 1.74–1.93 (m, 4H, -CH2CH2-), 2.54 (s, 2H, N-CH2), 2.67–2.71 (m, 4H, piperazine), 3.18–3.21 (m, 4H, piperazine), 3.94 (t, J = 6.0 Hz, 2H, CH2-N-PTZ), 6.81 (s, 1H, Ar), 6.89–6.94 (m, 4H, Ar), 7.14–7.17 (m, 4H, Ar), 7.40 (d, J = 12.0 Hz, 1H, Ar), 7.90 (d, J = 12.0 Hz, 1H, Ar), 8.04 (s, 1H, Ar), 8.71 (d, J = 6.0 Hz, 1H,Ar). 13C NMR: δ 23.5, 24.6, 47.0, 52.0, 52.9, 57.8, 109.1, 115.8, 122.0, 122.7, 125.2, 125.4, 126.3, 127.4, 127.7, 129.0, 135.1, 145.4, 150.2, 152.0. MS (ES) m/z: 502.1 (M + H).
- 10-(5-(4-(7-chloroquinolin-4-yl)piperazin-1-yl)pentyl)-10H-phenothiazine (3). Applying the procedure used for 2 and starting from 0.66 g (1.9 mmol, 1.0 eq) of 11, compound 3 was obtained. The crude was purified by flash column chromatography (PE/EtOAc (NH4OH 5%) 9:1) to obtain 0.30 g of 3 (yield 53%), m.p. 77–79 °C. 1H NMR: δ 1.58–1.48 (m, 4H, CH2CH2), 1.83–1.88 (m, 2H, CH2), 2.44 (t, J = 12Hz, 2H, N-CH2), 2.68–2.72 (m, 4H, piperazine), 3.19–3.21 (m, 4H, piperazine),, 3.89 (t, J = 6.0 Hz, 2H, CH2N-PTZ), 6.81–6.92 (m, 5H, Ar), 7.13–7.16 (m, 4H, Ar), 7.41 (d, J = 12.0 Hz, 1H, Ar), 7.92 (d, J = 12.0 Hz, 1H, Ar), 8.03 (s, 1H, Ar), 8.70 (d, J = 6.0 Hz, 1H, Ar). 13C NMR: δ 24.9, 26.6, 26.9, 47.2, 52.3, 55.2, 58.5, 109.1, 115.6, 122.0, 122.6, 125.4, 126.2, 127.3, 127.6, 129.0, 135.0, 145.5, 150.3, 152.1, 157.1. MS (ES) m/z: 515.1 (M + H).
- 7-chloro-4-(4-(3-chloropropyl)piperazin-1-yl)quinoline (13). A solution of 7-chloro-4-(piperazin-1-yl)quinoline (1.16 g, 4.7 mmol, 1 eq), 1-bromo-3-chloropropane (0.93 mL, 9.4 mmol, 2 eq), and TEA (0.7 mL, 4.7 mmol, 1 eq) in toluene (20 mL) was heated for 6 h under reflux. The mixture was cooled at r. t., diluted with DCM, washed with H2O, dried over anhydrous Na2SO4, and the solvent was evaporated to dryness, obtaining 1.00 g of 13 (66%), used for the subsequent step without further purification. 1H NMR: δ 1.97–2.04 (m, 2H, CH2-CH2-CH2), 2.59–2.65 (m, 2H, N-CH2-), 2.67–2.72 (m, 4H, piperazine), 3.18–3.22 (m, 4H, piperazine), 3.62–3.69 (m, 2H, -CH2Cl), 6.78–6.84 (m, 1H, Ar), 7.56 (d, J = 16.0 Hz, 1H, Ar), 7.87–7.93 (m, 1H, Ar), 8.03 (s, 1H, Ar); 8.69 (d, J = 4.0 Hz, 1H, Ar). MS (ES) m/z: 324.2 (M + H).
- 4-(4-(3-azidopropyl)piperazin-1-yl)-7-chloroquinoline (14). A mixture of 1.0 g (3.1 mmol) of 13 and 0.3 g (4.6 mmol) of NaN3 in 25 mL of DMSO was stirred at r.t. for 24 h, then quenched in ice and water, extracted with EtOAc, washed with H2O, dried over anhydrous Na2SO4 and the solvent was evaporated under vacuum to obtain 0.62 g (60%) of 14 used without purification. 1H NMR: δ 1.81–1.88 (m, 2H, CH2-CH2-CH2), 2.61 (t, J = 12.0 Hz, 2H, N-CH2), 2.69–2.75 (m, 4H, piperazine), 3.20–3.25 (m, 4H, piperazine), 3.53 (dt, J1 = 10.4 Hz, J2 = 12 Hz, 2H, CH2N3), 6.84 (d, J = 8.0 Hz, 1H, Ar), 7.42 (dd, J1 = 4.0 Hz, J2 = 8.0 Hz, 1H, Ar), 7.95 (d, J = 8.0 Hz, 1H, Ar), 8.05 (d, J = 4.0 Hz, 1H, Ar), 8.72 (d, J = 8.0 Hz, 1H, Ar). MS (ES) m/z: 331.2 (M + H).
- 2-(5-azidopentyl)isoindoline-1,3-dione (15). Applying the procedure used for 14 and starting from 0.5 g (1.7 mmol) of 2-(5-bromopentyl)isoindoline-1,3-dione and 0.16 g (2.5 mmol) of NaN3, 0.4 g of 15 was obtained (91%), as an oily compound. 1H NMR: δ 1.41–1.45 (m, 2H), 1.58–1.74 (m, 4H), 3.28 (t, J = 12.0 Hz, 2H, N-CH2), 3.70 (t, J = 12.0 Hz, 2H, CH2-N), 7.71–7.73 (m, 2H, Ar), 7.84–7.86 (m, 2H, Ar). MS (ES) m/z: 259.3 (M + H).
