Design and Synthesis of Novel Amino and Acetamidoaurones with Antimicrobial Activities
Abstract
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Biology
4.1.1. Microorganism Strains and Growth Conditions
4.1.2. Antimicrobial Activity Assay
4.1.3. Cytotoxic Assays
4.2. Chemistry
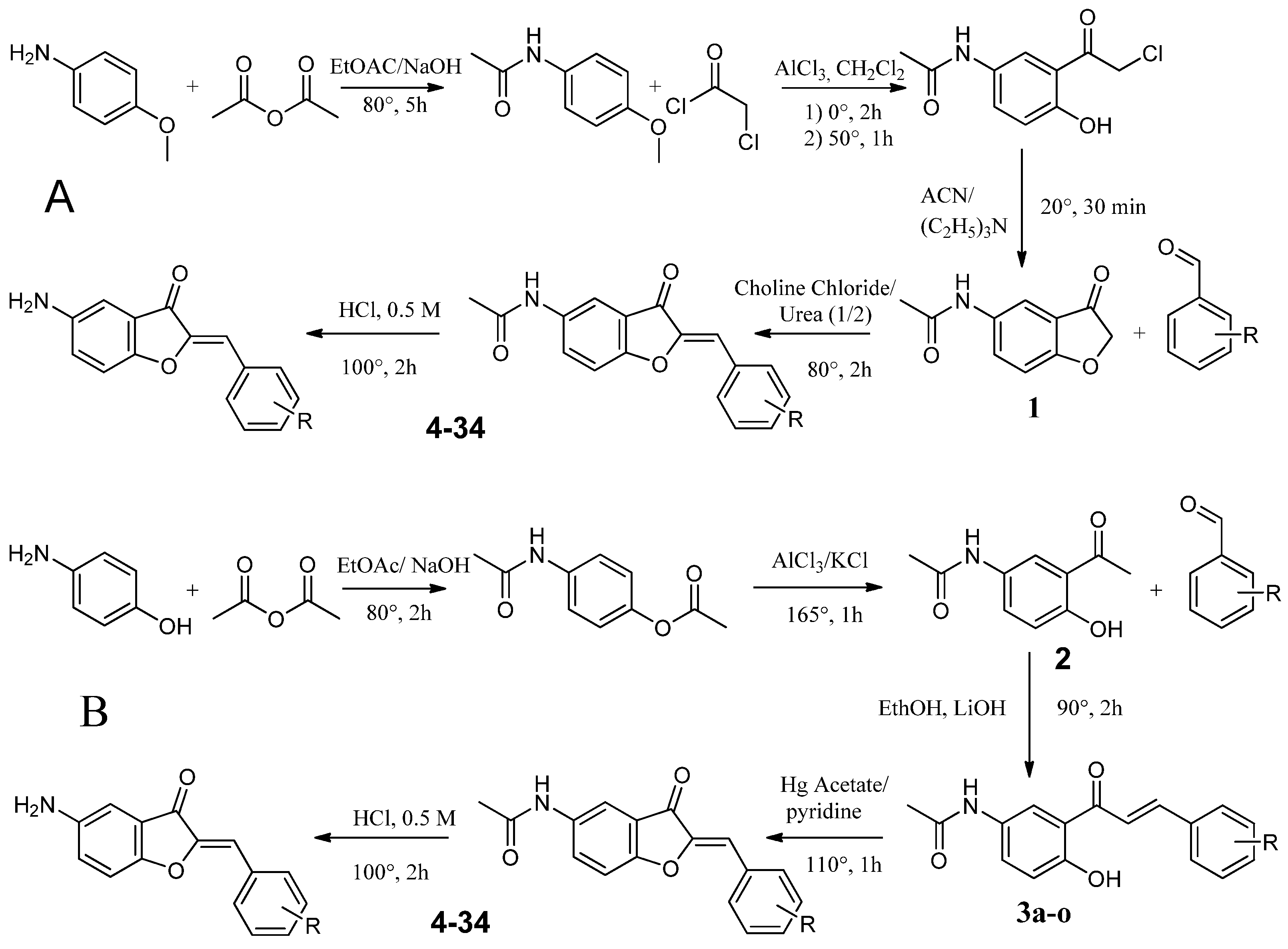
4.2.1. Synthesis Route A
Synthesis of N-(4-methoxyphenyl)acetamide (1a)
Synthesis of 2-Chloro-1-{2-hydroxy-5-[(1-hydroxyethyl)amino]cyclohexyl}ethan-1-one (2a)
Synthesis of N-(3-oxo-2,3-dihydro-1-benzofuran-5-yl)acetamide (3a)
Synthesis of Substituted 5-Acetamidoaurones
Synthesis of Substituted 5-Aminoaurone
4.2.2. Synthesis Route B
Synthesis of 4-Acetamidophenyl Acetate (1b)
Synthesis of N-(3-acetyl-4-hydroxyphenyl)acetamide (2b)
Synthesis of Substituted of 5-Acetamidochalcones (3b)
Synthesis of Substituted 5-Acetamidoaurones
Synthesis of Substituted 5-Aminoaurone
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- O’Neill, A. New Antibacterial Agents for Treating Infections Caused by Multi-Drug Resistant Gram-Negative Bacteria. Expert Opin. Investig. Drugs 2008, 17, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Abdeen, S.; Kunkle, T.; Salim, N.; Ray, A.-M.; Mammadova, N.; Summers, C.; Stevens, M.; Ambrose, A.J.; Park, Y.; Schultz, P.G.; et al. Sulfonamido-2-Arylbenzoxazole GroEL/ES Inhibitors as Potent Antibacterials against Methicillin-Resistant Staphylococcus Aureus (MRSA). J. Med. Chem. 2018, 61, 7345–7357. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Wang, L.; Lü, Y.; Yue, C. Development and Research Progress of Anti-Drug Resistant Fungal Drugs. J. Infect. Public Health 2022, 15, 986–1000. [Google Scholar] [CrossRef]
- Bastos, R.W.; Rossato, L.; Goldman, G.H.; Santos, D.A. Fungicide Effects on Human Fungal Pathogens: Cross-Resistance to Medical Drugs and Beyond. PLoS Pathog. 2021, 17, e1010073. [Google Scholar] [CrossRef] [PubMed]
