Abstract
The emergence and spread of antimicrobial resistance (AMR) among Enterobacteriaceae pose significant threats to global public health. In this study, we conducted a short-term surveillance effort in Southern Thailand hospitals to characterize the genomic diversity, AMR profiles, and virulence factors of Enterobacteriaceae strains. We identified 241 carbapenem-resistant Enterobacteriaceae, of which 12 were selected for whole-genome sequencing (WGS) and genome analysis. The strains included Proteus mirabilis, Serratia nevei, Klebsiella variicola, Klebsiella aerogenes, Klebsiella indica, Klebsiella grimontii, Phytobacter ursingii, Phytobacter palmae, Kosakonia spp., and Citrobacter freundii. The strains exhibited high levels of multidrug resistance, including resistance to carbapenem antibiotics. Whole-genome sequencing revealed a diverse array of antimicrobial resistance genes (ARGs), with strains carrying genes for ß-lactamase, efflux pumps, and resistance to other antibiotic classes. Additionally, stress response, metal tolerance, and virulence-associated genes were identified, highlighting the adaptability and pathogenic potential of these strains. A plasmid analysis identified several plasmid replicons, including IncA/C2, IncFIB(K), and Col440I, as well as several plasmids identical to those found globally, indicating the potential for the horizontal gene transfer of ARGs. Importantly, this study also identified a novel species of Kosakonia spp. PSU27, adding to the understanding of the genetic diversity and resistance mechanisms of Enterobacteriaceae in Southern Thailand. The results reported in this study highlight the critical importance of implementing effective antimicrobial management programs and developing innovative treatment approaches to urgently tackle AMR.
1. Introduction
The emergence and spread of antimicrobial resistance (AMR) among Enterobacteriaceae play significant roles to global public health. These bacteria are responsible for a wide range of infections, including urinary tract infections (UTIs), hospital-acquired and ventilator-associated pneumonia (HAP/VAP), complicated intra-abdominal infections (cIAIs), and bloodstream infections (BSIs) [1]. AMR is a critical issue that contributes to significant morbidity and mortality worldwide. According to the World Health Organization (WHO), AMR causes an estimated 700,000 deaths annually, and this number could rise to 10 million by 2050 if no effective measures are taken [2]. The economic burden is also substantial, with healthcare costs rising due to prolonged hospital stays, the need for more expensive treatments, and increased mortality rates. Infections caused by multidrug-resistant (MDR) Enterobacteriaceae are particularly challenging to manage and control, leading to significant treatment costs and posing a threat to public health [3].
In Southern Thailand, where infectious diseases are a major public health concern, understanding the genomic diversity, AMR profiles, and virulence factors of Enterobacteriaceae is crucial for guiding local treatment strategies and infection control measures. Despite the importance of this issue, there is a scarcity of studies focusing on the genomics of Enterobacteriaceae in this region. The ability of this bacterial family to rapidly acquire and disseminate resistance genes has made them difficult challengers in the clinical setting, often resulting in treatment failures and increased healthcare costs.
We obtained a diverse family of bacteria that includes several clinically significant genera, such as Proteus, Serratia, Klebsiella, Phytobacter, Kosakonia, and Citrobacter, from short-term survey of Enterobacteriaceae in Southern Thailand hospitals. Therefore, this study aims to address this gap by providing genomic insights into Enterobacteriaceae diversity, AMR, and virulence in Southern Thailand. By employing whole-genome sequencing and bioinformatic analyses, we seek to characterize the genetic diversity of Enterobacteriaceae isolates, identify AMR genes and mechanisms, and elucidate virulence factors contributing to their pathogenicity. The findings of this study are expected to have significant implications for clinical practice and public health in Southern Thailand. By providing a comprehensive understanding of the genomic landscape of Enterobacteriaceae in this region, we might be able to suggest local treatment guidelines, enhance antimicrobial deployment efforts, and contribute to global AMR surveillance efforts.
2. Results and Discussions
2.1. Clinical Data and Antimicrobial Susceptibility Testing Results
The clinical data and antimicrobial resistance susceptibility results are exhibited in Table 1, providing a detailed overview of Enterobacteriaceae strains isolated from various hospital sources and their resistance patterns to a range of antibiotics. The included strains are P. mirabilis, S. nevei, two isolates of K. variicola, K. aerogenes, K. indica, K. grimontii, P. ursingii, P. palmae, Kosakonia spp., and two isolates of C. freundii from different source of isolations, such as rectal, throat, endotracheal tube, and nasopharynx samples, highlighting the diverse nature of these pathogens and their potential to cause infections in different clinical contexts. All strains in this study present a resistance to carbapenem antibiotics (meropenem, imipenem, and ertapenem) and some types of third-generation cephalosporin (ceftriaxone and ceftazidime), indicating a high level of multidrug resistance among these pathogens. Previous studies have also reported the problem of carbapenem-resistant Enterobacteriaceae (CRE). For example, in 2017, Logan and Weinstein reported on the evolution and epidemiology of CRE globally, highlighting the rapid global dissemination of carbapenemase-producing Enterobacteriaceae [4]. Another study in 2021 emphasized the global challenge of multidrug-resistant (MDR) Enterobacteriaceae and the need for new treatment options to combat CRE infections [5]. In addition to resistance to ß-lactam drugs, the resistance profiles reveal variability among the strains. For instance, K. variicola PSU7 shows resistance to most antibiotics except amikacin, while K. variicola PSU16 exhibits resistance to all antibiotics tested. This bacterium is recognized as an emerging pathogen in humans and has been identified in various environments. It is a member of the K. pneumoniae complex and has been found to be a more serious pathogen, containing a broad range of resistance genes that confer the resistance phenotype. This indicates a particularly worrisome level of MDR, especially the ß-lactam class [5].

Table 1.
Antimicrobial susceptibility testing in Enterobacteriaceae strains in this study.