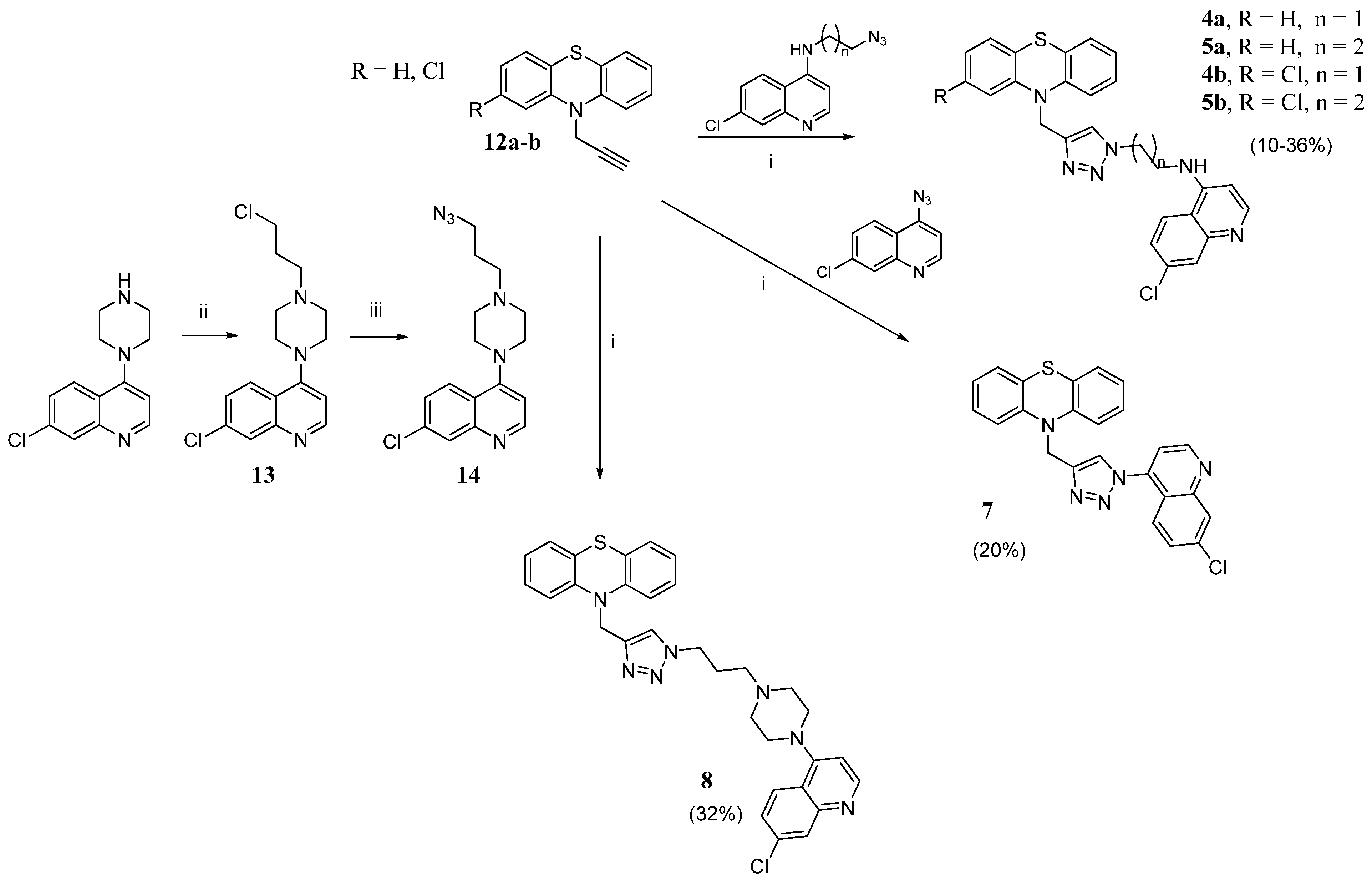
- General procedures for the click chemistry CuAAC reaction (4–5a,b, 7, 8, 16a,b). To a solution of the alkyne (1 eq) in DMSO, the selected azide (1.3 eq) and TEA (4.7 eq) were added. A solution of CuSO4 (0.1 eq) and sodium ascorbate (0.5 eq) in H2O was prepared and promptly added to the reaction mixture, which was stirred for 72 h at r.t. and then poured into ice. The mixture was extracted with EtOAc (3 × 40 mL), and the organic layer was washed with H2O, dried over Na2SO4, and evaporated to dryness. The obtained crude product was purified by flash chromatography or crystallization.
- N-(2-(4-((10H-phenothiazin-10-yl)methyl)-1H-1,2,3-triazol-1-yl)ethyl)-7-chloroquinolin-4-amine (4a). Using the previous procedure, 0.17 g (0.7 mmol) of N-(2-azidoethyl)-7-chloroquinolin-4-amine [28] was reacted with 0.13 g (0.53 mmol) of 12a. The crude material was purified by flash chromatography (DCM/MeOH 9.75:0.25), to obtain 0.07 g of 4a (yield 27%), m.p. 110–112 °C. 1H NMR: δ 3.78–3.81 (m, 2H, -CH2NH), 4.62 (t, J = 6.4 Hz, 2H, -CH2-N), 5.20 (s, 2H, -CH2-NPhen), 5.58 (broad, 1H, NH), 6.35 (s, 1H, Ar), 6.70 (d, J = 6.4 Hz, 2H, Ar), 6.86 (t, J = 6.4 Hz, 2H, Ar), 6.95 (t, J = 12.0 Hz, 2H, Ar), 7.10 (d, J = 12.0 Hz, 2H, Ar), 7.35 (s, 1H, Ar), 7.28 (dd, J1 = 12.0 Hz, J2 = 6.0 Hz, 1H, Ar), 7.56 (d, J = 6.0 Hz, 1H, Ar), 7.97 (s, 1H, triazole), 8.52 (d, J = 6.0 Hz, 1H, Ar). 13C NMR: δ 42.9, 44.9, 49.0, 99.0, 115.2, 117.3, 121.0, 123.1, 123.5, 124.1, 126.2, 127.4, 127.5, 129.0, 135.5, 144.2, 145.6, 149.0, 149.2, 152.0. MS (ES) m/z: 486.0 (M + H).
- 7-chloro-N-(2-(4-((2-chloro-10H-phenothiazin-10-yl)methyl)-1H-1,2,3-triazol-1-yl)ethyl)quinolin-4-amine (4b). Using the previous procedure, 0.21 g (0.85 mmol) of N-(2-azidoethyl)-7-chloroquinolin-4-amine [28] was reacted with 0.18 g (0.65 mmol) of 12b. The crude was purified by flash chromatography (EtOAc/EP 9:1), to obtain 0.03 g of 4b (yield 9%), m.p. 104–106 °C. 1H NMR (DMSO-d6): δ 3.72–3.75 (m, 2H, -CH2NH), 4.64 (t, J = 12.0 Hz, 2H, -CH2-N), 5.09 (s, 2H, -CH2-N-PTZ), 6.47 (d, J = 6.4 Hz, 1H, Ar), 6.84 (d, J = 12 Hz, 1H, Ar), 6.88–6.92 (m, 3H, Ar), 6.99 (t, J = 18.0 Hz, 1H, Ar), 7.06–7.09 (m, 2H, Ar), 7.37 (br, 1H, NH), 7.41 (dd, J1 = 12.0 Hz, J2 = 3.0 Hz, 1H, Ar), 7.78 (d, J = 3.0 Hz, 1H, Ar), 8.02 (s, 1H, triazole), 8.09 (d, J = 12.0 Hz, 1H, Ar), 8.36 (d, J = 6.0 Hz, 1H, Ar). 13C NMR: δ 42.4, 44.1, 48.0, 98.8, 115.5, 115.9, 117.4, 121.3, 122.0, 122.2, 123.0, 123.8, 123.9, 124.3, 126.7, 127.4, 127.5, 132.2, 133.0, 142.9, 143.3, 145.4, 149.7, 151.8. MS (ES) m/z: 519.2 (M + H).
- N-(3-(4-((10H-phenothiazin-10-yl)methyl)-1H-1,2,3-triazol-1-yl)propyl)-7-chloroquinolin-4-amine (5a). Using the previous procedure, 0.17 g (0.65 mmol) of N-(3-azidopropyl)-7-chloroquinolin-4-amine [28] was reacted with 0.12 g (0.5 mmol) of 12a. The crude was purified by flash chromatography (EtOAc/EP 9:1), to obtain 0.08 g of 5a (yield 32%), m.p. 98–100 °C. 1H NMR (Acetone-d): δ 2.31–2.36 (m, 2H, -CH2-), 3.36–3.38 (m, 2H, -CH2NH-), 4.57–4.60 (m, 2H, -CH2N); 5.17 (s, 2H, -CH2N-PTZ); 6.42 (d, J = 12.0 Hz, 1H, Ar); 6.63 (br, 1H, NH); 6.89–6.93 (m, 4H, Ar); 7.05–7.11 (m, 4H, Ar); 7.37 (d, J = 12.0 Hz, 1H, Ar), 7.80 (s, 1H, triazole), 7.82 (d, J = 6.0 Hz, 1H, Ar), 8.09 (d, J = 12.0 Hz, 1H, Ar), 8.41 (d, J = 6.0 Hz, 1H, Ar). 13C NMR (CDCl3): δ 28.4, 40.1, 45.0, 48.2, 99.0, 115.3, 117.3, 121.1, 123.1, 124.1, 125.8, 127.5, 129.1, 135.3, 144.3, 145.7, 149.2, 149.3, 152.1. MS (ES) m/s 500.0 (M + H).