- Sati, H.; Alastruey-Izquierdo, A.; Perfect, J.; Govender, N.P.; Harrison, T.S.; Chiller, T.; Sorrell, T.C.; Bongomin, F.; Oladele, R.; Chakrabarti, A.; et al. HIV and Fungal Priority Pathogens. Lancet HIV 2023, 10, e750–e754. [Google Scholar] [CrossRef] [PubMed]
- Bongomin, F.; Gago, S.; Oladele, R.O.; Denning, D.W. Global and Multi-National Prevalence of Fungal Diseases—Estimate Precision. J. Fungi 2017, 3, 57. [Google Scholar] [CrossRef] [PubMed]
- Haudecoeur, R.; Boumendjel, A. Recent Advances in the Medicinal Chemistry of Aurones. Curr. Med. Chem. 2012, 19, 2861–2875. [Google Scholar] [CrossRef] [PubMed]
- Boumendjel, A. Aurones: A Subclass of Flavones with Promising Biological Potential. Curr. Med. Chem. 2003, 10, 2621–2630. [Google Scholar] [CrossRef]
- Lazinski, L.M.; Royal, G.; Robin, M.; Maresca, M.; Haudecoeur, R. Bioactive Aurones, Indanones, and Other Hemiindigoid Scaffolds: Medicinal Chemistry and Photopharmacology Perspectives. J. Med. Chem. 2022, 65, 12594–12625. [Google Scholar] [CrossRef]
- Boucherle, B.; Peuchmaur, M.; Boumendjel, A.; Haudecoeur, R. Occurrences, Biosynthesis and Properties of Aurones as High-End Evolutionary Products. Phytochemistry 2017, 142, 92–111. [Google Scholar] [CrossRef]
- Pare, P.W.; Dmitrieva, N.; Mabry, T.J. Phytoalexin Aurone Induced in Cephalocereus Senilis Liquid Suspension Culture. Phytochemistry 1991, 30, 1133–1135. [Google Scholar] [CrossRef]
- Paré, P.W.; Mischke, C.F.; Edwards, R.; Dixon, R.A.; Norman, H.A.; Mabry, T.J. Induction of Phenylpropanoid Pathway Enzymes in Elicitor-Treated Cultures ofCephalocereus Senilis. Phytochemistry 1992, 31, 149–153. [Google Scholar] [CrossRef]
- Farag, M.A.; Deavours, B.E.; de Fátima, A.; Naoumkina, M.; Dixon, R.A.; Sumner, L.W. Integrated Metabolite and Transcript Profiling Identify a Biosynthetic Mechanism for Hispidol in Medicago Truncatula Cell Cultures. Plant Physiol. 2009, 151, 1096–1113. [Google Scholar] [CrossRef] [PubMed]