Surprisingly, we identified a novel species of Kosakonia spp. PSU27, isolated from the nasopharynx of a patient at Satun Hospital. The average nucleotide identity (ANI) shows a 94.55% similarity to the closest species, Kosakonia sacchari BO-1. We also collected data from the RefSeq database, which contains 99 isolates of Kosakonia genomes (Figure 1). Among these isolates, only five are clinical samples: one Kosakonia spp. from an oral metagenome, three Kosakonia radicincitans from blood, and one Kosakonia cawanii from an oral swab. Our Kosakonia spp. PSU27 is the first novel species reported in Thailand.
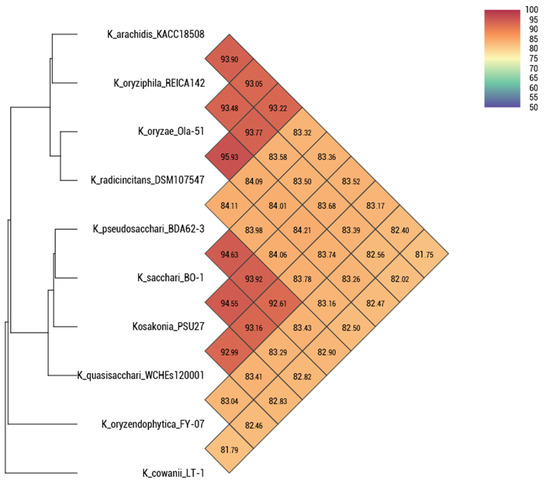
Figure 1.
Average nucleotide identity of nine Kosakonia isolates collected from RefSeq NCBI database and Kosakonia spp. PSU27 in this study. Percent nucleotide identity lower than 95% is considered as a novel species.
This discovery of a novel species, Kosakonia spp. PSU27, isolated from a patient’s nasopharynx at Satun Hospital, is significant for several reasons. Firstly, it expands the known diversity of the Kosakonia genus, which can have implications for understanding its ecological niche, pathogenicity, and potential biotechnological applications. The identification of this novel species was supported by an average nucleotide identity (ANI) analysis, which showed a 94.55% similarity to the closest known species, Kosakonia sacchari BO-1. This level of genetic divergence suggests that Kosakonia spp. PSU27 represents a distinct species within the genus. Furthermore, the rarity of clinical isolates of Kosakonia in public databases, as evidenced by the limited number of clinical samples among the 99 isolates of Kosakonia genomes in RefSeq NCBI [6], underscores the novelty of this finding. Among these isolates, only five were clinical samples, which indicates the importance of this discovery in expanding our understanding of the clinical relevance of Kosakonia species.
These antimicrobial profiles and the ANI provide valuable insights into the diversity and antimicrobial resistance patterns of Enterobacteriaceae strains in clinical settings. The inclusion of various strains from different hospital sources highlights the diverse nature of these pathogens and their potential to cause infections in different clinical contexts. The high level of multidrug resistance, including resistance to carbapenem antibiotics, highlights the urgent need for new treatment options and effective antimicrobial management programs. The variability in resistance profiles among strains further emphasizes the complexity of antimicrobial resistance mechanisms within Enterobacteriaceae. The discovery of a novel species, Kosakonia spp. PSU27, expands our understanding of the genus and raises awareness among clinical staff and researchers about the importance of ongoing surveillance and research to identify emerging pathogens.
2.2. Antimicrobial Resistance Genes in Enterobacteriaceae
The Enterobacteriaceae isolates in this study exhibit a wide range of antimicrobial resistance genes, with a total of 45 ARGs, which were categorized into 14 classes based on the antibiotics they confer resistance to (Figure 2). Among the isolates, Klebsiella and Citrobacter strains were found to carry several ß-lactamase genes (e.g., blaLEN, blaCMY, blaDHA, blaNDM, blaOXA, blaTEM, and blaVEB), indicating resistance to ß-lactam antibiotics. The presence of these genes suggests the enzymatic inactivation of beta-lactams. A study by Chen et al. found that blaTEM, blaCTX-M, and blaOXA were the most common ß-lactamase genes detected in Enterobacteriaceae isolates [7], aligning with our findings. Furthermore, our study identified additional resistance genes, including those for efflux pumps (e.g., kdeA, emrD, sdeB, sdeY, and smfY) in strains such as S. nevei PSU6 and some isolates of Klebsiella. Efflux pumps, regulated by these genes, play a crucial role in bacterial survival by expelling toxic substances, including antibiotics. Wang et al. similarly identified efflux pump genes, such as acrAB and oqxAB, in Enterobacteriaceae isolates, which contribute significantly to multidrug resistance [8].
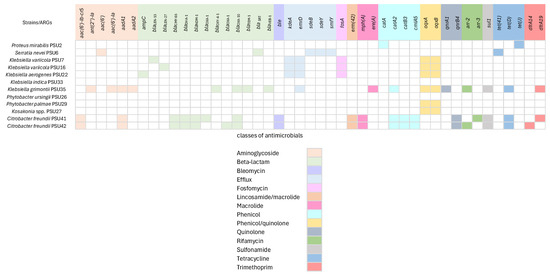
Figure 2.
Antimicrobial resistance genes (ARGs) in 13 Enterobacteriaceae isolates. Forty-five genes conferring antimicrobial resistance genes which were categorized into 14 classes. The presence of the genes is represented by different colors. Classes of related antimicrobial resistance genes are represented below.