- 7-chloro-N-(3-(4-((2-chloro-10H-phenothiazin-10-yl)methyl)-1H-1,2,3-triazol-1-yl)propyl)quinolin-4-amine (5b). Using the previous procedure, 0.22 g (0.85 mmol) of N-(3-azidopropyl)-7-chloroquinolin-4-amine [28] was reacted with 0.18 g (0.65 mmol) of 12b. The crude was purified by flash chromatography (EtOAc/EP 9:1), to obtain 0.13 g of 5b (yield 37%), m.p. 106–108 °C. 1H NMR: δ 2.26–2.28 (m, 2H, -CH2-), 3.30–3.33 (m, 2H, -CH2NH-), 4.46 (t, J = 6.0 Hz, 2H, -CH2N), 5.16 (s, 2H, -CH2N-PTZ), 5.31 (br, 1H, NH), 6.32 (d, J = 3.0 Hz, 1H, Ar), 6.69 (d, J = 6.0 Hz, 1H, Ar), 6.72 (s, 1H, Ar), 6.87–6,91 (m, 2H, Ar), 6.97 (t, J = 18.0 Hz, 1H, Ar), 7.01 (d, J = 12.0 Hz, 1H, Ar), 7.10 (d, J = 6.0 Hz, 1H, Ar), 7.34 (s, 1H, CHtri), 7.39 (d, J = 12.0 Hz, 1H, Ar), 7.65 (d, J = 12.0 Hz, 1H, Ar), 7.95 (s, 1H, Ar), 8.5 (d, J = 3.0 Hz, 1H, Ar). 13C NMR: δ 28.5, 39.9, 45.1, 48.1, 99.0, 115.6, 115.7, 117.3, 121.1, 122.6, 122.9, 123.5, 123.8, 124.1, 125.8, 127.4, 127.7, 127.9, 129.0, 133.4, 135.2, 143.6, 145.0, 145.6, 149.3, 152.0. MS (ES) m/z: 533.2 (M + H).
- 10-((1-(7-chloroquinolin-4-yl)-1H-1,2,3-triazol-4-yl)methyl)-10H-phenothiazine (7). Using the previous procedure, 0.5 g (2.10 mmol) of compound 12a was reacted with 0.43 g (2.74 mmol) of 4-azido-7-chloroquinoline [29]. The crude was purified by flash chromatography (EtOAc/EP 3:7), to obtain 0.2 g of 7 (yield 20%), m.p. 120–122 °C. 1H NMR: δ 5.46 (s, 2H, CH2N-PTZ), 6.90 (d, J = 18.0 Hz, 2H, Ar), 6.98 (t, J = 18.0 Hz, 2H, Ar), 7.12 (t, J = 18.0 Hz, 2H, Ar), 7.20 (d, J = 18.0 Hz, 2H, Ar), 7.42 (d, J = 6.0 Hz, 1H, Ar), 7.49 (d, J = 18.0 Hz, 1H, Ar), 7.64 (d, J = 12.0 Hz, 1H, Ar), 7.78 (s, 1H, CHtri), 8.20 (s, 1H, Ar), 9.00 (d, J = 6.0 Hz, 1H, Ar). 13C NMR: δ 44.9, 115.6, 116.2, 120.6, 123.3, 124.3, 124.5, 124.8, 127.6, 129.2, 129.6, 137.0, 140.9, 144.4, 146.0, 150.3, 151.5. MS (ES) m/z: 442.9 (M + H).
- 10-((1-(3-(4-(7-chloroquinolin-4-yl)piperazin-1-yl)propyl)-1H-1,2,3-triazol-4-yl)methyl)-10H-phenothiazine (8). Using the previous procedure, 0.62 g (1.90 mmol) of 14 was reacted with 0.34 g (1.45 mmol) of 12a. The crude material was purified by flash chromatography (DCM/MeOH 9.5:0.5), to obtain 0.26 g of 8 (yield 32%), m.p. 110–112 °C. 1H NMR: δ 2.06–2.11 (m, 2H), 2.26 (t, J = 6.6 Hz, 2H), 2.52–2.53 (m, 4H), 3.15 (s, 4H), 4.44 (t, J = 6.6 Hz, 2H), 5.26 (d, J = 0.8 Hz, 2H), 6.82–6.85 (m, 2+1H), 6.93 (t, J = 6.6 Hz, 2H), 7.09–7.12 (m, 2H), 7.14–7.16 (m, 2H), 7.42 (d, J = 0.8 Hz, 1H), 7.46 (dd, J = 8.9, 2.2 Hz, 1H), 7.93 (d, J = 9.0 Hz, 1H), 8.08 (d, J = 2.1 Hz, 1H), 8.76 (d, J = 5.0 Hz, 1H). 13C NMR: δ 27.0, 44.9, 47.9, 52.2, 52.8, 54.0, 109.1, 115.5, 122.0, 123.0, 123.4, 124.0, 125.2, 126.3, 127.4, 127.5, 129.0, 135.1, 144.5, 144.6, 150.3, 152.0, 156.9. MS (ES) m/z: 569.1 (M + H).