- Olleik, H.; Yahiaoui, S.; Roulier, B.; Courvoisier-Dezord, E.; Perrier, J.; Pérès, B.; Hijazi, A.; Baydoun, E.; Raymond, J.; Boumendjel, A.; et al. Aurone Derivatives as Promising Antibacterial Agents against Resistant Gram-Positive Pathogens. Eur. J. Med. Chem. 2019, 165, 133–141. [Google Scholar] [CrossRef]
- Tiwari, K.N.; Monserrat, J.-P.; Hequet, A.; Ganem-Elbaz, C.; Cresteil, T.; Jaouen, G.; Vessières, A.; Hillard, E.A.; Jolivalt, C. In Vitro Inhibitory Properties of Ferrocene-Substituted Chalcones and Aurones on Bacterial and Human Cell Cultures. Dalton Trans. 2012, 41, 6451–6457. [Google Scholar] [CrossRef] [PubMed]
- Hadj-esfandiari, N.; Navidpour, L.; Shadnia, H.; Amini, M.; Samadi, N.; Faramarzi, M.A.; Shafiee, A. Synthesis, Antibacterial Activity, and Quantitative Structure–Activity Relationships of New (Z)-2-(Nitroimidazolylmethylene)-3(2H)-Benzofuranone Derivatives. Bioorganic Med. Chem. Lett. 2007, 17, 6354–6363. [Google Scholar] [CrossRef] [PubMed]
- Kumar, G.; Lathwal, E.; Saroha, B.; Kumar, S.; Kumar, S.; Chauhan, N.S.; Kumar, T. Synthesis and Biological Evaluation of Quinoline-Based Novel Aurones. ChemistrySelect 2020, 5, 3539–3543. [Google Scholar] [CrossRef]
- Olleik, H.; Nicoletti, C.; Lafond, M.; Courvoisier-Dezord, E.; Xue, P.; Hijazi, A.; Baydoun, E.; Perrier, J.; Maresca, M. Comparative Structure-Activity Analysis of the Antimicrobial Activity, Cytotoxicity, and Mechanism of Action of the Fungal Cyclohexadepsipeptides Enniatins and Beauvericin. Toxins 2019, 11, 514. [Google Scholar] [CrossRef]
- Berthier, M.; Fauchère, J.-L.; Perrin, J.; Grignon, B.; Oriot, D. Fulminant Meningitis Due to Bacillus Anthracis in 11-Year-Old Girl during Ramadan. Lancet 1996, 347, 828. [Google Scholar] [CrossRef]
- Doll, J.M.; Zeitz, P.S.; Ettestad, P.; Bucholtz, A.L.; Davis, T.; Gage, K. Cat-Transmitted Fatal Pneumonic Plague in a Person Who Traveled from Colorado to Arizona. Am. J. Trop. Med. Hyg. 1994, 51, 109–114. [Google Scholar] [CrossRef]
