Antimicrobial and Antibiofilm Activities of Some Antioxidant 3,4-Dihydroxyphenyl-Thiazole-Coumarin Hybrid Compounds: In Silico and In Vitro Evaluation
Abstract
1. Introduction
2. Results
2.1. Antimicrobial Activity
2.2. Antibiofilm Activity
2.3. Molecular Docking
2.3.1. Target Selection
2.3.2. Superposition and Morphing
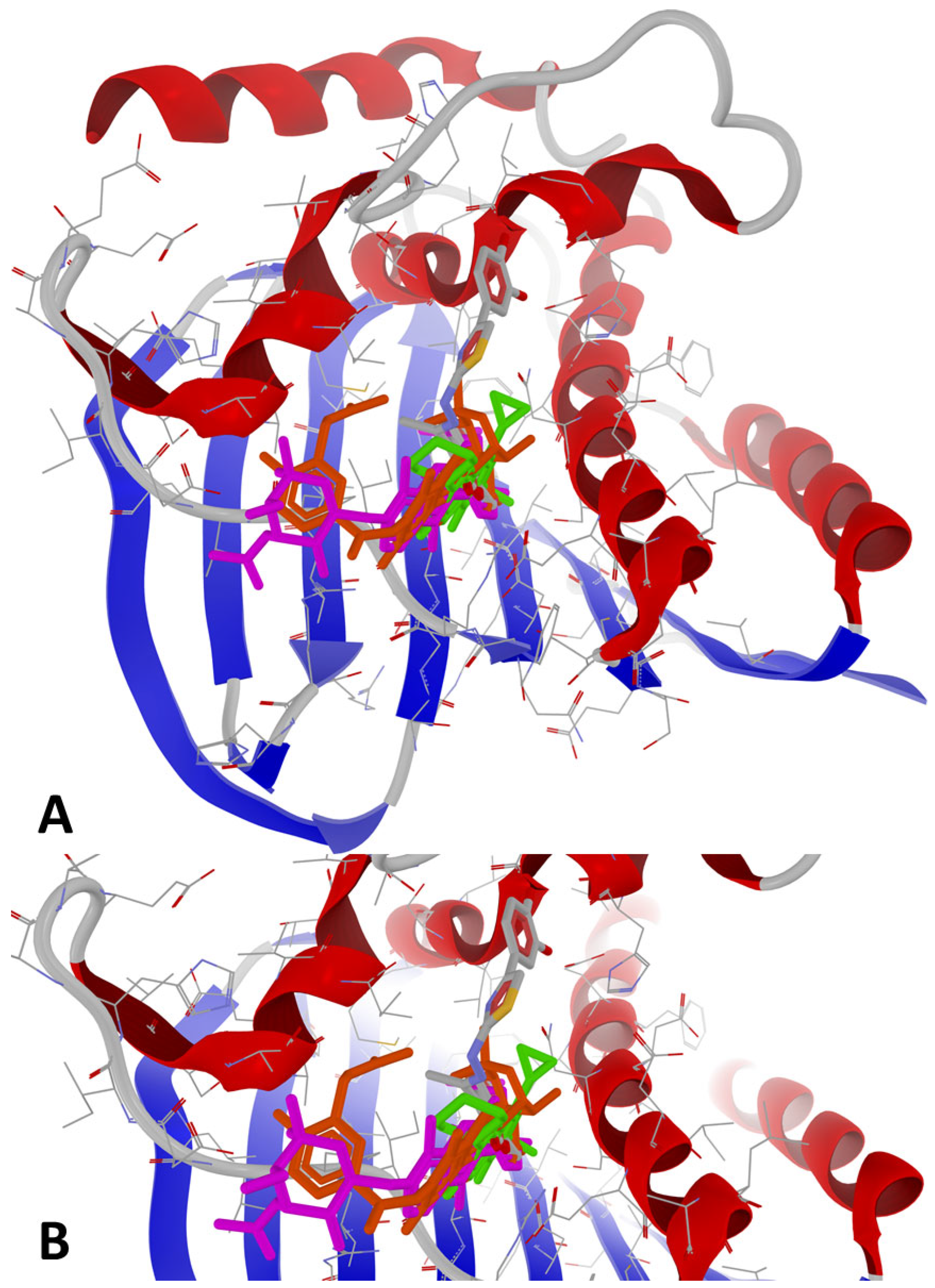
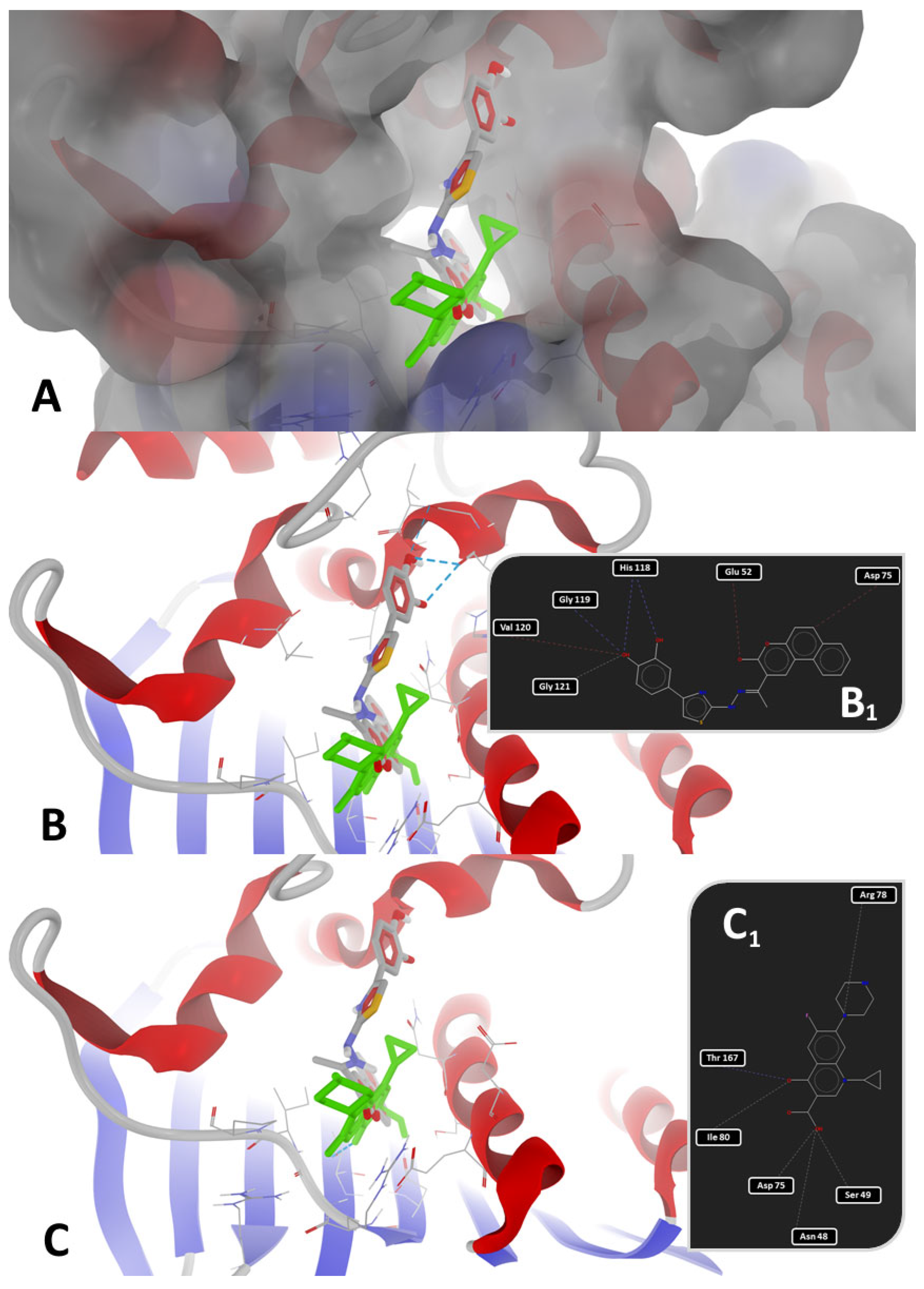
2.3.3. Protein-Ligand Docking
2.4. Molecular Dynamics Simulation
2.5. ADMETox Prediction
3. Discussion
3.1. Molecular Docking
3.1.1. Target Selection
3.1.2. Protein-Ligand Docking
3.2. Molecular Dynamics Simulation
3.3. Antimicrobial Activity
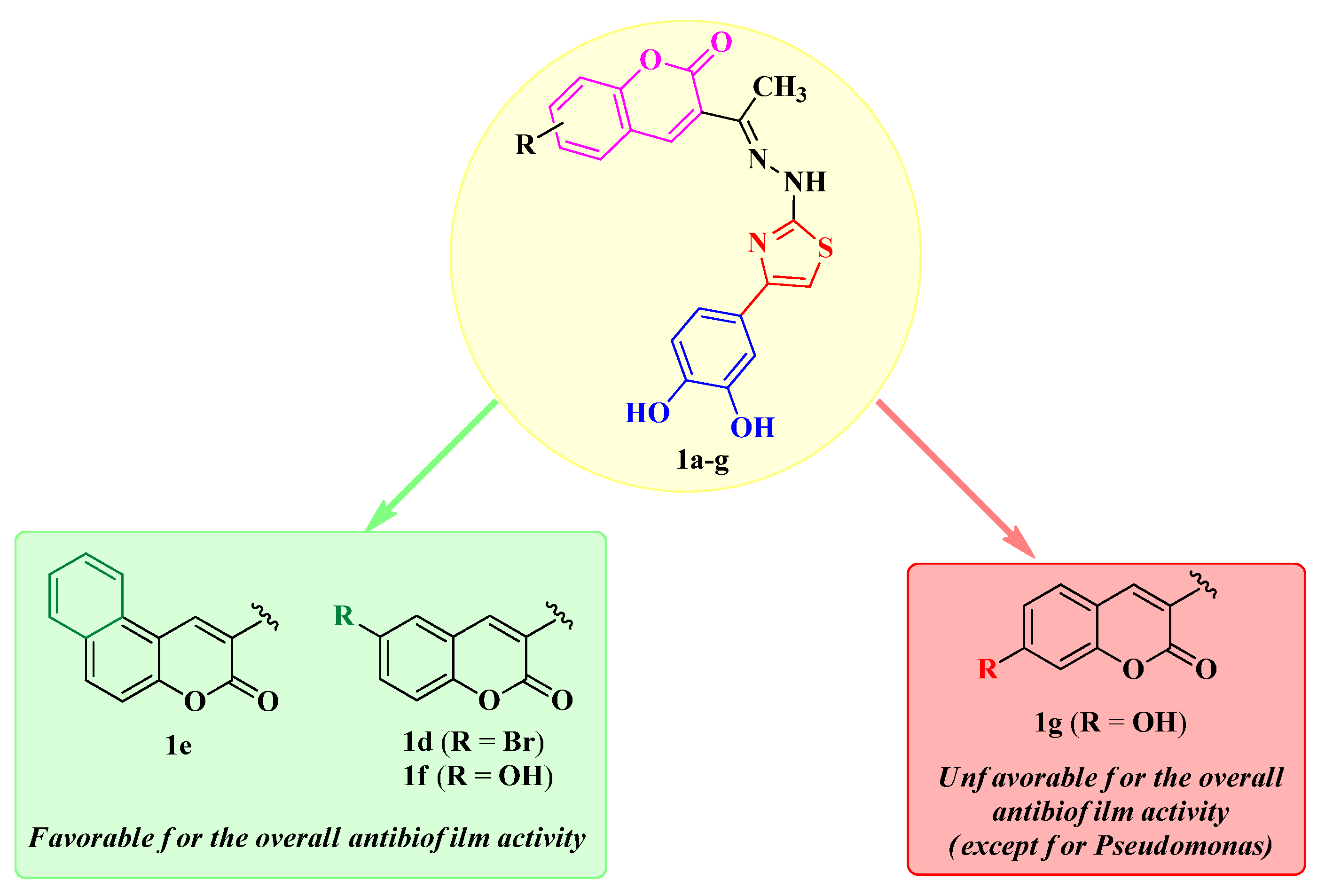
3.4. Antibiofilm Activity
3.5. ADMETox Prediction
4. Materials and Methods
4.1. Antimicrobial Activity
4.2. Antibiofilm Activity
4.3. Molecular Docking
4.3.1. Target Selection
4.3.2. Superposition and Morphing
4.3.3. Protein-Ligand Docking
4.4. Molecular Dynamics Simulation
4.5. ADMETox Prediction
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ROS | Reactive oxygen species |
| NO | Nitric oxide |
| NOS | Nitric oxide synthase |
| DNA | Deoxyribonucleic acid |
| BF | Biofilm |
| QS | Quorum sensing |
| MIC | Minimal inhibitory concentration |
| MBC | Minimal bactericidal concentration |
| MFC | Minimal fungicidal concentration |
| ATCC | American Type Culture Collection |
| n.d. | Not identified |
| GI | Gastrointestinal |
| BBB | Blood-brain barrier |
| P-gp | P-glycoprotein |
| CYP450 | Cytochrome P450 |
| UniProtKB | The UniProt Knowledgebase |
| RCSB PDB | The Research Collaboratory for Structural Bioinformatics Protein Data Bank |
| GyrB | DNA gyrase subunit B |
| CYP51 | Lanosterol 14α-demethylase cytochrome P450 |
| AA | Amino acids |
| P-LD | Protein-ligand docking |
| CID | PubChem Compound Identifier |
| CJC | PubChem CID 70699420 |
| N1N | PubChem CID 151595514 |
| CPF | Ciprofloxacin |
| NOV | Novobiocin |
| 1YN | Itraconazole |
| TPF | Fluconazole |
| VT1 | Oteseconazole |
| X2N | Posaconazole |
| T-LC | Target-ligand complex |
| BA | Binding affinity |
| RD | Re-docking of co-crystallized ligand |
| RMSD | Root mean square deviation |
| 4GGL | E. faecalis V583 |
| 6TCK | S. aureus |
| 7PTF | P. aeruginosa PAO1 |
| 7P2M | E. coli K-12 |
| 5VSZ | C. albicans SC5314 |
| 5FSA | C. albicans |
| 5TZI | C. albicans |
| 4WMZ | S. cerevisiae YJM784 |
| DMSO | Dimethylsulfoxide |
| M+ | Inoculum control |
| ND | Not determined |
| MLogP | Octanol-water partition coefficient implemented by Moriguchi |
| ESOL | Estimated solubility |
| VS | Virtual screening |
| CPK | Corey-Pauling-Koltun |
| SAR | Structure-activity relationship |
| IC50 | Half-maximal inhibitory concentration |
| DPPH• | 2,2-diphenyl-1-picrylhydrazyl |
| ABTS•+ | 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) |
| TAC | Total Antioxidant Capacity |
| RP | Reducing Power |
| FRAP | Ferric Reducing Antioxidant Potential |
| CUPRAC | Cupric Reducing Antioxidant Capacity |
| Eq | Equivalent molar activity |
| ID | Identifier |
| NB | Nutrient broth |
| MHB | Muller–Hinton broth |
| BHI | Brain heart infusion |
| OD | Optical density |
| GA | Genetic algorithm |
| BFGS | Broyden-Fletcher-Goldfarb-Shanno |
| CFU | Colony-forming unit |
Appendix A
Superposition and Morphing Analyses of the GyrB and CYP51 Crystal Structures


References
- Ungureanu, D.; Oniga, O.; Moldovan, C.; Ionuț, I.; Marc, G.; Stana, A.; Pele, R.; Duma, M.; Tiperciuc, B. An Insight into Rational Drug Design: The Development of In-House Azole Compounds with Antimicrobial Activity. Antibiotics 2024, 13, 763. [Google Scholar] [CrossRef]
- Ungureanu, D.; Tiperciuc, B.; Nastasă, C.; Ionuț, I.; Marc, G.; Oniga, I.; Oniga, O. An Overview of the Structure–Activity Relationship in Novel Antimicrobial Thiazoles Clubbed with Various Heterocycles (2017–2023). Pharmaceutics 2024, 16, 89. [Google Scholar] [CrossRef]
- McCollister, B.D.; Hoffman, M.; Husain, M.; Vázquez-Torres, A. Nitric Oxide Protects Bacteria from Aminoglycosides by Blocking the Energy-Dependent Phases of Drug Uptake. Antimicrob. Agents Chemother. 2011, 55, 2189–2196. [Google Scholar] [CrossRef]