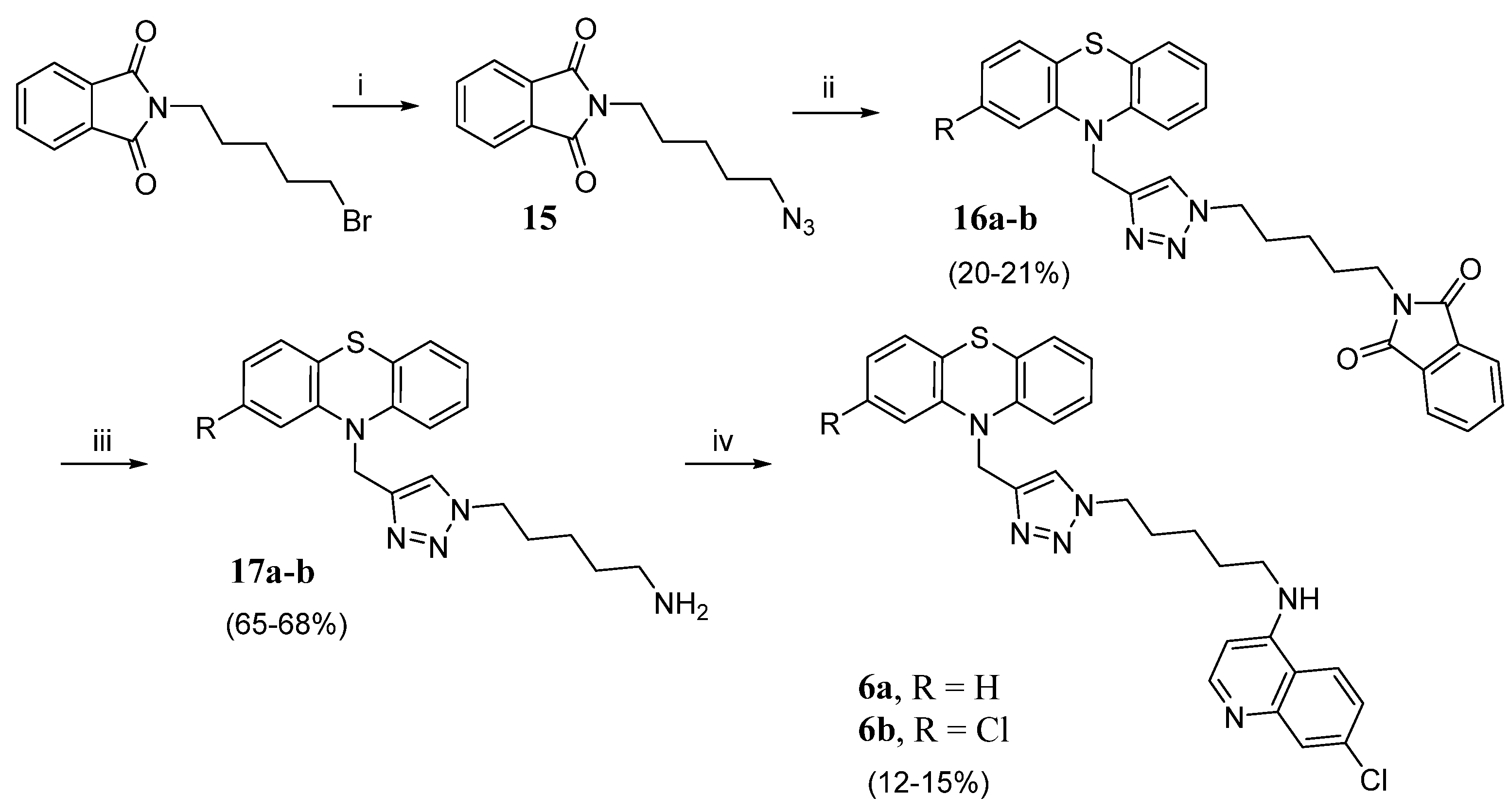
- 2-(5-(4-((10H-phenothiazin-10-yl)methyl)-1H-1,2,3-triazol-1-yl)pentyl)isoindoline-1,3-dione (16a). Using the previous procedure, 0.4 g (1.56 mmol) of 15 was reacted with 0.28 g (1.20 mmol) of 12a. The crude material was purified by flash chromatography (Toluene/EtOAc 9:1), to obtain 0.12 g of 16a (yield 20%), which was characterized by MS and used for the subsequent step without further purification. MS (ES) m/z: 496.3 (M + H).
- 2-(5-(4-((2-chloro-10H-phenothiazin-10-yl)methyl)-1H-1,2,3-triazol-1-yl)pentyl)isoindoline-1,3-dione (16b). Using the previous procedure, 0.4 g (1.56 mmol) of 15 was reacted with 0.33 g (1.20 mmol) of 12b in 25 mL of DMSO. The crude was purified by flash chromatography (EP/EtOAc 8:2), to obtain 0.13 g of 16b (yield 21%), which was characterized by MS and used for the subsequent step without further purification. MS (ES) m/z: 530.3 (M + H).
- 5-(4-((10H-phenothiazin-10-yl)methyl)-1H-1,2,3-triazol-1-yl)pentan-1-amine (17a). 0.12 g (0.24 mmol, 1 eq) of 16a was dissolved in 10 mL of ethanol, and 0.035 mL (0.72 mmol, 3 eq) of hydrazine hydrate was added. The mixture was heated under reflux for 3 h, cooled to r.t., and acidified with 6 drops of HCl 37%. The solution was stirred for 30 min., and the solvent was removed under reduced pressure. The residue was diluted with water, alkalinized with K2CO3, and the precipitate was collected by filtration. The crude material was purified by flash chromatography (Toluene/acetone 3:2) to obtain 60 mg of 17a (yield 68%). 1H NMR: δ 1.23–1.29 (m, 2H, -CH2-), 1.42–1.45 (m, 2H, -CH2-), 1.79–1.85 (m, 2H, -CH2-), 2.17–2.22 (m, 2H, -CH2-), 2.65 (br, 2H, -NH2), 4.26 (t, J = 8.0 Hz, 2H, N-CH2-), 5.20 (s, 2H, -CH2N-PTZ), 6.77 (d, J = 8.0 Hz, 2H, Ar), 6.90 (t, J = 8.0 Hz, 2H, Ar), 7.05 (t, J = 8.0 Hz, 2H, Ar), 7.12 (d, J = 8.0 Hz, 2H, Ar), 7.29 (s, 1H, CHtri). MS (ES) m/z: 366.3 (M + H).
- 5-(4-((2-chloro-10H-phenothiazin-10-yl)methyl)-1H-1,2,3-triazol-1-yl)pentan-1-amine (17b). Using the previous procedure, starting from 0.45 g (1.12 mmol) of 16b and 0.16 mL (3.36 mmol) of hydrazine hydrate, 0.3 g of 17b were obtained (yield 65%). 1H NMR: δ 1.21–1.27 (m, 2H, -CH2-), 1.43–1.49 (m, 2H, -CH2-), 1.82–1.89 (m, 4H, -CH2-CH2-), 2.68 (br, 2H, -NH2), 4.31 (t, J = 8.0 Hz, 2H, N-CH2-), 5.18 (s, 2H, -CH2N-PTZ), 6.75–6.80 (m, 2H, Ar), 6.87–6.94 (m, 2H, Ar), 6.99–7.27 (m, 3H, Ar), 7.31 (s, 1H, triazole). MS (ES) m/z: 400.3 (M + H).
- N-(5-(4-((10H-phenothiazin-10-yl)methyl)-1H-1,2,3-triazol-1-yl)pentyl)-7-chloroquinolin-4-amine (6a). 60 mg (0.164 mmol, 1 eq) of 17a were treated with 0.16 g (0.82 mmol, 5eq) of 4,7-dichloroquinoline, and the mixture was heated for 7 h at 120–130 °C under N2. The residue was diluted with water, and the precipitate formed was filtered and purified by flash chromatography (EtOAc/EP 4:1), to obtain 10 mg (yield 12%) of 6a, m.p. 104–106 °C. 1H NMR: δ 1.21–1.25 (m, 2H, -CH2-), 1.88–1.95 (m, 2H, -CH2-), 1.99–2.07 (m, 2H, -CH2-), 3.24–3.29 (m, 2H, -CH2-), 4.33 (t, J = 6.0 Hz, 2H, -NCH2-), 5.22 (s, 2H, -CH2N-PTZ), 6.35 (d, J = 4.0 Hz, 1H, Ar), 6.77 (d, J = 8.0 Hz, 2H, Ar), 6.89 (t, J = 8.0 Hz, 2H, Ar), 6.99–7.05 (m, 2H, Ar), 7.13 (d, J = 8.0 Hz, 2H, Ar), 7.32 (s, 1H, triazole), 7.38 (d, J = 4.0 Hz, 1H, Ar), 7.76 (d, J = 8.0 Hz, 1H, Ar); 7.97 (s, 1H, Ar), 8.49 (d, J = 4.0 Hz, 1H, Ar). 13C NMR: δ 24.1, 27.5, 29.6, 44.1, 45.9, 53.1, 99.3, 116.4, 116.7, 120.1, 121.8, 122.4, 122.8, 123.4, 123.6, 125.9, 126.6, 127.1, 127.9, 143.5, 144.2, 145.3. MS (ES) m/z: 527.2 (M + H).