For Fosfomycin resistance (fosA) gene detection, studies from Hong Kong and Japan have examined the prevalence of fosfomycin resistance in clinical isolates of Enterobacteriaceae, including Klebsiella species. These studies revealed that the fosA gene is commonly found among resistant isolates, indicating a widespread distribution of this resistance determinant. For example, a study in Hong Kong reported a high prevalence of fosA among fosfomycin-resistant clinical isolates, emphasizing the significance of this gene in conferring resistance [9]. Likewise, research from Japan identified fosA as a frequent contributor to fosfomycin resistance in Enterobacteriaceae [9]. The presence of fosA in our Klebsiella isolates aligns with these findings and suggests the gene’s role in resistance mechanisms and its potential impact on the efficacy of fosfomycin as a treatment option. This highlights the importance of monitoring the prevalence of fosA and other resistance genes to inform treatment strategies and combat antibiotic resistance. Additionally, genes associated with phenicol resistance (catA and catB), quinolone resistance (oqxA and oqxB), sulfonamide resistance (sul1), and tetracycline resistance (tet(41), tet(D) and tet(J)) were found in various strains. These identified ARGs are mostly related to AST results, indicating the presence of these genes correlates with observed resistance phenotypes.
There are some discrepancies observed in P. ursingii PSU26, which shows no ARGs, and P. palmae PSU29 and Kosakonia spp. PSU27, which contain only oqxA and oqxB. However, this species was shown to resist all tested antimicrobials. This could be explained by the capability of the efflux system regulated by oqxAB [10]. The absence of ARGs in P. ursingii PSU26 and K. indica PSU33 is particularly important and could suggest alternative mechanisms of resistance. One such mechanism is the presence of multidrug efflux systems, which are known to confer resistance by actively pumping out antibiotics from the bacterial cell, thus reducing their intracellular concentrations to sub-lethal levels [11]. Another possible mechanism could be modifications in the bacterial cell membrane permeability. Bacteria can alter their outer membrane porins, reducing antibiotic uptake, and, thereby, conferring resistance. Additionally, the enzymatic inactivation of antibiotics through less well-characterized mechanisms or the presence of chromosomal mutations that confer resistance could also play roles [12]. Furthermore, short-read sequencing tends to produce errors and limitations that may impact the accuracy of results, especially in complex genomic regions or when dealing with highly similar sequences [13]. In addition, the presence of only oqxA and oqxB in P. palmae PSU29 and Kosakonia spp. PSU27 raises questions about the mechanisms underlying their MDR. These isolates might rely on specific efflux pumps, encoded by these genes, to expel multiple antibiotics from their cells [14]. The results suggest the need for effective antibiotic management programs and the development of novel antimicrobial agents to combat infections caused by MDR bacteria.
2.3. Stress Response, Metal Tolerance, and Virulence-Associated Genes in Enterobacteriaceae
A comprehensive examination of stress response, metal tolerance, and virulence genes in Enterobacteriaceae was presented in Figure 3. This study targeted these specific genes because they play crucial roles in the survival and pathogenicity of bacteria, impacting both human health and environmental management. Stress response genes help bacteria adapt to various environmental challenges, including oxidative stress, osmotic stress, and heat shock, which are common in hospital and natural settings. These genes are involved in the cellular response to environmental stressors and are essential for bacterial survival in challenging conditions [15]. Metal tolerance genes are vital due to the increasing environmental contamination with metals, which not only affects bacterial survival but can also be linked to antibiotic resistance mechanisms, posing significant treatment challenges. Virulence genes are essential for understanding the pathogenic potential, as they contribute to the ability of bacteria to cause disease in hosts.
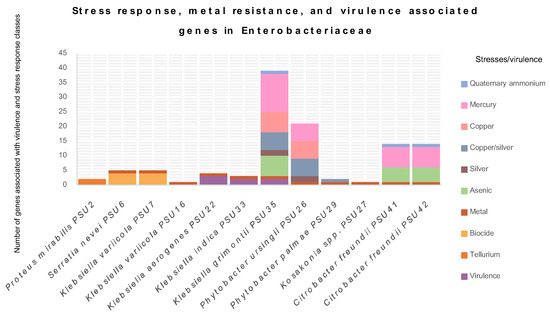
Figure 3.
The graph depicts the determination of stress response, metal tolerance, and virulence-associated genes in 12 Enterobacteriaceae isolates from this study. Different colors represent classes of stress or virulence.
The results reveal that all Enterobacteriaceae in this study contain at least one type of virulence or stress gene. P. mirabilis PSU2 possesses the terD and terZ genes, conferring tellurium resistance. S. nevei PSU6 exhibits biocide-regulating genes, such as smdA, smdB, sdeA, and ssmE, associated with multidrug efflux systems. These systems play a crucial role in bacterial resistance to antimicrobial compounds, including antibiotics, by pumping them out of the cell [16,17,18]. The strain PSU6 also carries fieF, which confers metal tolerance. This gene is identified in almost all isolates in this study, except for P. mirabilis PSU2. Generally, Enterobacteriaceae can exhibit intrinsic or acquired resistance mechanisms, often involving efflux pumps or metal-binding proteins [19]. Metal tolerance is concerning, as it can be related to antibiotic resistance, potentially leading to treatment challenges [20]. Moreover, environmental contamination with metals can further contribute to the development and spread of metal resistance in this family.