- Delattin, N.; Cammue, B.P.; Thevissen, K. Reactive Oxygen Species-Inducing Antifungal Agents and Their Activity Against Fungal Biofilms. Future Med. Chem. 2014, 6, 77–90. [Google Scholar] [CrossRef]
- Vaishampayan, A.; Grohmann, E. Antimicrobials Functioning through ROS-Mediated Mechanisms: Current Insights. Microorganisms 2022, 10, 61. [Google Scholar] [CrossRef]
- Delghandi, P.S.; Soleimani, V.; Fazly Bazzaz, B.S.; Hosseinzadeh, H. A Review on Oxidant and Antioxidant Effects of Antibacterial Agents: Impacts on Bacterial Cell Death and Division and Therapeutic Effects or Adverse Reactions in Humans. Naunyn Schmiedeberg’s Arch. Pharmacol. 2023, 396, 2667–2686. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, J.; Andleeb, A.; Ashraf, H.; Meer, B.; Mehmood, A.; Jan, H.; Zaman, G.; Nadeem, M.; Drouet, S.; Fazal, H.; et al. Potential Antimicrobial, Antidiabetic, Catalytic, Antioxidant and ROS/RNS Inhibitory Activities of Silybum Marianum Mediated Biosynthesized Copper Oxide Nanoparticles. RSC Adv. 2022, 12, 14069–14083. [Google Scholar] [CrossRef] [PubMed]
- Tamaian, R.; Moţ, A.; Silaghi-Dumitrescu, R.; Ionuţ, I.; Stana, A.; Oniga, O.; Nastasă, C.; Benedec, D.; Tiperciuc, B. Study of the Relationships between the Structure, Lipophilicity and Biological Activity of Some Thiazolyl-Carbonyl-Thiosemicarbazides and Thiazolyl-Azoles. Molecules 2015, 20, 22188–22201. [Google Scholar] [CrossRef]
- Lemilemu, F.; Bitew, M.; Demissie, T.B.; Eswaramoorthy, R.; Endale, M. Synthesis, Antibacterial and Antioxidant Activities of Thiazole-Based Schiff Base Derivatives: A Combined Experimental and Computational Study. BMC Chem. 2021, 15, 67. [Google Scholar] [CrossRef]
- Al-Majedy, Y.; Al-Duhaidahawi, D.; Al-Azawi, K.; Al-Amiery, A.; Kadhum, A.; Mohamad, A. Coumarins as Potential Antioxidant Agents Complemented with Suggested Mechanisms and Approved by Molecular Modeling Studies. Molecules 2016, 21, 135. [Google Scholar] [CrossRef] [PubMed]
- Al-Majedy, Y.; Al-Amiery, A.; Kadhum, A.A.; BakarMohamad, A. Antioxidant Activity of Coumarins. Syst. Rev. Pharm. 2016, 8, 24–30. [Google Scholar] [CrossRef]
- Login, C.C.; Bâldea, I.; Tiperciuc, B.; Benedec, D.; Vodnar, D.C.; Decea, N.; Suciu, Ş. A Novel Thiazolyl Schiff Base: Antibacterial and Antifungal Effects and In Vitro Oxidative Stress Modulation on Human Endothelial Cells. Oxidative Med. Cell. Longev. 2019, 2019, 1607903. [Google Scholar] [CrossRef]
- Dufour, D.; Leung, V.; Lévesque, C.M. Bacterial Biofilm: Structure, Function, and Antimicrobial Resistance. Endod. Top. 2010, 22, 2–16. [Google Scholar] [CrossRef]
- Todd, O.A.; Peters, B.M. Candida albicans and Staphylococcus aureus Pathogenicity and Polymicrobial Interactions: Lessons beyond Koch’s Postulates. J. Fungi 2019, 5, 81. [Google Scholar] [CrossRef]
- Asma, S.T.; Imre, K.; Morar, A.; Herman, V.; Acaroz, U.; Mukhtar, H.; Arslan-Acaroz, D.; Shah, S.R.A.; Gerlach, R. An Overview of Biofilm Formation–Combating Strategies and Mechanisms of Action of Antibiofilm Agents. Life 2022, 12, 1110. [Google Scholar] [CrossRef]
- Saverina, E.A.; Frolov, N.A.; Kamanina, O.A.; Arlyapov, V.A.; Vereshchagin, A.N.; Ananikov, V.P. From Antibacterial to Antibiofilm Targeting: An Emerging Paradigm Shift in the Development of Quaternary Ammonium Compounds (QACs). ACS Infect. Dis. 2023, 9, 394–422. [Google Scholar] [CrossRef] [PubMed]
- Melander, R.J.; Basak, A.K.; Melander, C. Natural Products as Inspiration for the Development of Bacterial Antibiofilm Agents. Nat. Prod. Rep. 2020, 37, 1454–1477. [Google Scholar] [CrossRef]
- Li, S.; Chen, S.; Fan, J.; Cao, Z.; Ouyang, W.; Tong, N.; Hu, X.; Hu, J.; Li, P.; Feng, Z.; et al. Anti-Biofilm Effect of Novel Thiazole Acid Analogs against Pseudomonas aeruginosa through IQS Pathways. Eur. J. Med. Chem. 2018, 145, 64–73. [Google Scholar] [CrossRef]
- More, P.G.; Karale, N.N.; Lawand, A.S.; Narang, N.; Patil, R.H. Synthesis and Anti-Biofilm Activity of Thiazole Schiff Bases. Med. Chem. Res. 2014, 23, 790–799. [Google Scholar] [CrossRef]