- 7-chloro-N-(5-(4-((2-chloro-10H-phenothiazin-10-yl)methyl)-1H-1,2,3-triazol-1-yl)pentyl)quinolin-4-amine (6b). Using the previous procedure, starting from 0.3 g (0.75 mmol) of 17b and 0.74 g (3.76 mmol) of 4,7-dichloroquinoline, 65 mg (yield 15%) of 6b were obtained, m.p. 111–113 °C. 1H NMR: δ 1.33–1.38 (m, 2H, -CH2-), 1.71–1.74 (m, 2H, -CH2-), 1.92–1.94 (m, 2H, -CH2-), 3.24–3.27 (m, 2H, -CH2-), 4.35 (t, J = 6.0 Hz, 2H, -NCH2-), 4.90 (br, 1H, NH), 5.21 (s, 2H, -CH2N-PTZ), 6.35 (d, J = 6.0 Hz, 1H, Ar), 6.73 (s, 1H, Ar), 6.76 (d, J = 6.0 Hz, 1H, Ar), 6.88 (d, J = 12.0 Hz, 1H, Ar), 6.91 (t, J = 6.0 Hz, 1H, Ar), 7.00–7.04 (m, 2H, Ar), 7.10 (d, J = 6.0 Hz, 1H, Ar), 7.31 (s, 1H, triazole), 7.37 (d, J = 6.0 Hz, 1H, Ar), 7.68 (d, J = 6.0 Hz, 1H, Ar), 7.96 (s, 1H, Ar), 8.52 (d, J = 6.0 Hz, 1H, Ar). 13C NMR: δ 23.8, 28.05, 29.8, 43.1, 45.3, 50.1, 99.1, 115.6, 115.7, 121.1, 122.5, 122.6, 122.8, 123.5, 123.7, 125.7, 127.4, 127.7, 127.9, 133.4, 143.7, 144.5, 145.6. MS (ES) m/z: 561.5 (M + H).
4.2. Biological Evaluations
4.2.1. PTZ-Quinoline Compounds and Reference Drugs
4.2.2. Bacterial Strains and Growth Conditions
4.2.3. Candida Albicans and Growth Conditions
4.2.4. Parasite Growth and Drug Susceptibility Assay
4.2.5. Antimicrobial Activity
4.2.6. Cell Viability and Proliferation Assay
4.2.7. Haemolytic Activity Assay
4.3. Computational Protocols
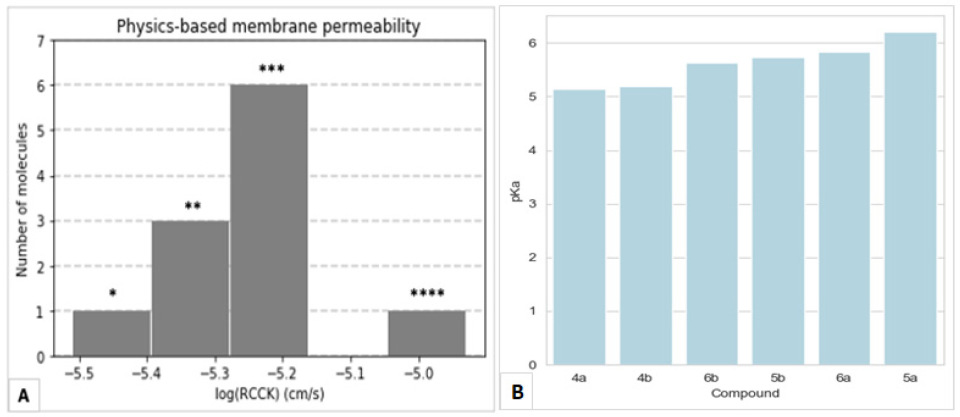
4.3.1. Ab Initio pKa Computation
4.3.2. Physics-Based Membrane Permeability Prediction
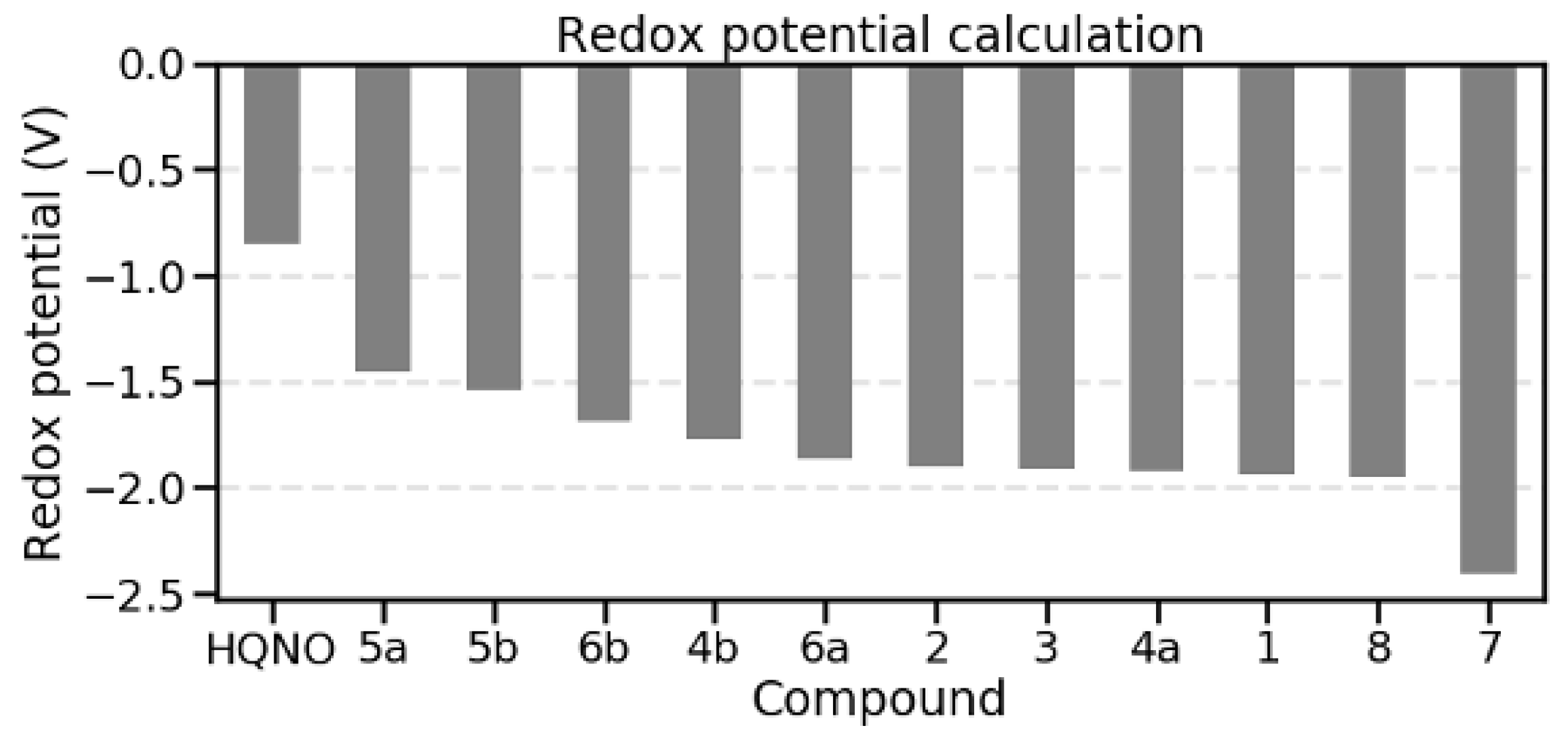
4.3.3. Redox Potential Computation
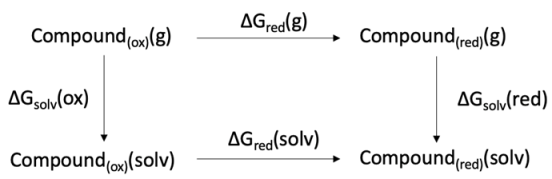
4.3.4. Molecular Modelling and Docking Simulations
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bergkessel, M.; Forte, B.; Gilbert, I.H. Small-Molecule Antibiotic Drug Development: Need and Challenges. ACS Infect. Dis. 2023, 9, 2062–2071. [Google Scholar] [CrossRef]
- Lu, L.; Åkerbladh, L.; Ahmad, S.; Konda, V.; Cao, S.; Vocat, A.; Maes, L.; Cole, S.T.; Hughes, D.; Larhed, M.; et al. Synthesis and In Vitro Biological Evaluation of Quinolinyl Pyrimidines Targeting Type II NADH-Dehydrogenase (NDH-2). ACS Infect. Dis. 2022, 8, 482–498. [Google Scholar] [CrossRef]
- Butler, M.S.; Vollmer, W.; Goodall, E.C.A.; Capon, R.J.; Henderson, I.R.; Blaskovich, M.A.T. A Review of Antibacterial Candidates with New Modes of Action. ACS Infect. Dis. 2024, 10, 3440–3474. [Google Scholar] [CrossRef]
- Wilairatana, P.; Mala, W.; Masangkay, F.R.; Kotepui, K.U.; Kotepui, M. The Prevalence of Malaria and Bacteremia Co-Infections among Febrile Patients: A Systematic Review and Meta-Analysis. Trop. Med. Infect. Dis. 2022, 7, 243. [Google Scholar] [CrossRef]
- Church, J.; Maitland, K. Invasive bacterial co-infection in African children with Plasmodium falciparum malaria: A systematic review. BMC Med. 2014, 12, 31. [Google Scholar] [CrossRef] [PubMed]
- Tibon, N.S.; Ng, C.H.; Cheong, S.L. Current progress in antimalarial pharmacotherapy and multi-target drug discovery. Eur. J. Med. Chem. 2020, 188, 111983. [Google Scholar] [CrossRef] [PubMed]
- Yadav, V.; Reang, J.; Sharma, V.; Majeed, J.; Sharma, P.C.; Sharma, K.; Giri, N.; Kumar, A.; Tonk, R.K. Quinoline-derivatives as privileged scaffolds for medicinal and pharmaceutical chemists: A comprehensive review. Chem. Biol. Drug Des. 2022, 100, 389–418. [Google Scholar] [CrossRef]
- Moor, L.F.E.; Vasconcelos, T.R.A.; Reis, R.d.R.; Pinto, L.S.S.; da Costa, T.M. Quinoline: An Attractive Scaffold in Drug Design. Mini-Rev. Med. Chem. 2021, 21, 2209–2226. [Google Scholar] [CrossRef]
- Kucharski, D.J.; Jaszczak, M.K.; Boratyński, P.J. A Review of Modifications of Quinoline Antimalarials: Mefloquine and (hydroxy)Chloroquine. Molecules 2022, 27, 1003. [Google Scholar] [CrossRef]
- Hu, Y.-Q.; Gao, C.; Zhang, S.; Xu, L.; Xu, Z.; Feng, L.-S.; Wu, X.; Zhao, F. Quinoline hybrids and their antiplasmodial and antimalarial activities. Eur. J. Med. Chem. 2017, 139, 22–47. [Google Scholar] [CrossRef] [PubMed]
- Teng, P.; Li, C.; Peng, Z.; Marie, V.A.; Nimmagadda, A.; Su, M.; Li, Y.; Sun, X.; Cai, J. Facilely accessible quinoline derivatives as potent antibacterial agents. Bioorg. Med. Chem. 2018, 26, 3573–3579. [Google Scholar] [CrossRef]