Other interesting virulence-associated genes detected were the iroB, iroC, and iroN genes, carried by K. aerogenes PSU22. These genes are important components of the iron acquisition system in Escherichia coli, related to the production, export, and uptake of the siderophore salmochelin, crucial for scavenging iron, an essential nutrient, particularly in environments with limited iron [21]. For ybtP, ybtQ, and the yersiniabactin ABC Transporter ATP-Binding/Permease Protein, detected in K. indica PSU33 and K. grimontii PSU35, they are key components of the yersiniabactin gene cluster in Enterobacteriaceae, contributing to the biosynthesis and uptake of the siderophore yersiniabactin, crucial for acquiring iron from the host environment [22]. The details of stress response, metal tolerance, and virulence-associated genes are displayed in Table S2. The presence of multidrug efflux genes in S. nevei PSU6 highlights its ability to resist various antimicrobial agents, which might explain the lack of sensitivity to all tested antimicrobials in this isolate. K. variicola PSU7 and P. mirabilis PSU2 exhibit genes for tellurite resistance, which is particularly interesting given the scarceness of tellurium resistance mechanisms in bacteria [23]. K. aerogenes PSU22 and K. indica PSU33 display genes associated with virulence, indicating their potential pathogenicity. K. grimontii PSU35 and PSU36 are notable for their extensive repertoire of stress response genes, suggesting their adaptability to diverse environmental stresses, including metals, arsenic, and biocides [24,25]. P. ursingii PSU26 and P. palmae PSU29 also exhibit stress response genes, highlighting their ability to survive in metal-contaminated environments. Kosakonia spp. PSU27 demonstrates a similar trend, indicating its adaptation to metal stress. C. freundii PSU41 and PSU42 show a combination of stress response genes related to metals, arsenic, and mercury, along with genes conferring resistance to quaternary ammonium compounds, reflecting their flexibility against multiple stressors [26]. Understanding these genes emphasizes the genetic diversity and adaptive capacity of bacteria, offering insights into their survival strategies and potential implications for human health and environmental management.
2.4. Plasmid Identification in Enterobacteriaceae
According to PlasmidFinder, various plasmid replicons were found in bacterial strains, along with their lengths. In this study, plasmid replicons were identified in five isolates: P. ursingii PSU26, P. palmae PSU29, K. grimontii PSU35, and C. freundii PSU42 and PSU41. The identified plasmids included IncFIB(K), Col440I, IncA/C2, IncFIB(pB171), IncFII(K), and RepA, ranging in length from 4332 to 70,146 base pairs (Figure 4). The most abundant plasmid replicon type found was IncA/C2, identified in C. freundii PSU41 and PSU42, and K. grimontii PSU35 (Figure 4). IncA/C2 is commonly found in Enterobacteriaceae and other Gram-negative bacteria, known for carrying multiple ARGs and other mobile genetic elements [27,28].
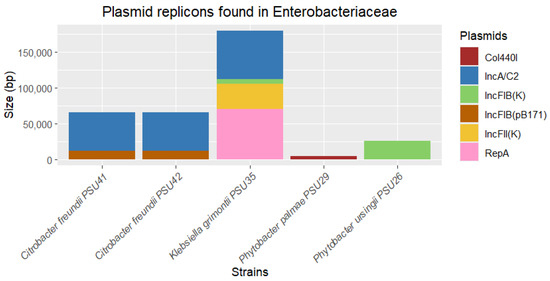
Figure 4.
Determination of plasmid replicons identified from five out of twelve Enterobacteriaceae in this study. The thickness of each stack indicates the size of sequences, and different colors represent the types of replicons.
A further analysis using the PLSDB tool confirmed that these isolates carry plasmids with a high sequence identity to those found in a variety of bacterial species globally. A PLSDB analysis identified several plasmids in Enterobacteriaceae isolates. These plasmids range from 1657 to 71,960 base pairs and are associated with strains from diverse geographic locations, including China, the USA, Canada, the UK, India, Japan, and Hong Kong. The same set of plasmids was identified in two isolates of C. freundii. These identified plasmids are associated with a variety of bacterial species, such as Enterobacter hormaechei, Citrobacter sp., Salmonella enterica, Klebsiella pneumoniae, Raoultella sp., Klebsiella quasipneumoniae, and Enterobacter kobei (Table 2). The presence of high-sequence-identity plasmids in these isolates probably suggests a potential for horizontal gene transfer and the global dissemination of plasmid-borne genes [29]. This is particularly significant for genes related to antimicrobial resistance and virulence, highlighting the role of plasmids in spreading resistance traits across different bacterial species and geographic regions. Moreover, the identical set of plasmids present in C. freundii collected from different samples might indicate a clonal spread or a common source of infection, suggesting that these plasmids confer a selective advantage that allows for their persistence and proliferation within and across different hosts [30]. The varying lengths of plasmid sequences underscore the diverse nature of plasmid genomes, which can carry different sets of genes that contribute to bacterial adaptability and survival. Taxonomically, the plasmids identified in this study are associated with several clinically significant bacterial species. Specially, species such as K. pneumoniae and S. enterica are known for their roles in hospital-acquired infections and their ability to acquire multidrug resistance [31].

Table 2.
Plasmid identification using PLSDB.
Despite identifying several ARGs and plasmid, no ARGs were found to be mediated by the plasmids. This could be because ARGs may be located on the bacterial chromosome rather than on plasmids. Furthermore, the ARGs may have integrated into the bacterial chromosome from plasmids through mechanisms such as transposition or recombination. The short-read sequencing used in this study could not obtain the complete genome of the bacteria, leading to limitations in identifying plasmid–ARG associations, such as sensitivity issues or incomplete plasmid databases [32,33]. The detection of these plasmids in our isolates might suggest that this study may uncover critical information regarding the spread and characteristics of resistance plasmids in different bacterial hosts.
3. Materials and Methods
3.1. Sample Collection
This study is part of a short-term surveillance effort conducted in ICU patients from five different hospitals in Southern Thailand over a 6-month period in 2019 [34,35,36]. The project identified 241 Enterobacteriaceae suspected of being resistant to carbapenem antimicrobials, all screened for resistance using MacConkey agar supplemented with 2 µg/mL of imipenem. Twelve out of 241 Enterobacteriaceae isolates were selected for this study. These included one Proteus mirabilis, one Serratia nevei, two Klebsiella variicola, one Klebsiella aerogenes, one Klebsiella indica, one Klebsiella grimontii, one Phytobacter ursingii, one Phytobacter palmae, one Kosakonia spp., and two Citrobacter freundii. They were chosen because they are uncommon in clinical samples but are found in outside communities or contaminated surfaces in the environment. These twelve isolates were collected from patients admitted to intensive care units (ICUs) in hospitals in southern Thailand, including Songklanagarind Hospital, Patthalung Hospital, Satun Hospital, Pattani Hospital, and Yala Hospital. The bacteria were isolated from various specimens, such as nasopharynx, swabs, endotracheal tubes, throats, and rectums. All patients had received prior antibiotics before their specimens were collected (Table S1).