- Olleik, H.; Yacoub, T.; Hoffer, L.; Gnansounou, S.M.; Benhaiem-Henry, K.; Nicoletti, C.; Mekhalfi, M.; Pique, V.; Perrier, J.; Hijazi, A.; et al. Synthesis and Evaluation of the Antibacterial Activities of 13-Substituted Berberine Derivatives. Antibiotics 2020, 9, 381. [Google Scholar] [CrossRef] [PubMed]
- Casanova, M.; Olleik, H.; Hdiouech, S.; Roblin, C.; Cavalier, J.-F.; Point, V.; Jeannot, K.; Caron, B.; Perrier, J.; Charriau, S.; et al. Evaluation of the Efficiency of Random and Diblock Methacrylate-Based Amphiphilic Cationic Polymers against Major Bacterial Pathogens Associated with Cystic Fibrosis. Antibiotics 2023, 12, 120. [Google Scholar] [CrossRef] [PubMed]
- Loupias, P.; Laumaillé, P.; Morandat, S.; Mondange, L.; Guillier, S.; El Kirat, K.; Da Nascimento, S.; Biot, F.; Taudon, N.; Dassonville-Klimpt, A.; et al. Synthesis and Study of New Siderophore Analog-Ciprofloxacin Conjugates with Antibiotic Activities against Pseudomonas aeruginosa and Burkholderia spp. Eur. J. Med. Chem. 2023, 245, 114921. [Google Scholar] [CrossRef]
- Broncano-Lavado, A.; Senhaji-Kacha, A.; Santamaría-Corral, G.; Esteban, J.; García-Quintanilla, M. Alternatives to Antibiotics against Mycobacterium Abscessus. Antibiotics 2022, 11, 1322. [Google Scholar] [CrossRef] [PubMed]
- Madani, A.; Ridenour, J.N.; Martin, B.P.; Paudel, R.R.; Abdul Basir, A.; Le Moigne, V.; Herrmann, J.-L.; Audebert, S.; Camoin, L.; Kremer, L.; et al. Cyclipostins and Cyclophostin Analogues as Multitarget Inhibitors That Impair Growth of Mycobacterium Abscessus. ACS Infect. Dis. 2019, 5, 1597–1608. [Google Scholar] [CrossRef]
- Benkhaled, B.T.; Hadiouch, S.; Olleik, H.; Perrier, J.; Ysacco, C.; Guillaneuf, Y.; Gigmes, D.; Maresca, M.; Lefay, C. Elaboration of Antimicrobial Polymeric Materials by Dispersion of Well-Defined Amphiphilic Methacrylic SG1-Based Copolymers. Polym. Chem. 2018, 9, 3127–3141. [Google Scholar] [CrossRef]