- Yao, H.; Cui, L.; Liu, H.; Li, X.; Shen, L.; Yang, R.; Qin, S.; Guo, Y. Quinoline-based anti-MRSA agents: Current development, structure-activity relationships, and mechanisms. Chin. Chem. Lett. 2023, 35, 108511. [Google Scholar] [CrossRef]
- Sabt, A.; Abdelraof, M.; Hamissa, M.F.; Noamaan, M.A. Antibacterial Activity of Quinoline-Based Derivatives against Methicillin-Resistant Staphylococcus aureus and Pseudomonas aeruginosa: Design, Synthesis, DFT and Molecular Dynamic Simulations. Chem. Biodivers. 2023, 20, e202300804. [Google Scholar] [CrossRef]
- Kumar, P. A review on quinoline derivatives as anti-methicillin resistant Staphylococcus aureus (MRSA) agents. BMC Chem. 2020, 14, 1–14. [Google Scholar] [CrossRef]
- Edinoff, A.N.; Armistead, G.; A Rosa, C.; Anderson, A.; Patil, R.; Cornett, E.M.; Murnane, K.S.; Kaye, A.M.; Kaye, A.D. Phenothiazines and their Evolving Roles in Clinical Practice: A Narrative Review. Health Psychol. Res. 2022, 10, 38930. [Google Scholar] [CrossRef] [PubMed]
- Montoya, M.C.; DiDone, L.; Heier, R.F.; Meyers, M.J.; Krysan, D.J. Antifungal Phenothiazines: Optimization, Characterization of Mechanism, and Modulation of Neuroreceptor Activity. ACS Infect. Dis. 2017, 4, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Grimsey, E.M.; Piddock, L.J.V. Do phenothiazines possess antimicrobial and efflux inhibitory properties? FEMS Microbiol. Rev. 2019, 43, 577–590. [Google Scholar] [CrossRef]
- González-González, A.; Vazquez-Jimenez, L.K.; Paz-González, A.D.; Bolognesi, M.L.; Rivera, G. Recent Advances in the Medicinal Chemistry of Phenothiazines, New Anticancer and Antiprotozoal Agents. Curr. Med. Chem. 2021, 28, 7910–7936. [Google Scholar] [CrossRef]
- de Sena Murteira Pinheiro, P.; Franco, L.S.; Montagnoli, T.L.; Fraga, C.A.M. Molecular hybridization: A powerful tool for multitarget drug discovery. Expert Opin. Drug Discov. 2024, 19, 451–470. [Google Scholar] [CrossRef]
- Posso, M.C.; Domingues, F.C.; Ferreira, S.; Silvestre, S. Development of Phenothiazine Hybrids with Potential Medicinal Interest: A Review. Molecules 2022, 27, 276. [Google Scholar] [CrossRef]
- Reddyrajula, R.; Dalimba, U.; Kumar, S.M. Molecular hybridization approach for phenothiazine incorporated 1,2,3-triazole hybrids as promising antimicrobial agents: Design, synthesis, molecular docking and in silico ADME studies. Eur. J. Med. Chem. 2019, 168, 263–282. [Google Scholar] [CrossRef]
- Nakatani, Y.; Shimaki, Y.; Dutta, D.; Muench, S.P.; Ireton, K.; Cook, G.M.; Jeuken, L.J.C. Unprecedented Properties of Phenothiazines Unraveled by a NDH-2 Bioelectrochemical Assay Platform. J. Am. Chem. Soc. 2019, 142, 1311–1320. [Google Scholar] [CrossRef]
- Heikal, A.; Nakatani, Y.; Dunn, E.; Weimar, M.R.; Day, C.L.; Baker, E.N.; Lott, J.S.; Sazanov, L.A.; Cook, G.M. Structure of the bacterial type II NADH dehydrogenase: A monotopic membrane protein with an essential role in energy generation. Mol. Microbiol. 2014, 91, 950–964. [Google Scholar] [CrossRef]
- Sena, F.V.; Batista, A.P.; Catarino, T.; Brito, J.A.; Archer, M.; Viertler, M.; Madl, T.; Cabrita, E.J.; Pereira, M.M. Type-II NADH: Quinone oxidoreductase from Staphylococcus aureus has two distinct binding sites and is rate limited by quinone reduction. Mol. Microbiol. 2015, 98, 272–288. [Google Scholar] [CrossRef]
- Goodman, C.D.; Buchanan, H.D.; McFadden, G.I. Is the Mitochondrion a Good Malaria Drug Target? Trends Parasitol. 2017, 33, 185–193. [Google Scholar] [CrossRef]
- Godoy-Hernandez, A.; Tate, D.J.; McMillan, D.G.G. Revealing the Membrane-Bound Catalytic Oxidation of NADH by the Drug Target Type-II NADH Dehydrogenase. Biochemistry 2019, 58, 4272–4275. [Google Scholar] [CrossRef] [PubMed]
- Petri, J.; Shimaki, Y.; Jiao, W.; Bridges, H.R.; Russell, E.R.; Parker, E.J.; Aragão, D.; Cook, G.M.; Nakatani, Y. Structure of the NDH-2—HQNO inhibited complex provides molecular insight into quinone-binding site inhibitors. Biochim. et Biophys. Acta (BBA) Bioenerg. 2018, 1859, 482–490. [Google Scholar] [CrossRef]
- Chu, X.-M.; Wang, C.; Wang, W.-L.; Liang, L.-L.; Liu, W.; Gong, K.-K.; Sun, K.-L. Triazole derivatives and their antiplasmodial and antimalarial activities. Eur. J. Med. Chem. 2019, 166, 206–223. [Google Scholar] [CrossRef]