Initial species identification of the twelve strains was initially conducted using biochemical tests based on Bergey’s Manual of Systematic Bacteriology [37], with further confirmation accomplished using Matrix-Assisted Laser Desorption/Ionization–Time of Flight (MALDI-TOF) mass spectrometry (MS). However, recognizing the limitations of MALDI-TOF MS in distinguishing between species within the Klebsiella pneumoniae complex, we confirmed the identification of our selected. All of them were stored at −20 °C for further identification and analysis using WGS.
3.2. Antimicrobial Susceptibility Testing
All isolates underwent antimicrobial susceptibility testing (AST) using the disk diffusion method, according to Kirby–Bauer disk diffusion susceptibility test protocol [38]. The study utilized antimicrobial disks, including ciprofloxacin (5 µg), levofloxacin (5 µg), amikacin (10 µg), gentamicin (10 µg), imipenem (10 µg), meropenem (10 µg), tazocin (100/10 µg), ertapenem (30 µg), ceftriaxone (30 µg), ceftazidime (30 µg), and sulperazon (100/10 µg). Escherichia coli ATCC® 25922 (for co-trimoxazole) and Pseudomonas aeruginosa ATCC® 27853 were used as quality controls. AST results were interpreted according to the Clinical & Laboratory Standards Institute (CLSI) standard [39].
3.3. DNA Extraction and Sequencing
The genomic DNA from all Enterobacteriaceae isolates were obtained by the TIANamp Bacterial DNA Kit (Tiangen, Beijing, China), followed the manufacturer’s guidelines [40]. The DNA concentrations were assessed using a NanoDrop™ 2000/2000c Spectrophotometer (Thermo Scientific, Norristown, PA, USA), while the integrity and purity of the DNA were verified through Agarose Gel Electrophoresis. Subsequently, the DNA samples were sent to the Beijing Genomics Institute (BGI) for short-read WGS with 150 bp paired-end reads using MGISEG-2000 platform.
3.4. Bioinformatics and Sequence Analysis
The BacSeq pipeline (Accession date: 18 February 2024) [41], a program-based bioinformatic tool for analyzing bacterial genomes, was used for processing Enterobacteriaceae genomes. All analyses and programs used in this study were performed using default parameters. Firstly, FastQC was utilized for quality control of the sequences [42]. The sequences that passed quality control were then assembled using SPAdes [43]. The assembly quality and completeness were checked using Quast and BUSCO [44,45], respectively. Genome annotation was performed using Prokka [46], and all annotated results were employed for downstream analysis. For the downstream analysis, AMRFinderPlus (Accession date: 1 April 2024) [47] was utilized for the detection of antimicrobial resistance, stress response, and virulence genes, while plasmid replicon and plasmid were identified using PlasmidFinder (Accession date: 6 April 2024) and PLSDB (Accession date: 20 May 2024) [48,49]. RStudio version 4.3.2 (Accession date: 22 April 2024) was used for generating figures [50]. To confirm the novel species of Kosakonia spp. PSU27, OrthoANI (Accession date: 12 April 2024) [51] was used to evaluate average nucleotide identity (ANI) of Kosakonia spp. PSU27 and other nine representative clinical isolates of Kosakonia obtained from RefSeq NCBI. An identity value lower than 95% is considered indicative of a novel species. The complete plasmid was selected for comparison with highly similar plasmids available in the NCBI database and was visualized using Proksee (Accession date: 21 April 2024) [52].
4. Conclusions
This study provides valuable insights into the genomic characteristics, antimicrobial resistance profiles, and virulence factors of Enterobacteriaceae strains isolated from hospitals in Southern Thailand. The findings highlight the diversity and complexity of these pathogens, emphasizing their potential to cause a wide range of infections and the challenges they pose to clinical management. The high prevalence of multidrug resistance, particularly to carbapenem antibiotics, is a major concern and underscores the urgent need for effective antimicrobial management programs and the development of new treatment options. The variability in resistance profiles among strains further emphasizes the need for tailored treatment strategies based on accurate identification and susceptibility testing. This necessitates the regular monitoring of resistance patterns and the implementation of precise diagnostic methods, such as whole-genome sequencing, to guide the appropriate therapy. The discovery of a novel species, Kosakonia spp. PSU27, expands our understanding of the genus and highlights the importance of ongoing surveillance for emerging pathogens. Moreover, the presence of various stress response, metal tolerance, and virulence genes underscores the adaptability and pathogenic potential of these bacteria. These genetic traits contribute to their survival in harsh environments and their ability to evade host immune responses, posing significant challenges in both clinical and environmental settings.
Furthermore, we identified plasmids with a high sequence identity to those found in various bacterial species globally, highlighting the extensive horizontal gene transfer occurring across diverse geographic locations and bacterial species. The identification of multiple plasmids in isolates from this study, which match those detected in other regions, emphasizes the global challenge posed by plasmid-mediated resistance. This finding points to the need for robust monitoring systems and effective strategies to limit the dissemination of resistance genes within microbial populations. Our study underscores the critical need for continuous surveillance and increased awareness among healthcare staff regarding these rare, high-virulence, and multidrug-resistant pathogens. Understanding the local epidemiology of these pathogens is vital for informing regional public health strategies and improving infection control practices. The data collected from hospitals in Southern Thailand provide a representative snapshot of the current challenges faced in this area, highlighting the necessity for an enhanced healthcare infrastructure to better manage and mitigate the risks posed by these dangerous pathogens.