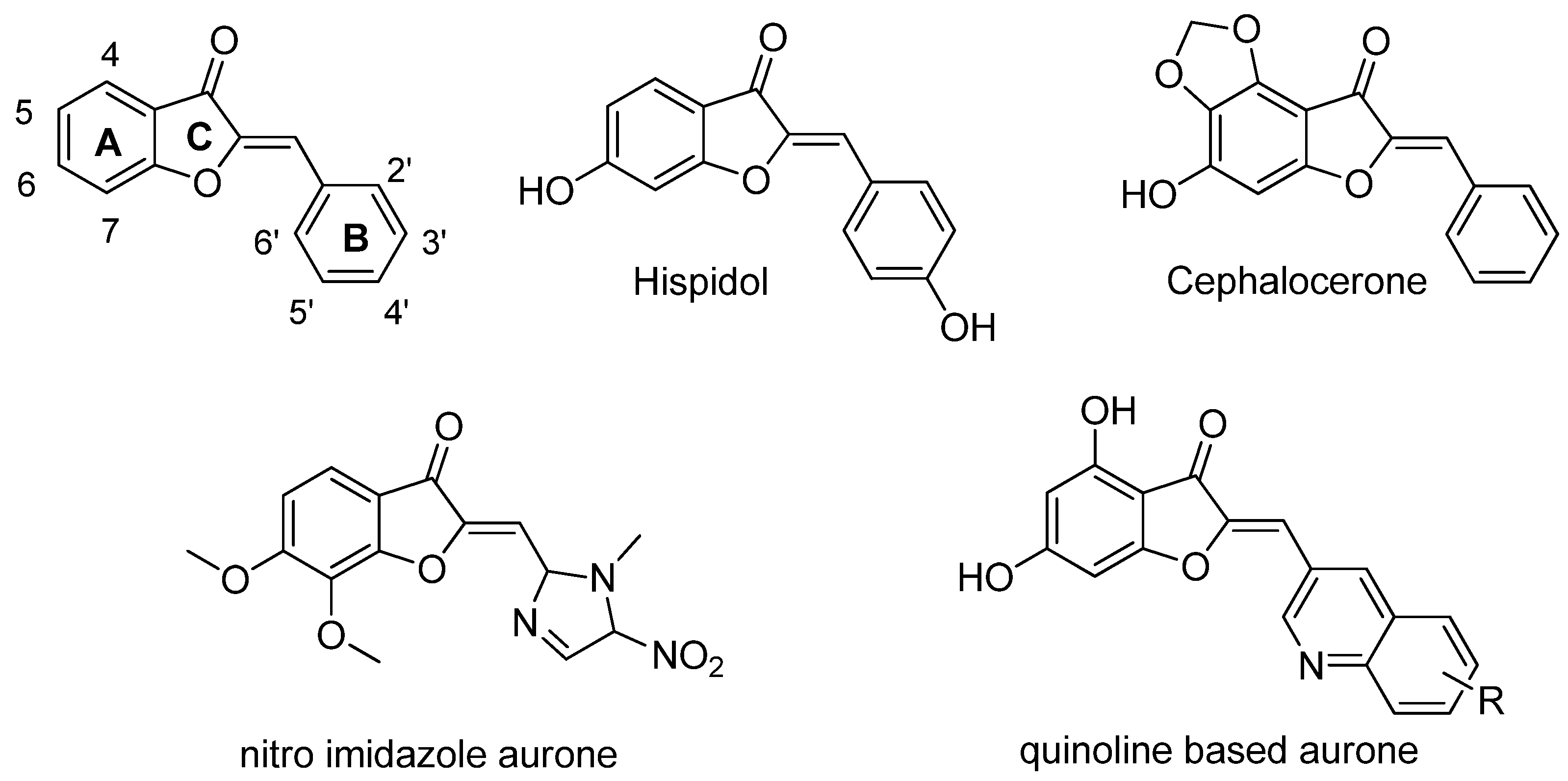

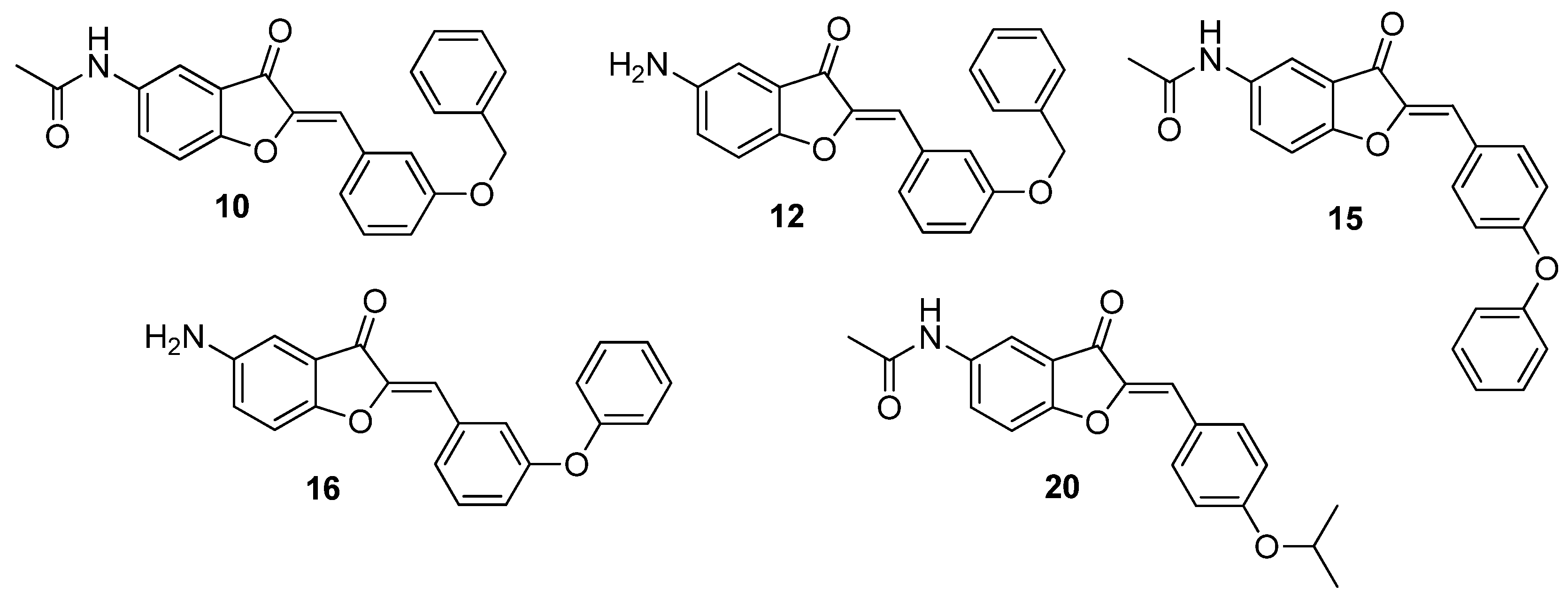

| Compound | 5 | 2′ | 3′ | 4′ |
|---|---|---|---|---|
| 4 | NHCOCH3 | OCH3 | H | H |
| 5 | NHCOCH3 | H | OCH3 | H |
| 6 | NHCOCH3 | H | H | OCH3 |
| 7 | NH2 | OCH3 | H | H |
| 8 | NH2 | H | OCH3 | H |
| 9 | NH2 | H | H | OCH3 |
| 10 | NHCOCH3 | H | OBn | H |
| 11 | NHCOCH3 | H | H | OBn |
| 12 | NH2 | H | OBn | H |
| 13 | NH2 | H | H | OBn |
| 14 | NHCOCH3 | H | OPh | H |
| 15 | NHCOCH3 | H | H | OPh |
| 16 | NH2 | H | OPh | H |
| 17 | NH2 | H | H | OPh |
| 18 | NHCOCH3 | OiPr | H | H |
| 19 | NHCOCH3 | H | OiPr | H |
| 20 | NHCOCH3 | H | H | OiPr |
| 21 | NH2 | OiPr | H | H |
| 22 | NH2 | H | OiPr | H |
| 23 | NH2 | H | H | OiPr |
| 24 | NHCOCH3 | F | H | H |
| 25 | NHCOCH3 | H | F | H |
| 26 | NH2 | F | H | H |
| 27 | NH2 | H | F | H |
| 28 | NH2 | H | H | F |
| 29 | NHCOCH3 | H | CF3 | H |
| 30 | NH2 | H | CF3 | H |
| 31 | NHCOCH3 | H | H | COOH |
| 32 | NHCOCH3 | H | OH | H |
| 33 | NH2 | H | OH | H |
| 34 | NHCOCH3 | H | H | H |
| Gram-Positive | Gram-Negative | Mycobacteria | Fungi | |||
|---|---|---|---|---|---|---|
| B. subtilis ATCC6633 | S. aureus ATCC6538P | E. coli ATCC8739 | P. aeruginosa ATCC9027 | M. smegmatis ATCC700084 | C. albicans DSM10697 | |
| Gemifloxacin | 0.03 | 0.06 | 0.06 | 0.25 | 2 | - |
| Amphotericin B | - | - | - | - | - | 0.62 |
| 4 | 50 | 100 | >100 | >100 | >100 | >100 |
| 5 | >100 | 100 | >100 | >100 | 100 | >100 |
| 6 | >100 | >100 | >100 | >100 | >100 | >100 |
| 7 | >100 | >100 | >100 | >100 | >100 | >100 |
| 8 | >100 | >100 | >100 | >100 | >100 | >100 |
| 9 | >100 | >100 | >100 | >100 | >100 | >100 |
| 10 | 3.12 | 12.5 | 12.5 | 25 | 50 | 50 |
| 11 | >100 | >100 | >100 | >100 | >100 | >100 |
| 12 | 25 | 100 | >100 | >100 | 50 | >100 |
| 13 | 100 | >100 | >100 | >100 | >100 | >100 |