- Reen, F.J.; Gutiérrez-Barranquero, J.A.; Parages, M.L.; O’Gara, F. Coumarin: A Novel Player in Microbial Quorum Sensing and Biofilm Formation Inhibition. Appl. Microbiol. Biotechnol. 2018, 102, 2063–2073. [Google Scholar] [CrossRef]
- Razaviamri, S.; Wang, K.; Liu, B.; Lee, B.P. Catechol-Based Antimicrobial Polymers. Molecules 2021, 26, 559. [Google Scholar] [CrossRef] [PubMed]
- Ungureanu, D.; Marc, G.; Tiperciuc, B.; Moldovan, C.; Ionuț, I.; Stana, A.; Oniga, I.; Vlase, L.; Pîrnău, A.; Oniga, O. Novel 3,4-Dihydroxyphenyl-Thiazole-Coumarin Hybrid Compounds: Synthesis, In Silico and In Vitro Evaluation of Their Antioxidant Activity. Antioxidants 2025, 14, 636. [Google Scholar] [CrossRef] [PubMed]
- Gurram, S.R.; Azam, M.A. GyrB Inhibitors as Potential Antibacterial Agents: A Review. Monatshefte Chem. Chem. Mon. 2021, 152, 725–744. [Google Scholar] [CrossRef]
- Chauhan, S.S.; Thaseen, E.A.; Parthasarathi, R. Computational Discovery of Potent Escherichia Coli DNA Gyrase Inhibitor: Selective and Safer Novobiocin Analogues. Comput. Toxicol. 2024, 29, 100302. [Google Scholar] [CrossRef]
- Tokarzewski, S.; Ziółkowska, G.; Nowakiewicz, A. Susceptibility Testing of Aspergillus Niger Strains Isolated from Poultry to Antifungal Drugs—A Comparative Study of the Disk Diffusion, Broth Microdilution (M 38-A) and Etest® Methods. Pol. J. Vet. Sci. 2012, 15, 125–133. [Google Scholar] [CrossRef]
- Gellert, M.; O’Dea, M.H.; Itoh, T.; Tomizawa, J. Novobiocin and Coumermycin Inhibit DNA Supercoiling Catalyzed by DNA Gyrase. Proc. Natl. Acad. Sci. USA 1976, 73, 4474–4478. [Google Scholar] [CrossRef]
- Nastasă, C.; Vodnar, D.; Ionuţ, I.; Stana, A.; Benedec, D.; Tamaian, R.; Oniga, O.; Tiperciuc, B. Antibacterial Evaluation and Virtual Screening of New Thiazolyl-Triazole Schiff Bases as Potential DNA-Gyrase Inhibitors. Int. J. Mol. Sci. 2018, 19, 222. [Google Scholar] [CrossRef]
- Stana, A.; Vodnar, D.C.; Tamaian, R.; Pîrnău, A.; Vlase, L.; Ionuț, I.; Oniga, O.; Tiperciuc, B. Design, Synthesis and Antifungal Activity Evaluation of New Thiazolin-4-Ones as Potential Lanosterol 14α-Demethylase Inhibitors. Int. J. Mol. Sci. 2017, 18, 177. [Google Scholar] [CrossRef]
- Abo-Ashour, M.F.; Eldehna, W.M.; George, R.F.; Abdel-Aziz, M.M.; Elaasser, M.M.; Abou-Seri, S.M.; Abdel Gawad, N.M. Synthesis and Biological Evaluation of 2-Aminothiazole-Thiazolidinone Conjugates as Potential Antitubercular Agents. Future Med. Chem. 2018, 10, 1405–1419. [Google Scholar] [CrossRef]
- Nyerges, A.; Tomašič, T.; Durcik, M.; Revesz, T.; Szili, P.; Draskovits, G.; Bogar, F.; Skok, Ž.; Zidar, N.; Ilaš, J.; et al. Rational Design of Balanced Dual-Targeting Antibiotics with Limited Resistance. PLoS Biol. 2020, 18, e3000819. [Google Scholar] [CrossRef]
- Cotman, A.E.; Durcik, M.; Benedetto Tiz, D.; Fulgheri, F.; Secci, D.; Sterle, M.; Možina, Š.; Skok, Ž.; Zidar, N.; Zega, A.; et al. Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter Baumannii and Pseudomonas aeruginosa. J. Med. Chem. 2023, 66, 1380–1425. [Google Scholar] [CrossRef]
- Keniya, M.V.; Sabherwal, M.; Wilson, R.K.; Woods, M.A.; Sagatova, A.A.; Tyndall, J.D.A.; Monk, B.C. Crystal Structures of Full-Length Lanosterol 14α-Demethylases of Prominent Fungal Pathogens Candida Albicans and Candida Glabrata Provide Tools for Antifungal Discovery. Antimicrob. Agents Chemother. 2018, 62, e01134-18. [Google Scholar] [CrossRef]
- Hargrove, T.Y.; Friggeri, L.; Wawrzak, Z.; Qi, A.; Hoekstra, W.J.; Schotzinger, R.J.; York, J.D.; Guengerich, F.P.; Lepesheva, G.I. Structural Analyses of Candida Albicans Sterol 14α-Demethylase Complexed with Azole Drugs Address the Molecular Basis of Azole-Mediated Inhibition of Fungal Sterol Biosynthesis. J. Biol. Chem. 2017, 292, 6728–6743. [Google Scholar] [CrossRef]
- Sagatova, A.A.; Keniya, M.V.; Wilson, R.K.; Monk, B.C.; Tyndall, J.D.A. Structural Insights into Binding of the Antifungal Drug Fluconazole to Saccharomyces Cerevisiae Lanosterol 14α-Demethylase. Antimicrob. Agents Chemother. 2015, 59, 4982–4989. [Google Scholar] [CrossRef] [PubMed]