Further research is needed to elucidate the mechanisms underlying resistance and virulence in these strains. This includes studying the regulation and expression of resistance and virulence genes, as well as investigating the ecological and evolutionary pressures that drive their emergence and persistence. Understanding these mechanisms will be crucial for developing innovative strategies to combat the spread of multidrug-resistant Enterobacteriaceae.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/antibiotics13060531/s1, Table S1: Demographic data of isolates in this study. Table S2: Stress response, metal resistance, and virulence-associated genes.
Author Contributions
Conceptualization, T.Y., K.S. (Komwit Surachat) and K.S. (Kamonnut Singkhamanan); methodology, T.Y., S.S. and M.W.; software, T.Y., S.S. and K.S. (Komwit Surachat); validation, K.S. (Komwit Surachat), T.Y. and R.P.; formal analysis, T.Y. and K.S. (Komwit Surachat); investigation, T.Y. and K.S. (Komwit Surachat); resources, S.C. and R.P.; data curation, T.Y. and K.S. (Komwit Surachat); writing—original draft preparation, T.Y., S.S., K.S. (Komwit Surachat) and K.S. (Kamonnut Singkhamanan); writing—review and editing, T.Y., K.S. (Komwit Surachat) and K.S. (Kamonnut Singkhamanan); visualization, T.Y. and K.S. (Komwit Surachat); supervision, K.S. (Komwit Surachat) and K.S. (Kamonnut Singkhamanan); project administration, K.S. (Komwit Surachat); funding acquisition, K.S. (Komwit Surachat) and S.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research has received funding support from the NSRF via the Program Management Unit for Human Resources & Institutional Development, Research Innovation (Grant No. B13F670075). In addition, this research was supported by the Postdoctoral Fellowship from Prince of Songkla University, Thailand, and has also received funding support from the NSRF via the Program Management Unit for Human Resources & Institutional Development, Research and Innovation, grant number B13F660074.
Institutional Review Board Statement
This study was conducted in accordance with the Declaration of Helsinki and approved by the Human Research Ethics Committee (HREC) of Prince of Songkla University (protocol code: 64-284-14-1, date of approval: 9 June 2021).
Informed Consent Statement
According to retrospective reviews, the ethical committee allowed the waiver of consent forms.
Data Availability Statement
The assembled genomes of all Enterobacteriaceae isolates in this study have been deposited in the NCBI GenBank under BioProject number PRJNA1080727, with BioSample numbers SAMN40146458 to SAMN40146472.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Bologna, E.; Licari, L.C.; Manfredi, C.; Ditonno, F.; Cirillo, L.; Fusco, G.M.; Abate, M.; Passaro, F.; Di Mauro, E.; Crocetto, F.; et al. Carbapenem-Resistant Enterobacteriaceae in Urinary Tract Infections: From Biological Insights to Emerging Therapeutic Alternatives. Medicina 2024, 60, 214. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.W.K.; Millar, B.C.; Moore, J.E. Antimicrobial Resistance (AMR). Br. J. Biomed. Sci. 2023, 80, 11387. [Google Scholar] [CrossRef] [PubMed]
- Poudel, A.N.; Zhu, S.; Cooper, N.; Little, P.; Tarrant, C.; Hickman, M.; Yao, G. The economic burden of antibiotic resistance: A systematic review and meta-analysis. PLoS ONE 2023, 18, e0285170. [Google Scholar] [CrossRef] [PubMed]
- Logan, L.K.; Weinstein, R.A. The Epidemiology of Carbapenem-Resistant Enterobacteriaceae: The Impact and Evolution of a Global Menace. J. Infect. Dis. 2017, 215, S28–S36. [Google Scholar] [CrossRef] [PubMed]
- Tilahun, M.; Kassa, Y.; Gedefie, A.; Ashagire, M. Emerging Carbapenem-Resistant Enterobacteriaceae Infection, Its Epidemiology and Novel Treatment Options: A Review. Infect. Drug Resist. 2021, 14, 4363–4374. [Google Scholar] [CrossRef] [PubMed]
- National Center for Biotechnology Information. Kosakonia Assemblies. RefSeq NCBI. Available online: https://www.ncbi.nlm.nih.gov/assembly/?term=Kosakonia (accessed on 12 April 2024).
- Chen, L.; Todd, R.; Kiehlbauch, J.; Walters, M.; Kallen, A. Notes from the Field: Pan-Resistant New Delhi Metallo-Beta-Lactamase-Producing Klebsiella pneumoniae—Washoe County, Nevada, 2016. MMWR. Morb. Mortal. Wkly. Rep. 2017, 66, 33. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, H.; Zhang, Y.; Wang, Q.; Zhao, C.; Li, H.; He, W.; Zhang, F.; Wang, Z.; Li, S.; et al. Genetic characterisation of clinical Klebsiella pneumoniae isolates with reduced susceptibility to tigecycline: Role of the global regulator RamA and its local repressor RamR. Int. J. Antimicrob. Agents 2015, 45, 635–640. [Google Scholar] [CrossRef] [PubMed]
- Ho, P.L.; Chan, J.; Lo, W.U.; Lai, E.L.; Cheung, Y.Y.; Lau, T.C.K.; Chow, K.H. Prevalence and molecular epidemiology of plasmid-mediated fosfomycin resistance genes among blood and urinary Escherichia coli isolates. J. Med. Microbiol. 2013, 62, 1707–1713. [Google Scholar] [CrossRef]