| 14 | >100 | >100 | >100 | >100 | >100 | >100 |
| 15 | 50 | 100 | >100 | >100 | 50 | >100 |
| 16 | 25 | 25 | 25 | >100 | 50 | >100 |
| 17 | >100 | >100 | >100 | >100 | >100 | >100 |
| 18 | >100 | >100 | >100 | >100 | >100 | >100 |
| 19 | >100 | >100 | >100 | 25 | >100 | >100 |
| 20 | 25 | 12.5 | 25 | >100 | 50 | 50 |
| 21 | >100 | >100 | >100 | >100 | >100 | >100 |
| 22 | >100 | >100 | >100 | >100 | >100 | >100 |
| 23 | >100 | >100 | >100 | >100 | >100 | >100 |
| 24 | >100 | >100 | >100 | >100 | >100 | >100 |
| 25 | 50 | >100 | >100 | >100 | 100 | >100 |
| 26 | 50 | >100 | >100 | >100 | >100 | >100 |
| 27 | >100 | >100 | >100 | >100 | >100 | >100 |
| 28 | >100 | >100 | >100 | >100 | >100 | >100 |
| 29 | >100 | >100 | >100 | >100 | >100 | >100 |
| 30 | >100 | >100 | >100 | >100 | >100 | >100 |
| 31 | 50 | 100 | >100 | >100 | 100 | >100 |
| 32 | >100 | >100 | >100 | >100 | >100 | >100 |
| 33 | >100 | >100 | >100 | >100 | >100 | >100 |
| 34 | 50 | >100 | >100 | >100 | >100 | >100 |
| Compound | 10 | 12 | 15 | 16 | 20 |
|---|---|---|---|---|---|
| A498 | 398.2 +/− 164.6 | 152.4 +/− 31.4 | 145.7 +/− 17.3 | 452.3 +/− 147.9 | 453.0 +/− 46.4 |
| BEAS-2B | 169.0 +/− 28.3 | 74.6 +/− 12.1 | 109.5 +/− 17.4 | 129.6 +/− 18.3 | 125.9 +/− 17.2 |
| Caco-2 | 199.6 +/− 33.5 | 111.7 +/− 15.0 | 136.8 +/− 7.2 | 131.7 +/− 18.0 | 186.5 +/− 27.8 |
| HaCaT | 268.5 +/− 51.6 | 51.3 +/− 9.5 | >500 | 80.4 +/− 17.8 | 322.9 +/− 73.2 |
| HepG2 | 472.4 +/− 145.9 | 343.4 +/− 68.0 | >500 | 397.8 +/− 94.1 | >500 |
| IMR-90 | 421.4 +/− 119.3 | 42.4 +/− 9.6 | >500 | 116.5 +/− 29.4 | 437.6 +/− 128 |
| Mean CC50 | 321.5 | 129.3 | 130.6 | 218.0 | 305.1 |
| Compound | 10 | 12 | 15 | 16 | 20 |
|---|---|---|---|---|---|
| MIC on S. aureus | 12.5 | 100 | 100 | 25 | 12.5 |
| Lowest CC50 | 169.0 | 42.4 | 109.5 | 80.4 | 125.9 |
| Highest CC50 | 472.4 | 343.4 | >500 | 452.3 | >500 |
| Lowest TI | 13.5 | 0.4 | 1.0 | 3.2 | 10.0 |