- Tari, L.W.; Trzoss, M.; Bensen, D.C.; Li, X.; Chen, Z.; Lam, T.; Zhang, J.; Creighton, C.J.; Cunningham, M.L.; Kwan, B.; et al. Pyrrolopyrimidine Inhibitors of DNA Gyrase B (GyrB) and Topoisomerase IV (ParE). Part I: Structure Guided Discovery and Optimization of Dual Targeting Agents with Potent, Broad-Spectrum Enzymatic Activity. Bioorg. Med. Chem. Lett. 2013, 23, 1529–1536. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Nastasă, C.; Tamaian, R.; Oniga, O.; Tiperciuc, B. 5-Arylidene(Chromenyl-Methylene)-Thiazolidinediones: Potential New Agents against Mutant Oncoproteins K-Ras, N-Ras and B-Raf in Colorectal Cancer and Melanoma. Medicina 2019, 55, 85. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Zoete, V. A BOILED-Egg To Predict Gastrointestinal Absorption and Brain Penetration of Small Molecules. ChemMedChem 2016, 11, 1117–1121. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A Free Web Tool to Evaluate Pharmacokinetics, Drug-Likeness and Medicinal Chemistry Friendliness of Small Molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Li, X.; Du, Z.; Wang, J.; Wu, Z.; Li, W.; Liu, G.; Shen, X.; Tang, Y. In Silico Estimation of Chemical Carcinogenicity with Binary and Ternary Classification Methods. Mol. Inform. 2015, 34, 228–235. [Google Scholar] [CrossRef]
- Wang, Q.; Li, X.; Yang, H.; Cai, Y.; Wang, Y.; Wang, Z.; Li, W.; Tang, Y.; Liu, G. In Silico Prediction of Serious Eye Irritation or Corrosion Potential of Chemicals. RSC Adv. 2017, 7, 6697–6703. [Google Scholar] [CrossRef]
- Di, P.; Yin, Y.; Jiang, C.; Cai, Y.; Li, W.; Tang, Y.; Liu, G. Prediction of the Skin Sensitising Potential and Potency of Compounds via Mechanism-Based Binary and Ternary Classification Models. Toxicol. Vitr. 2019, 59, 204–214. [Google Scholar] [CrossRef]
- Mulliner, D.; Schmidt, F.; Stolte, M.; Spirkl, H.-P.; Czich, A.; Amberg, A. Computational Models for Human and Animal Hepatotoxicity with a Global Application Scope. Chem. Res. Toxicol. 2016, 29, 757–767. [Google Scholar] [CrossRef]
- Wang, Z.; Zhao, P.; Zhang, X.; Xu, X.; Li, W.; Liu, G.; Tang, Y. In Silico Prediction of Chemical Respiratory Toxicity via Machine Learning. Comput. Toxicol. 2021, 18, 100155. [Google Scholar] [CrossRef]
- Jiang, C.; Yang, H.; Di, P.; Li, W.; Tang, Y.; Liu, G. In Silico Prediction of Chemical Reproductive Toxicity Using Machine Learning. J. Appl. Toxicol. 2019, 39, 844–854. [Google Scholar] [CrossRef]
- Yang, H.; Lou, C.; Sun, L.; Li, J.; Cai, Y.; Wang, Z.; Li, W.; Liu, G.; Tang, Y. AdmetSAR 2.0: Web-Service for Prediction and Optimization of Chemical ADMET Properties. Bioinformatics 2019, 35, 1067–1069. [Google Scholar] [CrossRef] [PubMed]
- Patlewicz, G.; Jeliazkova, N.; Safford, R.J.; Worth, A.P.; Aleksiev, B. An Evaluation of the Implementation of the Cramer Classification Scheme in the Toxtree Software. SAR QSAR Environ. Res. 2008, 19, 495–524. [Google Scholar] [CrossRef] [PubMed]
- Knox, C.; Wilson, M.; Klinger, C.M.; Franklin, M.; Oler, E.; Wilson, A.; Pon, A.; Cox, J.; Chin, N.E.; Strawbridge, S.A.; et al. DrugBank 6.0: The DrugBank Knowledgebase for 2024. Nucleic Acids Res. 2024, 52, D1265–D1275. [Google Scholar] [CrossRef]
- Pietsch, F.; Bergman, J.M.; Brandis, G.; Marcusson, L.L.; Zorzet, A.; Huseby, D.L.; Hughes, D. Ciprofloxacin Selects for RNA Polymerase Mutations with Pleiotropic Antibiotic Resistance Effects. J. Antimicrob. Chemother. 2017, 72, 75–84. [Google Scholar] [CrossRef]
- Zhang, G.-F.; Liu, X.; Zhang, S.; Pan, B.; Liu, M.-L. Ciprofloxacin Derivatives and Their Antibacterial Activities. Eur. J. Med. Chem. 2018, 146, 599–612. [Google Scholar] [CrossRef]
- Determination That ALBAMYCIN (Novobiocin Sodium) Capsule, 250 Milligrams, Was Withdrawn from Sale for Reasons of Safety or Effectiveness. Available online: https://www.federalregister.gov/documents/2011/01/19/2011-1000/determination-that-albamycin-novobiocin-sodium-capsule-250-milligrams-was-withdrawn-from-sale-for (accessed on 11 March 2025).
- 21 CFR 216.24—Drug Products Withdrawn or Removed from the Market for Reasons of Safety or Effectiveness. Available online: https://www.ecfr.gov/current/title-21/part-216/section-216.24 (accessed on 11 March 2025).