- Moosavian, M.; Khoshkholgh Sima, M.; Ahmad Khosravi, N.; Abbasi Montazeri, E. Detection of OqxAB Efflux Pumps, a Multidrug-Resistant Agent in Bacterial Infection in Patients Referring to Teaching Hospitals in Ahvaz, Southwest of Iran. Int. J. Microbiol. 2021, 2021, 2145176. [Google Scholar] [CrossRef] [PubMed]
- Kerluku, M.; Ratkova Manovska, M.; Prodanov, M.; Stojanovska-Dimzoska, B.; Hajrulai-Musliu, Z.; Jankuloski, D.; Blagoevska, K. Phenotypic and Genotypic Analysis of Antimicrobial Resistance of Commensal Escherichia coli from Dairy Cows’ Feces. Processes 2023, 11, 1929. [Google Scholar] [CrossRef]
- Bobate, S.; Mahalle, S.; Dafale, N.A.; Bajaj, A. Emergence of environmental antibiotic resistance: Mechanism, monitoring and management. Environ. Adv. 2023, 13, 100409. [Google Scholar] [CrossRef]
- Li, K.; Xu, P.; Wang, J.; Yi, X.; Jiao, Y. Identification of errors in draft genome assemblies at single-nucleotide resolution for quality assessment and improvement. Nat. Commun. 2023, 14, 6556. [Google Scholar] [CrossRef]
- Amereh, F.; Arabestani, M.R.; Shokoohizadeh, L. Relationship of OqxAB efflux pump to antibiotic resistance, mainly fluoroquinolones in Klebsiella pneumoniae, isolated from hospitalized patients. Iran. J. Basic Med. Sci. 2023, 26, 93–98. [Google Scholar] [CrossRef]
- da Cruz Nizer, W.S.; Inkovskiy, V.; Versey, Z.; Strempel, N.; Cassol, E.; Overhage, J. Oxidative Stress Response in Pseudomonas aeruginosa. Pathogens 2021, 10, 1187. [Google Scholar] [CrossRef]
- Matsuo, T.; Chen, J.; Minato, Y.; Ogawa, W.; Mizushima, T.; Kuroda, T.; Tsuchiya, T. SmdAB, a heterodimeric ABC-Type multidrug efflux pump, in Serratia marcescens. J. Bacteriol. 2008, 190, 648–654. [Google Scholar] [CrossRef]
- Dalvi, S.D.; Worobec, E.A. Gene expression analysis of the SdeAB multidrug efflux pump in antibiotic-resistant clinical isolates of Serratia marcescens. Indian J. Med. Microbiol. 2012, 30, 302–307. [Google Scholar] [CrossRef]
- Minato, Y.; Shahcheraghi, F.; Ogawa, W.; Kuroda, T.; Tsuchiya, T. Functional gene cloning and characterization of the SsmE multidrug efflux pump from Serratia marcescens. Biol. Pharm. Bull. 2008, 31, 516–519. [Google Scholar] [CrossRef]
- Nguyen, T.H.T.; Nguyen, H.D.; Le, M.H.; Nguyen, T.T.H.; Nguyen, T.D.; Nguyen, D.L.; Nguyen, Q.H.; Nguyen, T.K.O.; Michalet, S.; Dijoux-Franca, M.G.; et al. Efflux Pump Inhibitors in Controlling Antibiotic Resistance: Outlook under a Heavy Metal Contamination Context. Molecules 2023, 28, 2912. [Google Scholar] [CrossRef]
- Vats, P.; Kaur, U.J.; Rishi, P. Heavy metal-induced selection and proliferation of antibiotic resistance: A review. J. Appl. Microbiol. 2022, 132, 4058–4076. [Google Scholar] [CrossRef]
- Mohsen, Y.; Tarchichi, N.; Barakat, R.; Kawtharani, I.; Ghandour, R.; Ezzeddine, Z.; Ghssein, G. The Different Types of Metallophores Produced by Salmonella enterica: A Review. Microbiol. Res. 2023, 14, 1457–1469. [Google Scholar] [CrossRef]
- Koh, E.I.; Hung, C.S.; Henderson, J.P. The Yersiniabactin-Associated ATP Binding Cassette Proteins YbtP and YbtQ Enhance Escherichia coli Fitness during High-Titer Cystitis. Infect. Immun. 2016, 84, 1312–1319. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.; Wang, Y.; Fu, Y.; Deng, Z.; Lin, S.; Liang, R. Characterization of the Tellurite-Resistance Properties and Identification of the Core Function Genes for Tellurite Resistance in Pseudomonas citronellolis SJTE-3. Microorganisms 2022, 10, 95. [Google Scholar] [CrossRef] [PubMed]
- Kovács, J.K.; Felső, P.; Horváth, G.; Schmidt, J.; Dorn, Á.; Ábrahám, H.; Cox, A.; Márk, L.; Emődy, L.; Kovács, T.; et al. Stress Response and Virulence Potential Modulating Effect of Peppermint Essential Oil in Campylobacter jejuni. BioMed Res. Int. 2019, 2019, 2971741. [Google Scholar] [CrossRef] [PubMed]
- Mourão, J.; Magalhães, M.; Ribeiro-Almeida, M.; Rebelo, A.; Novais, C.; Peixe, L.; Novais, Â.; Antunes, P. Decoding Klebsiella pneumoniae in poultry chain: Unveiling genetic landscape, antibiotic resistance, and biocide tolerance in non-clinical reservoirs. Front. Microbiol. 2024, 15, 1365011. [Google Scholar] [CrossRef] [PubMed]
- Abd Elnabi, M.K.; Elkaliny, N.E.; Elyazied, M.M.; Azab, S.H.; Elkhalifa, S.A.; Elmasry, S.; Mouhamed, M.S.; Shalamesh, E.M.; Alhorieny, N.A.; Abd Elaty, A.E.; et al. Toxicity of Heavy Metals and Recent Advances in Their Removal: A Review. Toxics 2023, 11, 580. [Google Scholar] [CrossRef] [PubMed]
- Papagiannitsis, C.C.; Dolejska, M.; Izdebski, R.; Giakkoupi, P.; Skálová, A.; Chudějová, K.; Dobiasova, H.; Vatopoulos, A.C.; Derde, L.P.; Bonten, M.J.; et al. Characterisation of IncA/C2 plasmids carrying an In416-like integron with the blaVIM-19 gene from Klebsiella pneumoniae ST383 of Greek origin. Int. J. Antimicrob. Agents 2016, 47, 158–162. [Google Scholar] [CrossRef] [PubMed]