- Basco, L.K.; Le Bras, J. In vitro activities of chloroquine in combination with chlorpromazine or prochlorperazine against isolates of Plasmodium falciparum. Antimicrob. Agents Chemother. 1992, 36, 209–213. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Youngsaye, W.; Vincent, B.; Hartland, C.L.; Morgan, B.J.; Buhrlage, S.J.; Johnston, S.; Bittker, J.A.; MacPherson, L.; Dandapani, S.; Palmer, M.; et al. Piperazinyl quinolines as chemosensitizers to increase fluconazole susceptibility of Candida albicans clinical isolates. Bioorg. Med. Chem. Lett. 2011, 21, 5502–5505. [Google Scholar] [CrossRef] [PubMed]
- Bisi, A.; Meli, M.; Gobbi, S.; Rampa, A.; Tolomeo, M.; Dusonchet, L. Multidrug resistance reverting activity and antitumor profile of new phenothiazine derivatives. Bioorg. Med. Chem. 2008, 16, 6474–6482. [Google Scholar] [CrossRef]
- Manohar, S.; Khan, S.I.; Rawat, D.S. Synthesis of 4-aminoquinoline-1,2,3-triazole and 4-aminoquinoline-1,2,3-triazole-1,3,5-triazine Hybrids as Potential Antimalarial Agents. Chem. Biol. Drug Des. 2011, 78, 124–136. [Google Scholar] [CrossRef]
- Rosado-Solano, D.N.; Barón-Rodríguez, M.A.; Florez, P.L.S.; Luna-Parada, L.K.; Puerto-Galvis, C.E.; Zorro-González, A.F.; Kouznetsov, V.V.; Vargas-Méndez, L.Y. Synthesis, Biological Evaluation and In Silico Computational Studies of 7-Chloro-4-(1H-1,2,3-triazol-1-yl)quinoline Derivatives: Search for New Controlling Agents against Spodoptera frugiperda (Lepidoptera: Noctuidae) Larvae. J. Agric. Food Chem. 2019, 67, 9210–9219. [Google Scholar] [CrossRef]
- Kushwaha, K.; Kaushik, N.; Lata; Jain, S.C. Design and synthesis of novel 2H-chromen-2-one derivatives bearing 1,2,3-triazole moiety as lead antimicrobials. Bioorg. Med. Chem. Lett. 2014, 24, 1795–1801. [Google Scholar] [CrossRef] [PubMed]
- Sæbø, I.P.; Bjørås, M.; Franzyk, H.; Helgesen, E.; Booth, J.A. Optimization of the Hemolysis Assay for the Assessment of Cytotoxicity. Int. J. Mol. Sci. 2023, 24, 2914. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Schurig-Briccio, L.A.; Yano, T.; Rubin, H.; Gennis, R.B. Characterization of the type 2 NADH:menaquinone oxidoreductases from Staphylococcus aureus and the bactericidal action of phenothiazines. Biochim. Biophys. Acta (BBA) Bioenerg. 2014, 1837, 954–963. [Google Scholar] [CrossRef]
- Speelman, A.L.; Gillmore, J.G. Efficient Computational Methods for Accurately Predicting Reduction Potentials of Organic Molecules. J. Phys. Chem. A 2008, 112, 5684–5690. [Google Scholar] [CrossRef]
- Lynch, E.J.; Speelman, A.L.; Curry, B.A.; Murillo, C.S.; Gillmore, J.G. Expanding and Testing a Computational Method for Predicting the Ground State Reduction Potentials of Organic Molecules on the Basis of Empirical Correlation to Experiment. J. Org. Chem. 2012, 77, 6423–6430. [Google Scholar] [CrossRef] [PubMed]
- Sousa, F.M.; Sena, F.V.; Batista, A.P.; Athayde, D.; Brito, J.A.; Archer, M.; Oliveira, A.S.F.; Soares, C.M.; Catarino, T.; Pereira, M.M. The key role of glutamate 172 in the mechanism of type II NADH: Quinone oxidoreductase of Staphylococcus aureus. Biochim. Biophys. Acta (BBA) Bioenerg. 2017, 1858, 823–832. [Google Scholar] [CrossRef]
- Yang, Y.; Yu, Y.; Li, X.; Li, J.; Wu, Y.; Yu, J.; Ge, J.; Huang, Z.; Jiang, L.; Rao, Y.; et al. Target Elucidation by Cocrystal Structures of NADH-Ubiquinone Oxidoreductase of Plasmodium falciparum (PfNDH2) with Small Molecule To Eliminate Drug-Resistant Malaria. J. Med. Chem. 2017, 60, 1994–2005. [Google Scholar] [CrossRef]
- Vallières, C.; Fisher, N.; Antoine, T.; Al-Helal, M.; Stocks, P.; Berry, N.G.; Lawrenson, A.S.; Ward, S.A.; O’NEill, P.M.; Biagini, G.A.; et al. HDQ, a Potent Inhibitor of Plasmodium falciparum Proliferation, Binds to the Quinone Reduction Site of the Cytochrome bc 1 Complex. Antimicrob. Agents Chemother. 2012, 56, 3739–3747. [Google Scholar] [CrossRef]
- Trager, W.; Jensen, J.B. Human Malaria Parasites in Continuous Culture. Science 1976, 193, 673–675. [Google Scholar] [CrossRef] [PubMed]
- Makler, M.T.; Ries, J.M.; Williams, J.A.; Bancroft, J.E.; Piper, R.C.; Gibbins, B.L.; Hinrichs, D.J. Parasite Lactate Dehydrogenase as an Assay for Plasmodium falciparum Drug Sensitivity. Am. J. Trop. Med. Hyg. 1993, 48, 739–741. [Google Scholar] [CrossRef]