- Constable, P.D.; Hinchcliff, K.W.; Done, S.H.; Grünberg, W. Practical Antimicrobial Therapeutics. In Veterinary Medicine; Elsevier: Amsterdam, The Netherlands, 2017; pp. 153–174. [Google Scholar]
- Maxwell, A. The Interaction between Coumarin Drugs and DNA Gyrase. Mol. Microbiol. 1993, 9, 681–686. [Google Scholar] [CrossRef]
- Gormley, N.A.; Orphanides, G.; Meyer, A.; Cullis, P.M.; Maxwell, A. The Interaction of Coumarin Antibiotics with Fragments of the DNA Gyrase B Protein. Biochemistry 1996, 35, 5083–5092. [Google Scholar] [CrossRef] [PubMed]
- Tabary, X.; Moreau, N.; Dureuil, C.; Le Goffic, F. Effect of DNA Gyrase Inhibitors Pefloxacin, Five Other Quinolones, Novobiocin, and Clorobiocin on Escherichia coli Topoisomerase I. Antimicrob. Agents Chemother. 1987, 31, 1925–1928. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhang, Y.; Zhao, D.; Yu, X.; Shen, X.; Zhou, Y.; Wang, S.; Qiu, Y.; Chen, Y.; Zhu, F. TTD: Therapeutic Target Database Describing Target Druggability Information. Nucleic Acids Res. 2024, 52, D1465–D1477. [Google Scholar] [CrossRef] [PubMed]
- Kowalska-Krochmal, B.; Dudek-Wicher, R. The Minimum Inhibitory Concentration of Antibiotics: Methods, Interpretation, Clinical Relevance. Pathogens 2021, 10, 165. [Google Scholar] [CrossRef]
- de Castro, R.D.; de Souza, T.M.P.A.; Bezerra, L.M.D.; Ferreira, G.L.S.; de Brito Costa, E.M.M.; Cavalcanti, A.L. Antifungal Activity and Mode of Action of Thymol and Its Synergism with Nystatin against Candida Species Involved with Infections in the Oral Cavity: An in Vitro Study. BMC Complement. Altern. Med. 2015, 15, 417. [Google Scholar] [CrossRef]
- Martelli, G.; Giacomini, D. Antibacterial and Antioxidant Activities for Natural and Synthetic Dual-Active Compounds. Eur. J. Med. Chem. 2018, 158, 91–105. [Google Scholar] [CrossRef]
- Chua, S.L.; Ding, Y.; Liu, Y.; Cai, Z.; Zhou, J.; Swarup, S.; Drautz-Moses, D.I.; Schuster, S.C.; Kjelleberg, S.; Givskov, M.; et al. Reactive Oxygen Species Drive Evolution of Pro-Biofilm Variants in Pathogens by Modulating Cyclic-Di-GMP Levels. Open Biol. 2016, 6, 160162. [Google Scholar] [CrossRef]
- Rajakumari, K.; Aravind, K.; Balamugundhan, M.; Jagadeesan, M.; Somasundaram, A.; Brindha Devi, P.; Ramasamy, P. Comprehensive Review of DNA Gyrase as Enzymatic Target for Drug Discovery and Development. Eur. J. Med. Chem. Rep. 2024, 12, 100233. [Google Scholar] [CrossRef]
- Peralta, M.A.; Ortega, M.G.; Cabrera, J.L.; Paraje, M.G. The Antioxidant Activity of a Prenyl Flavonoid Alters Its Antifungal Toxicity on Candida Albicans Biofilms. Food Chem. Toxicol. 2018, 114, 285–291. [Google Scholar] [CrossRef]
- Peralta, M.A.; da Silva, M.A.; Ortega, M.G.; Cabrera, J.L.; Paraje, M.G. Antifungal Activity of a Prenylated Flavonoid from Dalea Elegans against Candida Albicans Biofilms. Phytomedicine 2015, 22, 975–980. [Google Scholar] [CrossRef] [PubMed]
- Hemmati, F.; Salehi, R.; Ghotaslou, R.; Kafil, H.S.; Hasani, A.; Gholizadeh, P.; Rezaee, M.A. The Assessment of Antibiofilm Activity of Chitosan-Zinc Oxide-Gentamicin Nanocomposite on Pseudomonas aeruginosa and Staphylococcus aureus. Int. J. Biol. Macromol. 2020, 163, 2248–2258. [Google Scholar] [CrossRef]
- Morita, Y.; Tomida, J.; Kawamura, Y. Responses of Pseudomonas aeruginosa to Antimicrobials. Front. Microbiol. 2014, 4, 422. [Google Scholar] [CrossRef]
- Torres, N.S.; Montelongo-Jauregui, D.; Abercrombie, J.J.; Srinivasan, A.; Lopez-Ribot, J.L.; Ramasubramanian, A.K.; Leung, K.P. Antimicrobial and Antibiofilm Activity of Synergistic Combinations of a Commercially Available Small Compound Library with Colistin Against Pseudomonas aeruginosa. Front. Microbiol. 2018, 9, 2541. [Google Scholar] [CrossRef]
- Bonincontro, G.; Scuderi, S.A.; Marino, A.; Simonetti, G. Synergistic Effect of Plant Compounds in Combination with Conventional Antimicrobials against Biofilm of Staphylococcus aureus, Pseudomonas aeruginosa, and Candida spp. Pharmaceuticals 2023, 16, 1531. Pharmaceuticals 2023, 16, 1531. [Google Scholar] [CrossRef]
- Scarpignato, C.; Pelosini, I. Rifaximin, a Poorly Absorbed Antibiotic: Pharmacology and Clinical Potential. Chemotherapy 2005, 51, 36–66. [Google Scholar] [CrossRef]
- Miners, J.O.; Birkett, D.J. Cytochrome P4502C9: An Enzyme of Major Importance in Human Drug Metabolism. Br. J. Clin. Pharmacol. 1998, 45, 525–538. [Google Scholar] [CrossRef] [PubMed]
- Dodd-Butera, T.; Broderick, M. Ciprofloxacin. In Encyclopedia of Toxicology; Elsevier: Amsterdam, The Netherlands, 2014; Volume 1, pp. 966–968. ISBN 9780123864543. [Google Scholar]
- Campbell, K.B. Antimicrobial Agents and Torsades de Pointes. In Torsades de Pointes; Elsevier: Amsterdam, The Netherlands, 2022; pp. 231–266. [Google Scholar]
- Bikobo, D.S.N.; Vodnar, D.C.; Stana, A.; Tiperciuc, B.; Nastasă, C.; Douchet, M.; Oniga, O. Synthesis of 2-Phenylamino-Thiazole Derivatives as Antimicrobial Agents. J. Saudi Chem. Soc. 2017, 21, 861–868. [Google Scholar] [CrossRef]
- Mohammed, T.K.; Aqel, N.; Al-Dujaili, E.A.S. Antimicrobial Activity of Liquid Residues of Cymbopogon Citratus Oil Extracts. J. Phys. Conf. Ser. 2020, 1660, 012006. [Google Scholar] [CrossRef]
- O’Toole, G.A. Microtiter Dish Biofilm Formation Assay. J. Vis. Exp. 2011, 47, e2437. [Google Scholar] [CrossRef]
- Kadiri, F.; Ezaouine, A.; Blaghen, M.; Bennis, F.; Chegdani, F. Antibiofilm Potential of Biosurfactant Produced by Bacillus Aerius against Pathogen Bacteria. Biocatal. Agric. Biotechnol. 2024, 56, 102995. [Google Scholar] [CrossRef]
- Akinboye, A.O.; Makhubu, F.N.; Karzis, J.; Petzer, I.-M.; McGaw, L.J. In Vitro Antibiofilm and Quorum Sensing Inhibition Activities of Selected South African Plants with Efficacy against Bovine Mastitis Pathogens. S. Afr. J. Bot. 2024, 166, 455–465. [Google Scholar] [CrossRef]
- Bateman, A.; Martin, M.-J.; Orchard, S.; Magrane, M.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; Bye-A-Jee, H.; Cukura, A.; et al. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [Google Scholar] [CrossRef]