- Pauly, N.; Hammerl, J.A.; Grobbel, M.; Käsbohrer, A.; Tenhagen, B.A.; Malorny, B.; Schwarz, S.; Meemken, D.; Irrgang, A. Identification of a bla(VIM-1)-Carrying IncA/C(2) Multiresistance Plasmid in an Escherichia coli Isolate Recovered from the German Food Chain. Microorganisms 2020, 9, 29. [Google Scholar] [CrossRef] [PubMed]
- von Wintersdorff, C.J.; Penders, J.; van Niekerk, J.M.; Mills, N.D.; Majumder, S.; van Alphen, L.B.; Savelkoul, P.H.; Wolffs, P.F. Dissemination of Antimicrobial Resistance in Microbial Ecosystems through Horizontal Gene Transfer. Front. Microbiol. 2016, 7, 173. [Google Scholar] [CrossRef] [PubMed]
- Marí-Almirall, M.; Ferrando, N.; Fernández, M.J.; Cosgaya, C.; Viñes, J.; Rubio, E.; Cuscó, A.; Muñoz, L.; Pellice, M.; Vergara, A.; et al. Clonal Spread and Intra- and Inter-Species Plasmid Dissemination Associated with Klebsiella pneumoniae Carbapenemase-Producing Enterobacterales During a Hospital Outbreak in Barcelona, Spain. Front. Microbiol. 2021, 12, 781127. [Google Scholar] [CrossRef]
- Peleg, A.Y.; Hooper, D.C. Hospital-acquired infections due to gram-negative bacteria. N. Engl. J. Med. 2010, 362, 1804–1813. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, H.; Yu, S.; Li, D.; Gillings, M.R.; Ren, H.; Mao, D.; Guo, J.; Luo, Y. Inter-plasmid transfer of antibiotic resistance genes accelerates antibiotic resistance in bacterial pathogens. ISME J. 2024, 18, wrad032. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Xu, Y.; Wang, M.; Li, X.; Liu, Z.; Kuang, D.; Deng, Z.; Ou, H.-Y.; Qu, J. Mobilizable plasmids drive the spread of antimicrobial resistance genes and virulence genes in Klebsiella pneumoniae. Genome Med. 2023, 15, 106. [Google Scholar] [CrossRef]
- Yaikhan, T.; Chukamnerd, A.; Singkhamanan, K.; Nokchan, N.; Chintakovid, N.; Chusri, S.; Pomwised, R.; Wonglapsuwan, M.; Surachat, K. Genomic Characterization of Mobile Genetic Elements Associated with Multidrug-Resistant Acinetobacter Non-baumannii Species from Southern Thailand. Antibiotics 2024, 13, 149. [Google Scholar] [CrossRef] [PubMed]
- Chukamnerd, A.; Pomwised, R.; Chusri, S.; Singkhamanan, K.; Chumtong, S.; Jeenkeawpiam, K.; Sakunrang, C.; Saroeng, K.; Saengsuwan, P.; Wonglapsuwan, M.; et al. Antimicrobial Susceptibility and Molecular Features of Colonizing Isolates of Pseudomonas aeruginosa and the Report of a Novel Sequence Type (ST) 3910 from Thailand. Antibiot. Antibiot. 2023, 12, 165. [Google Scholar] [CrossRef] [PubMed]
- Chukamnerd, A.; Singkhamanan, K.; Chongsuvivatwong, V.; Palittapongarnpim, P.; Doi, Y.; Pomwised, R.; Sakunrang, C.; Jeenkeawpiam, K.; Yingkajorn, M.; Chusri, S.; et al. Whole-genome analysis of carbapenem-resistant Acinetobacter baumannii from clinical isolates in Southern Thailand. Comput. Struct. Biotechnol. J. 2022, 20, 545–558. [Google Scholar] [CrossRef] [PubMed]
- Paul De Vos, E. Bergey’s Manual of Systematic Bacteriology. Volume Three, The Firmicutes, 2nd ed.; Springer: Dordrecht, The Netherlands; New York, NY, USA, 2009. [Google Scholar]
- Hudzicki, J. Kirby-Bauer Disk Diffusion Susceptibility Test Protocol; American Society for Microbiology: Washington, DC, USA, 2009. [Google Scholar]
- Weinstein Melvin, P.; Lewis James, S. The Clinical and Laboratory Standards Institute Subcommittee on Antimicrobial Susceptibility Testing: Background, Organization, Functions, and Processes. J. Clin. Microbiol. 2020, 58, 3. [Google Scholar] [CrossRef] [PubMed]
- TIANamp Bacterial DNA Kit: User Manual; Tiangen Biotech: Bejing, China, 2021.
- Chukamnerd, A.; Jeenkeawpiam, K.; Chusri, S.; Pomwised, R.; Singkhamanan, K.; Surachat, K. BacSeq: A User-Friendly Automated Pipeline for Whole-Genome Sequence Analysis of Bacterial Genomes. Microorganisms 2023, 11, 1769. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 18 February 2024).
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. J. Comput. Mol. Cell Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Feldgarden, M.; Brover, V.; Gonzalez-Escalona, N.; Frye, J.G.; Haendiges, J.; Haft, D.H.; Hoffmann, M.; Pettengill, J.B.; Prasad, A.B.; Tillman, G.E.; et al. AMRFinderPlus and the Reference Gene Catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci. Rep. 2021, 11, 12728. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Galata, V.; Fehlmann, T.; Backes, C.; Keller, A. PLSDB: A resource of complete bacterial plasmids. Nucleic Acids Res. 2019, 47, D195–D202. [Google Scholar] [CrossRef] [PubMed]
- R Studio Team. RStudio: Integrated Development for R; RStudio, PBC: Boston, MA, USA, 2022. [Google Scholar]
- Lee, I.; Ouk Kim, Y.; Park, S.C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.-Y.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).