- Yu, H.S.; Watson, M.A.; Bochevarov, A.D. Weighted Averaging Scheme and Local Atomic Descriptor for pKa Prediction Based on Density Functional Theory. J. Chem. Inf. Model. 2018, 58, 271–286. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Frisch, Gaussian 16, Revision C.01; Gaussian Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Barone, V.; Cossi, M. Quantum Calculation of Molecular Energies and Energy Gradients in Solution by a Conductor Solvent Model. J. Phys. Chem. A 1998, 102, 1995–2001. [Google Scholar] [CrossRef]
- Cossi, M.; Rega, N.; Scalmani, G.; Barone, V. Energies, structures, and electronic properties of molecules in solution with the C-PCM solvation model. J. Comput. Chem. 2003, 24, 669–681. [Google Scholar] [CrossRef] [PubMed]
- Sastry, G.M.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Johnston, R.C.; Yao, K.; Kaplan, Z.; Chelliah, M.; Leswing, K.; Seekins, S.; Watts, S.; Calkins, D.; Elk, J.C.; Jerome, S.V.; et al. Epik: pKa and Protonation State Prediction through Machine Learning. J. Chem. Theory Comput. 2023, 19, 2380–2388. [Google Scholar] [CrossRef]
- Lu, C.; Wu, C.; Ghoreishi, D.; Chen, W.; Wang, L.; Damm, W.; Ross, G.A.; Dahlgren, M.K.; Russell, E.; Von Bargen, C.D.; et al. OPLS4: Improving Force Field Accuracy on Challenging Regimes of Chemical Space. J. Chem. Theory Comput. 2021, 17, 4291–4300. [Google Scholar] [CrossRef]
- Halgren, T.A.; Murphy, R.B.; Friesner, R.A.; Beard, H.S.; Frye, L.L.; Pollard, W.T.; Banks, J.L. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 2. Enrichment Factors in Database Screening. J. Med. Chem. 2004, 47, 1750–1759. [Google Scholar] [CrossRef] [PubMed]
- Sherman, W.; Day, T.; Jacobson, M.P.; Friesner, R.A.; Farid, R. Novel Procedure for Modeling Ligand/Receptor Induced Fit Effects. J. Med. Chem. 2005, 49, 534–553. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]

| Compound | D10 (CQ-S) | W2 (CQ-R) | ATCC 25923 |
|---|---|---|---|
| 1 | 2.52 ± 0.84 | 2.32 ± 1.05 | n.d. 1 |
| 2 | n.d. | n.d. | n.d. |
| 3 | 2.34 ± 0.012 | 0.990 ± 0.245 | n.d. |
| 4a | 0.142 ± 0.02 | 0.200 ± 0.03 | 7.49 ± 0.15 |
| 4b | 0.156 ± 0.018 | 0.154 ± 0.002 | 4.86 ± 0.89 |
| 5a | 0.292 ± 0.078 | 0.174 ± 0.025 | 6.73 ± 1.32 |
| 5b | 0.146 ± 0.006 | 0.135 ± 0.017 | 5.03 ± 1.72 |
| 6a | 0.326 ± 0.12 | 0.377 ± 0.066 | n.d. |
| 6b | 0.602 ± 0.314 | 0.184 ± 0.093 | 25.05 ± 1.76 |
| 7 | n.d. | n.d. | n.d. |
| 8 | 12.60 ± 0.241 | 2.17 ± 0.243 | n.d. |
| CQ 2 | 0.025 ± 0.007 | 0.42 ± 0.103 | n.t. 3 |
| GEN 2 | n.t. | n.t. | 4.00 ± 0.05 |
| AMP 2 | n.t. | n.t. | 2.59 ± 0.13 |
| Compound | CC50 | SI, Vero/S. aureus | SI, Vero/D10 | SI, Vero/W2 |
|---|---|---|---|---|
| 1 | >100 | n.d. 1 | >39.7 | >43.1 |
| 2 | >100 | n.d. | n.d. | n.d. |
| 3 | >100 | n.d. | >42.7 | >101.0 |
| 4a | 17.91 ± 3.23 | 2.4 | 126.1 | 89.6 |
| 4b | 12.65 ± 3.51 | 2.6 | 81.1 | 82.1 |
| 5a | 11.78 ± 0.14 | 1.8 | 40.3 | 67.7 |
| 5b | 22.13 ± 0.66 | 4.4 | 151.6 | 163.9 |
| 6a | 48.27 ± 0.90 | n.d. | 148.1 | 128.6 |
| 6b | 34.34 ± 0.52 | 1.4 | 57.0 | 186.6 |
| 7 | >100 | n.d. | n.d. | n.d. |
| 8 | >100 | n.d. | >7.9 | >46.1 |
| CisPt 2 | 66.12 ± 8.65 | n.d. | n.d. | n.d. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gianibbi, B.; Corina, R.; Basilico, N.; Spiga, O.; Gobbi, S.; Belluti, F.; Gentilomi, G.A.; Parapini, S.; Bonvicini, F.; Bisi, A. STOP Strategy to Inhibit P. falciparum and S. aureus Growth: Molecular Mechanism Studies on Purposely Designed Hybrids. Antibiotics 2025, 14, 991. https://doi.org/10.3390/antibiotics14100991
Gianibbi B, Corina R, Basilico N, Spiga O, Gobbi S, Belluti F, Gentilomi GA, Parapini S, Bonvicini F, Bisi A. STOP Strategy to Inhibit P. falciparum and S. aureus Growth: Molecular Mechanism Studies on Purposely Designed Hybrids. Antibiotics. 2025; 14(10):991. https://doi.org/10.3390/antibiotics14100991
Chicago/Turabian StyleGianibbi, Beatrice, Riccardo Corina, Nicoletta Basilico, Ottavia Spiga, Silvia Gobbi, Federica Belluti, Giovanna Angela Gentilomi, Silvia Parapini, Francesca Bonvicini, and Alessandra Bisi. 2025. "STOP Strategy to Inhibit P. falciparum and S. aureus Growth: Molecular Mechanism Studies on Purposely Designed Hybrids" Antibiotics 14, no. 10: 991. https://doi.org/10.3390/antibiotics14100991
APA StyleGianibbi, B., Corina, R., Basilico, N., Spiga, O., Gobbi, S., Belluti, F., Gentilomi, G. A., Parapini, S., Bonvicini, F., & Bisi, A. (2025). STOP Strategy to Inhibit P. falciparum and S. aureus Growth: Molecular Mechanism Studies on Purposely Designed Hybrids. Antibiotics, 14(10), 991. https://doi.org/10.3390/antibiotics14100991