- Bateman, A.; Martin, M.-J.; Orchard, S.; Magrane, M.; Adesina, A.; Ahmad, S.; Bowler-Barnett, E.H.; Bye-A-Jee, H.; Carpentier, D.; Denny, P.; et al. UniProt: The Universal Protein Knowledgebase in 2025. Nucleic Acids Res. 2025, 53, D609–D617. [Google Scholar] [CrossRef]
- Berman, H.M. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Berman, H.M. The Protein Data Bank. In Leadership in Science and Technology: A Reference Handbook; SAGE Publications, Inc.: Thousand Oaks, CA, USA, 2003; pp. 661–667. [Google Scholar]
- Park, H.; Lee, H.; Seok, C. High-Resolution Protein–Protein Docking by Global Optimization: Recent Advances and Future Challenges. Curr. Opin. Struct. Biol. 2015, 35, 24–31. [Google Scholar] [CrossRef]
- McGovern, S.L.; Shoichet, B.K. Information Decay in Molecular Docking Screens against Holo, Apo, and Modeled Conformations of Enzymes. J. Med. Chem. 2003, 46, 2895–2907. [Google Scholar] [CrossRef]
- Rueda, M.; Bottegoni, G.; Abagyan, R. Recipes for the Selection of Experimental Protein Conformations for Virtual Screening. J. Chem. Inf. Model. 2010, 50, 186–193. [Google Scholar] [CrossRef]
- Roy, K.; Kar, S.; Das, R.N. Other Related Techniques. In Understanding the Basics of QSAR for Applications in Pharmaceutical Sciences and Risk Assessment; Elsevier: Amsterdam, The Netherlands, 2015; pp. 357–425. [Google Scholar]
- Stanzione, F.; Giangreco, I.; Cole, J.C. Use of Molecular Docking Computational Tools in Drug Discovery. In Progress in Medicinal Chemistry; Elsevier: Amsterdam, The Netherlands, 2021; pp. 273–343. [Google Scholar]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure Visualization for Researchers, Educators, and Developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Meng, E.C.; Goddard, T.D.; Pettersen, E.F.; Couch, G.S.; Pearson, Z.J.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Tools for Structure Building and Analysis. Protein Sci. 2023, 32, e4792. [Google Scholar] [CrossRef]
- Dallakyan, S.; Olson, A.J. Small-Molecule Library Screening by Docking with PyRx. In Methods in Molecular Biology; Human Press: New York, NY, USA, 2015; pp. 243–250. [Google Scholar]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Holland, J.H. Adaptation in Natural and Artificial Systems: An Introductory Analysis with Applications to Biology, Control, and Artificial Intelligence; The MIT Press: Cambridge, MA, USA, 1992; ISBN 9780262275552. [Google Scholar]
- Nocedal, J.; Wright, S.J. Sequential Quadratic Programming. In Numerical Optimization; Springer: New York, NY, USA, 1999; pp. 526–573. [Google Scholar]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Vanommeslaeghe, K.; Hatcher, E.; Acharya, C.; Kundu, S.; Zhong, S.; Shim, J.; Darian, E.; Guvench, O.; Lopes, P.; Vorobyov, I.; et al. CHARMM General Force Field: A Force Field for Drug-like Molecules Compatible with the CHARMM All-atom Additive Biological Force Fields. J. Comput. Chem. 2010, 31, 671–690. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Lv, Z.; Wang, H.S.; Niu, X.D. Molecular Dynamics Simulations Reveal Insight into Key Structural Elements of Aaptamines as Sortase Inhibitors with Free Energy Calculations. Chem. Phys. Lett. 2013, 585, 171–177. [Google Scholar] [CrossRef]
- Jin, H.; Zhou, Z.; Wang, D.; Guan, S.; Han, W. Molecular Dynamics Simulations of Acylpeptide Hydrolase Bound to Chlorpyrifosmethyl Oxon and Dichlorvos. Int. J. Mol. Sci. 2015, 16, 6217–6234. [Google Scholar] [CrossRef]
- Crișan, O.; Marc, G.; Nastasă, C.; Oniga, S.D.; Vlase, L.; Pîrnău, A.; Oniga, O. Synthesis and In Silico Approaches of New Symmetric Bis-Thiazolidine-2,4-Diones as Ras and Raf Oncoproteins Inhibitors. Farmacia 2023, 71, 254–263. [Google Scholar] [CrossRef]
- Manolov, S.; Bojilov, D.; Ivanov, I.; Marc, G.; Bataklieva, N.; Oniga, S.; Oniga, O.; Nedialkov, P. Synthesis, Molecular Docking, Molecular Dynamics Studies, and In Vitro Biological Evaluation of New Biofunctional Ketoprofen Derivatives with Different N-Containing Heterocycles. Processes 2023, 11, 1837. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Li, W.; Zhou, Y.; Shen, J.; Wu, Z.; Liu, G.; Lee, P.W.; Tang, Y. AdmetSAR: A Comprehensive Source and Free Tool for Assessment of Chemical ADMET Properties. J. Chem. Inf. Model. 2012, 52, 3099–3105. [Google Scholar] [CrossRef] [PubMed]
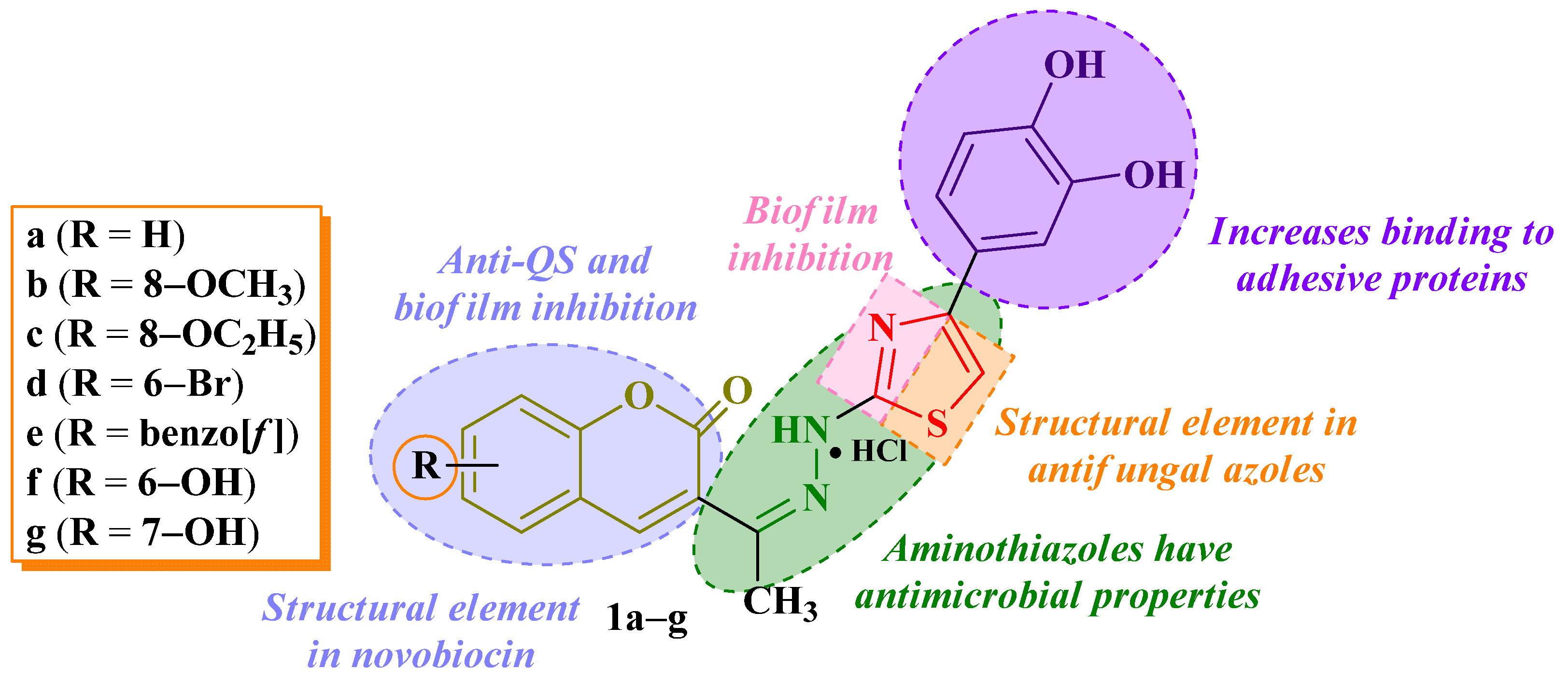




| Comp. | E. coli (ATCC 25922) | S. enteritidis (ATCC 13076) | S. typhimurium (ATCC 14028) | S. typhimurium (Food Isolate) | P. aeruginosa (ATCC 27853) | L. monocytogenes (ATCC 13932) | E. faecalis (ATCC 29212) | S. aureus (ATCC 6538P) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | |
| 1a | 15.62 | 31.25 | 62.5 | 62.5 | 62.5 | 62.5 | 125 | 62.5 | 31.25 | 31.25 | 31.25 | 31.25 | 62.5 | 125 | 15.62 | 15.62 |
| 1b | 125 | 125 | 125 | 125 | 125 | 250 | 62.5 | 125 | 31.25 | 62.5 | 31.25 | 62.5 | 125 | 250 | 15.62 | 31.25 |
| 1c | 125 | 125 | 125 | 250 | 125 | 125 | 62.5 | 125 | 31.25 | 62.5 | 31.25 | 31.25 | 125 | 125 | 15.62 | 31.25 |
| 1d | 62.5 | 125 | 62.5 | 125 | 62.5 | 125 | 31.25 | 62.5 | 31.25 | 62.5 | 31.25 | 62.5 | 62.5 | 62.5 | 15.62 | 31.25 |
| 1e | 62.5 | 62.5 | 62.5 | 125 | 62.5 | 125 | 31.25 | 62.5 | 31.25 | 62.5 | 31.25 | 62.5 | 62.5 | 125 | 15.62 | 31.25 |
| 1f | 62.5 | 125 | 62.5 | 125 | 62.5 | 125 | 31.25 | 62.5 | 31.25 | 31.25 | 31.25 | 62.5 | 62.5 | 125 | 15.62 | 31.25 |
| 1g | 125 | 62.5 | 125 | 250 | 125 | 250 | 31.25 | 62.5 | 15.62 | 31.25 | 31.25 | 31.25 | 125 | 250 | 31.25 | 62.5 |
| D | Bacterial growth in all wells | |||||||||||||||
| M+ | + | + | + | + | + | + | + | + | ||||||||
| CPF | 15.62 | 31.25 | 15.62 | 31.25 | 15.62 | 31.25 | 15.62 | 31.25 | 31.25 | 62.5 | 15.62 | 31.25 | 125 | 250 | 15.62 | 31.25 |
| Compound | MBC/MIC Ratio | |||||||
|---|---|---|---|---|---|---|---|---|