| Highest TI | 37.7 | 3.4 | >5 | 18.0 | >40 |
| 10 | 20 | |
|---|---|---|
| Gram-positive | ||
| Bacillus anthracis (CNR-charbon_04022) | 12.5 | 6.25 |
| Bacillus cereus (DSM31) | 12.5 | 25 |
| Bacillus subtilis (ATCC6633) | 3.12 | 25 |
| Clostridium botulinum (DSM1985) | 0.78 | 3.12 |
| Clostridium difficile (DSM1296) | 12.5 | 3.12 |
| Clostridium perfringens (ATCC13124) | >100 | >100 |
| Enterococcus faecalis (DSM2570) | 50 | 100 |
| Enterococcus faecium (DSM20477) | 100 | 25 |
| Listeria monocytogenes (DSM20600) | 3.12 | 6.25 |
| Propionibacterium acnes (ATCC6919) | 100 | >100 |
| Staphylococcus aureus (ATCC6538P) | 12.5 | 12.5 |
| MRSA (ATCCBAA-1717) | 12.5 | 25 |
| Streptococcus pyogenes (DSM20565) | 50 | 50 |
| Gram-negative | ||
| Acinetobacter baumannii (DSM30007) | 12.5 | 25 |
| Brucella melitensis (NR-256) | 25 | 12.5 |
| Enterobacter cloacae (DSM30054) | >100 | >100 |
| Escherichia coli (ATCC8739) | 12.5 | 25 |
| Francisella tularensis (NR-643) | 50 | 12.5 |
| Helicobacter pylori (ATCC43504) | 12.5 | 12.5 |
| Klebsiella pneumonia (DSM26371) | >100 | >100 |
| Pseudomonas aeruginosa (ATCC9027) | 25 | >100 |
| Salmonella enterica (CIP80.39) | 25 | 50 |
| Shigella flexneri (ATCC12022) | 25 | 50 |
| Vibrio alginolyticus (DSM2171) | 25 | 50 |
| Yersinia pestis (NR-641) | 12.5 | 12.5 |
| Mycobacteria | ||
| Mycobacterium abscessus S (CIP 104536T) | >100 | >100 |
| Mycobacterium abscessus R(CIP 104536T) | >100 | >100 |
| Mycobacterium smegmatis (ATCC700084) | 50 | 50 |
| Mycobacterium tuberculosis H37Rv (mc26230) | >100 | >100 |
| Filamentous fungi | ||
| Aspergillus fumigatus (DSM819) | >100 | >100 |
| Fusarium oxysporum (DSM62316) | 25 | 25 |
| Yeasts | ||
| Candida albicans (DSM10697) | 50 | 50 |
| Candida auris (DSM21092) | 50 | 12.5 |
| Candida glabrata (DSM11226) | 50 | 50 |
| Candida tropicalis (DSM9419) | 100 | 100 |
| Cryptococcus neoformans (DSM11959) | 25 | 25 |