| E. coli (ATCC 25922) | S. enteritidis (ATCC 13076) | S. typhimurium (ATCC 14028) | S. typhimurium (Food Isolate) | P. aeruginosa (ATCC 27853) | L. monocytogenes (ATCC 13932) | E. faecalis (ATCC 29212) | S. aureus (ATCC 6538P) | |
| 1a | 2 | 1 | 1 | 0.5 | 1 | 1 | 2 | 1 |
| 1b | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 |
| 1c | 1 | 2 | 1 | 2 | 2 | 1 | 1 | 2 |
| 1d | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 2 |
| 1e | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| 1f | 2 | 2 | 2 | 2 | 1 | 2 | 2 | 2 |
| 1g | 0.5 | 2 | 2 | 2 | 2 | 1 | 2 | 2 |
| CPF | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Compound | C. albicans (ATCC 10231) | A. brasiliensis (ATCC 16404) | ||||
|---|---|---|---|---|---|---|
| MIC | MFC | MFC/MIC | MIC | MFC | MFC/MIC | |
| 1a | 15.62 | 15.62 | 1 | 31.25 | 31.25 | 1 |
| 1b | 7.81 | 15.62 | 2 | 15.62 | 31.25 | 2 |
| 1c | 15.62 | 31.25 | 2 | 31.25 | 62.5 | 2 |
| 1d | 15.62 | 31.25 | 2 | 31.25 | 62.5 | 2 |
| 1e | 15.62 | 15.62 | 1 | 31.25 | 62.5 | 2 |
| 1f | 15.62 | 31.25 | 2 | 31.25 | 62.5 | 2 |
| 1g | 7.81 | 15.62 | 2 | 15.62 | 31.25 | 2 |
| DMSO | Fungal growth in all wells | |||||
| M+ | + | + | ||||
| Fluconazole | 15.62 | 31.25 | 2 | >250 | >250 | ND |
| Itraconazole [25] | - | - | - | 4 | - | - |
| Concentration (μg/mL) | C1 = 500 | C2 = 250 | C3 = 125 | C4 = 62.50 | C5 = 31.25 | C6 = 15.62 | C7 = 7.81 | C8 = 2.60 | C9 = 1.30 | C10 = 0.60 | C11 = 0.20 | C12 = 0.10 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Compound | BF inhibition (%)—E. faecalis ATCC 29212 | |||||||||||
| 1a | 13.61 | – | – | – | – | – | – | – | – | – | – | – |
| 1b | 15.18 | 19.90 | 21.47 | – | – | – | – | – | – | – | – | – |
| 1c | 10.47 | 18.32 | 19.90 | 12.04 | – | – | – | – | – | – | – | – |
| 1d | 4.19 | 10.47 | 16.75 | – | – | – | – | – | – | – | – | – |
| 1e | 13.61 | 26.18 | 26.18 | – | – | – | – | – | – | – | – | – |
| 1f | 27.75 | 24.61 | 24.61 | – | – | – | – | – | – | – | – | – |
| 1g | – | 12.04 | 18.32 | – | – | – | – | – | – | – | – | – |
| Gentamicin | 23.04 | 21.47 | 19.90 | 15.18 | – | – | – | – | – | – | – | – |
| BF inhibition (%)—P. aeruginosa ATCC 27583 | ||||||||||||
| 1a | 96.24 | 96.24 | 96.31 | 94.95 | 71.81 | – | – | – | – | – | – | – |
| 1b | 95.27 | 96.63 | 96.57 | 94.56 | 75.18 | 56.07 | 55.16 | – | – | – | – | – |
| 1c | 96.70 | 96.76 | 96.57 | 83.22 | 47.97 | 43.50 | 43.76 | 49.59 | – | – | – | – |
| 1d | 95.14 | 96.37 | 96.37 | 77.51 | 58.40 | 41.62 | 25.48 | 49.07 | 45.83 | 71.81 | 67.86 | 54.45 |
| 1e | 96.37 | 96.70 | 96.50 | 81.40 | 51.66 | 11.74 | 49.98 | 28.20 | 54.32 | 53.73 | 64.49 | 64.62 |
| 1f | 96.70 | 96.70 | 95.66 | 93.00 | 69.35 | 40.26 | 6.04 | 46.09 | 31.44 | 55.61 | 53.28 | 46.02 |
| 1g | 96.18 | 96.44 | 95.20 | 95.85 | 84.38 | 70.97 | 81.92 | 14.14 | 80.63 | 26.32 | 76.54 | 23.93 |
| Gentamicin | 96.50 | 96.70 | 96.70 | 96.57 | 88.73 | 94.95 | 95.79 | 95.20 | 94.95 | 96.11 | 50.82 | 3.77 |
| BF inhibition (%)—E. coli ATCC 25922 | ||||||||||||
| 1a | 81.06 | 75.62 | 83.92 | 72.76 | – | – | – | – | – | – | – | – |
| 1b | 82.36 | 83.92 | 86.25 | 73.28 | – | – | – | – | – | – | – | – |
| 1c | 84.17 | 85.73 | 85.99 | 83.40 | – | – | – | – | – | – | – | – |
| 1d | 81.06 | 84.17 | 86.77 | 77.43 | – | – | – | – | – | – | – | – |
| 1e | 84.69 | 86.25 | 86.77 | 78.73 | – | – | – | – | – | – | – | – |
| 1f | 85.47 | 86.51 | 86.25 | 83.66 | – | – | – | – | – | – | – | – |
| 1g | 84.69 | 84.43 | 85.73 | 82.88 | – | – | – | – | – | – | – | – |
| Gentamicin | 85.99 | 87.03 | 86.77 | 87.03 | 82.88 | 85.21 | 83.40 | 83.40 | 84.43 | 79.25 | 80.28 | 15.71 |
| BF inhibition (%)—S. typhimurium ATCC 14028 | ||||||||||||
| 1a | 25.64 | 15.35 | 38.23 | 27.93 | 5.05 | 7.34 | 13.06 | 18.78 | 16.49 | 14.20 | 13.06 | 7.34 |
| 1b | 37.08 | 31.36 | 35.94 | 29.08 | 13.06 | 15.35 | 22.21 | 22.21 | 25.64 | 26.79 | 18.78 | 18.78 |
| 1c | 34.80 | 35.94 | 41.66 | 32.51 | 13.06 | 15.35 | 23.36 | 23.36 | 24.50 | 32.51 | 22.21 | 22.21 |
| 1d | 26.79 | 24.50 | 35.94 | 32.51 | 9.63 | 3.91 | 24.50 | 27.93 | 26.79 | 32.51 | 24.50 | 21.07 |
| 1e | 40.51 | 39.37 | 42.80 | 39.37 | 3.91 | 3.91 | 10.77 | 22.21 | 26.79 | 16.49 | 25.64 | 5.05 |
| 1f | 45.09 | 40.51 | 46.23 | 39.37 | 26.79 | 23.36 | 32.51 | 25.64 | 30.22 | 27.93 | 29.08 | 10.77 |
| 1g | 30.22 | 30.22 | 17.64 | 29.08 | – | – | – | – | – | – | – | – |
| Gentamicin | 43.95 | 38.23 | 45.09 | 40.51 | 43.95 | 35.94 | 41.66 | 38.23 | 39.37 | 39.37 | 29.08 | 27.93 |
| Bacterial Target: DNA Gyrase Subunit B (GyrB) | |||
|---|---|---|---|
| UniProtKB | Annotation | Organism | RCSB PDB |
| Q839Z1 | reviewed | E. faecalis (strain ATCC 700802/V583) | Yes |
| P0A0K8 | reviewed | S. aureus | Yes |
| Q9I7C2 | reviewed | P. aeruginosa (strain ATCC 15692/DSM 22644/CIP 104116/JCM 14847/LMG 12228/1C/PRS 101/PAO1) | Yes |
| P0AES6 | reviewed | E. coli (strain K12; ATCC 23724) | Yes |
| A0A4U8JAX8 | unreviewed | S. enteritidis | No |
| P0A2I3 | reviewed | S. typhimurium (strain LT2/SGSC1412/ATCC 700720) | No |
| Q8YAV7 | unreviewed | L. monocytogenes serovar 1/2a (strain ATCC BAA-679/EGD-e) | No |
| Fungal target: lanosterol 14α-demethylase cytochrome P450 (CYP51) | |||
| UniProtKB | Annotation | Organism | RCSB PDB |
| P10613 | reviewed | C. albicans (strain SC5314/ATCC MYA-2876) | Yes |
| A6ZSR0 | reviewed | S. cerevisiae YJM789 | Yes |
| A0A1L9U4P7 (AA: 524) | unreviewed | A. brasiliensis (strain CBS 101740/IMI 381727/IBT 21946) | No |
| A0A9W6DJU7 (AA: 382) | unreviewed | A. brasiliensis | No |
| Bacterial Target: DNA Gyrase Subunit B (GyrB) | |||||
|---|---|---|---|---|---|
| IDs (Reference) | Organism | Resolution (Å) | Seq. Length (AA Pos) | Mutation (Mut Pos) | Co-Cry Lig (ID) |
| 4GGL/ Q839Z1 [29] | E. faecalis V583 | 1.69 | 642 AA (18–224) | 0 | PubChem CID 70699420 (CJC) DI |
| 6TCK/ P0A0K8 [30] | S. aureus | 1.60 | 644 AA (2–234) | 0 | PubChem CID 151595514 (N1N) |
| 7PTF/ Q9I7C2 [31] | P. aeruginosa PAO1 | 1.32 | 806 AA (1–221) | 0 | Novobiocin (NOV) |
| 7P2M/ P0AES6 [31] | E. coli K-12 | 1.16 | 804 AA (1–220) | 0 | PubChem CID 151595514 (N1N) |
| Fungal Target: Lanosterol 14α-Demethylase Cytochrome P450 (CYP51) | |||||
| IDs (Reference) | Organism | Resolution (Å) | Seq. Length (AA Pos) | Mutation (AA Pos) | Co-Cry Lig (ID) |
| 5V5Z/ P10613 [32] | C. albicans SC5314 | 2.90 | 528 AA (1–528) | 0 | Itraconazole (1YN) |
| 5FSA/ P10613 [33] | C. albicans | 2.86 | 528 AA (48–528) | 1 (221) | Posaconazole (X2N) |
| 5TZ1/ P10613 [33] | C. albicans | 2.00 | 528 AA (48–528) | 2 (6, 221) | Oteseconazole (VT1) |
| 4WMZ/ A6ZSR0 [34] | S. cerevisiae YJM789 | 2.05 | 530 AA (1–530) | 0 | Fluconazole (TPF) |
| 4GGL (E. faecalis V583) | 6TCK (S. aureus) | 7PTF (P. aeruginosa PAO1) | 7P2M (E. coli K12) | ||||
|---|---|---|---|---|---|---|---|
| T-LC | BA | T-LC | BA | T-LC | BA | T-LC | BA |
| 1a | −8.2 | 1a | −9.5 | 1a | −9.4 | 1a | −8.3 |
| 1b | −8.4 | 1b | −8.7 | 1b | −9.1 | 1b | −8.4 |
| 1c | −8.2 | 1c | −8.6 | 1c | −8.5 | 1c | −8.4 |
| 1d | −8.9 | 1d | −8.2 | 1d | −9.2 | 1d | −8.5 |
| 1e | −9.0 | 1e | −9.3 | 1e | −9.6 | 1e | −9.1 |
| 1f | −8.3 | 1f | −8.8 | 1f | −8.9 | 1f | −8.2 |
| 1g | −8.0 | 1g | −8.3 | 1g | −9.1 | 1g | −8.2 |
| Ciprofloxacin | −8.4 | Ciprofloxacin | −7.6 | Ciprofloxacin | −7.7 | Ciprofloxacin | −7.4 |
| Novobiocin | −3.4 | Novobiocin | −7.0 | Novobiocin RD | −7.6 | Novobiocin | −6.0 |
| 5V5Z (C. albicans SC5314) | 5FSA (C. albicans) | 5TZ1 (C. albicans) | 4WMZ (S. cerevisiae YJM789) | ||||
|---|---|---|---|---|---|---|---|
| T-LC | BA | T-LC | BA | T-LC | BA | T-LC | BA |
| 1a | −10.3 | 1a | −10.6 | 1a | −10.3 | 1a | −10.2 |
| 1b | −10.2 | 1b | −10.8 | 1b | −10.3 | 1b | −10.2 |
| 1c | −10.4 | 1c | −10.4 | 1c | −10.3 | 1c | −10.1 |
| 1d | −9.9 | 1d | −10.1 | 1d | −9.9 | 1d | −8.8 |
| 1e | −11.8 | 1e | −11.8 | 1e | −11.0 | 1e | −11.6 |
| 1f | −10.2 | 1f | −10.6 | 1f | −10.1 | 1f | −9.9 |
| 1g | −10.2 | 1g | −10.4 | 1g | −9.9 | 1g | −9.7 |
| Fluconazole | −7.3 | Fluconazole | −7.2 | Fluconazole | −7.0 | Fluconazole RD | −7.5 |
| Complex | Average Ligand RMSD (nm) | Average Backbone RMSD (nm) | Average Radius of Gyration (nm) | Average Ligand-Protein Hydrogen Bonds (No/Ns) |
|---|---|---|---|---|
| 1a-7PTF | 0.20 | 0.18 | 1.70 | 3.52 |
| 1b-7PTF | 0.37 | 0.20 | 1.70 | 0.46 |
| 1c-7PTF | 0.75 | 0.24 | 1.71 | 0.49 |
| 1d-7PTF | 0.16 | 0.19 | 1.70 | 3.84 |
| 1e-7PTF | 0.46 | 0.19 | 1.71 | 1.96 |
| 1f-7PTF | 0.20 | 0.19 | 1.70 | 3.30 |
| 1g-7PTF | 0.27 | 0.23 | 1.70 | 3.90 |
| NOV-7PTF | 0.21 | 0.17 | 1.70 | 4.31 |
| Apo 7PTF | N/A | 0.21 | 1.70 | N/A |
| Compound | GI Absorption | BBB Permeation | P-Gp Substrate | CYP1A2 Inhibitor | CYP2C19 Inhibitor | CYP2C9 Inhibitor | CYP2D6 Inhibitor | CYP3A4 Inhibitor |
|---|---|---|---|---|---|---|---|---|
| 1a | Low | No | No | No | No | Yes | No | No |
| 1b | Low | No | No | No | No | Yes | No | No |
| 1c | Low | No | No | No | No | Yes | No | No |
| 1d | Low | No | No | No | No | Yes | No | No |
| 1e | Low | No | No | No | No | Yes | No | No |
| 1f | Low | No | No | No | No | No | No | No |
| 1g | Low | No | No | No | No | No | No | No |
| Compound | Carcinogenicity | Eye Irritation | Skin Irritation | Hepatotoxicity | Respiratory Toxicity | Reproductive Toxicity | Nephrotoxicity | Acute Oral Toxicity |
|---|---|---|---|---|---|---|---|---|
| 1a | No | No | No | Yes | Yes | Yes | No | Class III |
| 1b | No | No | No | Yes | Yes | Yes | No | Class III |
| 1c | No | No | No | Yes | Yes | Yes | No | Class III |
| 1d | No | No | No | Yes | Yes | Yes | No | Class III |
| 1e | No | No | No | Yes | Yes | Yes | No | Class III |
| 1f | No | No | No | Yes | Yes | Yes | No | Class III |
| 1g | No | No | No | Yes | Yes | Yes | No | Class III |
| Compound | Antiradical Assays | Electron Transfer Capacity Assays | |||||
|---|---|---|---|---|---|---|---|
| IC50 DPPH• (μM) | IC50 ABTS•+ (μM) | TAC | RP | FRAP | CUPRAC | ||
| Eq Ascorbic Acid | Eq Ascorbic Acid | Eq Trolox | Eq Trolox | Eq Trolox | |||
| 1a | 29.90 | 12.62 | 1.53 | 1.96 | 1.52 | 1.37 | 3.09 |
| 1b | 29.54 | 11.77 | 1.53 | 1.84 | 1.43 | 1.40 | 3.16 |
| 1c | 29.88 | 11.60 | 1.47 | 1.83 | 1.42 | 1.38 | 3.21 |
| 1d | 33.49 | 14.04 | 1.61 | 1.08 | 0.84 | 1.14 | 2.27 |
| 1e | 28.60 | 10.88 | 1.33 | 1.71 | 1.33 | 1.20 | 2.99 |
| 1f | 24.57 | 8.38 | 2.20 | 2.54 | 1.98 | 1.45 | 3.63 |
| 1g | 23.84 | 7.06 | 2.18 | 2.59 | 2.02 | 1.42 | 3.60 |
| Ascorbic acid | 50.17 | - | |||||
| Trolox | 36.69 | 16.57 | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ungureanu, D.; Marc, G.; Duma, M.N.; Tamaian, R.; Vodnar, D.C.; Tiperciuc, B.; Moldovan, C.; Ionuț, I.; Stana, A.; Oniga, O. Antimicrobial and Antibiofilm Activities of Some Antioxidant 3,4-Dihydroxyphenyl-Thiazole-Coumarin Hybrid Compounds: In Silico and In Vitro Evaluation. Antibiotics 2025, 14, 943. https://doi.org/10.3390/antibiotics14090943
Ungureanu D, Marc G, Duma MN, Tamaian R, Vodnar DC, Tiperciuc B, Moldovan C, Ionuț I, Stana A, Oniga O. Antimicrobial and Antibiofilm Activities of Some Antioxidant 3,4-Dihydroxyphenyl-Thiazole-Coumarin Hybrid Compounds: In Silico and In Vitro Evaluation. Antibiotics. 2025; 14(9):943. https://doi.org/10.3390/antibiotics14090943
Chicago/Turabian StyleUngureanu, Daniel, Gabriel Marc, Mihaela Niculina Duma, Radu Tamaian, Dan Cristian Vodnar, Brîndușa Tiperciuc, Cristina Moldovan, Ioana Ionuț, Anca Stana, and Ovidiu Oniga. 2025. "Antimicrobial and Antibiofilm Activities of Some Antioxidant 3,4-Dihydroxyphenyl-Thiazole-Coumarin Hybrid Compounds: In Silico and In Vitro Evaluation" Antibiotics 14, no. 9: 943. https://doi.org/10.3390/antibiotics14090943
APA StyleUngureanu, D., Marc, G., Duma, M. N., Tamaian, R., Vodnar, D. C., Tiperciuc, B., Moldovan, C., Ionuț, I., Stana, A., & Oniga, O. (2025). Antimicrobial and Antibiofilm Activities of Some Antioxidant 3,4-Dihydroxyphenyl-Thiazole-Coumarin Hybrid Compounds: In Silico and In Vitro Evaluation. Antibiotics, 14(9), 943. https://doi.org/10.3390/antibiotics14090943