| A498 | BEAS-2B | Caco-2 | HaCaT | HepG-2 | IMR-90 | |
|---|---|---|---|---|---|---|
| CC50 (µM) | 398.2 | 169.0 | 199.6 | 268.5 | 472.4 | 421.4 |
| TI with MIC of 0.78 µM | 510.5 | 216.6 | 255.8 | 344.2 | 605.6 | 540.2 |
| A498 | BEAS-2B | Caco-2 | HaCaT | HepG-2 | IMR-90 | |
|---|---|---|---|---|---|---|
| IC50 (µM) | 453.0 | 125.9 | 186.5 | 322.9 | >500 | 437.6 |
| TI with MIC of 3.12 µM | 145.1 | 40.3 | 59.7 | 103.4 | >160.2 | 140.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Maio, A.; Olleik, H.; Courvoisier-Dezord, E.; Guillier, S.; Neulat-Ripoll, F.; Haudecoeur, R.; Bolla, J.-M.; Casanova, M.; Cavalier, J.-F.; Canaan, S.; et al. Design and Synthesis of Novel Amino and Acetamidoaurones with Antimicrobial Activities. Antibiotics 2024, 13, 300. https://doi.org/10.3390/antibiotics13040300
Di Maio A, Olleik H, Courvoisier-Dezord E, Guillier S, Neulat-Ripoll F, Haudecoeur R, Bolla J-M, Casanova M, Cavalier J-F, Canaan S, et al. Design and Synthesis of Novel Amino and Acetamidoaurones with Antimicrobial Activities. Antibiotics. 2024; 13(4):300. https://doi.org/10.3390/antibiotics13040300
Chicago/Turabian StyleDi Maio, Attilio, Hamza Olleik, Elise Courvoisier-Dezord, Sophie Guillier, Fabienne Neulat-Ripoll, Romain Haudecoeur, Jean-Michel Bolla, Magali Casanova, Jean-François Cavalier, Stéphane Canaan, and et al. 2024. "Design and Synthesis of Novel Amino and Acetamidoaurones with Antimicrobial Activities" Antibiotics 13, no. 4: 300. https://doi.org/10.3390/antibiotics13040300
APA StyleDi Maio, A., Olleik, H., Courvoisier-Dezord, E., Guillier, S., Neulat-Ripoll, F., Haudecoeur, R., Bolla, J.-M., Casanova, M., Cavalier, J.-F., Canaan, S., Pique, V., Charmasson, Y., Baydoun, E., Hijazi, A., Perrier, J., Maresca, M., & Robin, M. (2024). Design and Synthesis of Novel Amino and Acetamidoaurones with Antimicrobial Activities. Antibiotics, 13(4), 300. https://doi.org/10.3390/antibiotics13040300







