Abstract
The development of new and effective antimicrobial compounds is urgent due to the emergence of resistant bacteria. Natural plant flavonoids are known to be effective molecules, but their activity and selectivity have to be increased. Based on previous aurone potency, we designed new aurone derivatives bearing acetamido and amino groups at the position 5 of the A ring and managing various monosubstitutions at the B ring. A series of 31 new aurone derivatives were first evaluated for their antimicrobial activity with five derivatives being the most active (compounds 10, 12, 15, 16, and 20). The evaluation of their cytotoxicity on human cells and of their therapeutic index (TI) showed that compounds 10 and 20 had the highest TI. Finally, screening against a large panel of pathogens confirmed that compounds 10 and 20 possess large spectrum antimicrobial activity, including on bioweapon BSL3 strains, with MIC values as low as 0.78 µM. These results demonstrate that 5-acetamidoaurones are far more active and safer compared with 5-aminoaurones, and that benzyloxy and isopropyl substitutions at the B ring are the most promising strategy in the exploration of new antimicrobial aurones.
1. Introduction
The development of novel antibacterial molecules is a major necessity in the upcoming decades, due to the increasing emergence of multi-drug resistant bacterial strains. This is leading to an elevating mortality rate by infectious disease that may reach more than 10 billion deaths by 2050, according to World Health Organization (WHO) [1]. Among these strains, mycobacteria, in particular M. tuberculosis, the pathogenic agent of tuberculosis, are still responsible for 10 million new cases every year worldwide and killed almost 1.5 million patients in 2022. More alarmingly, the number of strains resistant and ultra-resistant to cocktails of antibiotics currently used to treat infection is constantly rising. One of the major concerns is about methicillin resistant Staphylococcus aureus (MRSA), but also other Gram-positive and Gram-negative species such as Acinetobacter baumannii, Pseudomonas aeruginosa or Klebsiella pneumoniae [2]. In addition to bacteria, fungi (filamentous fungi such as Aspergillus fumigatus, as well as yeasts such as Candida species and Cryptococcus neoformans) are also responsible for deadly infections, particularly in HIV-infected patients, but also in immunocompetent ones, affecting billions of patients and causing more than 1.5 million deaths per year [3,4,5,6].
Some natural plant molecules and/or their derivatives, including flavonoids, have been reported to possess strong antimicrobial activity. For decades, the biological effect of flavonoids has been studied, focusing on the major subclasses such as flavones, flavonols, flavanones and chalcones. However, in the past ten years, the aurone subclass has been demonstrated to display strong biological effects in diverse fields, such as oncology, dermatology, and infectiology [7,8,9]. The natural occurrences of aurones is limited to a limited number of advanced plant species where they play a variety of key roles as flower pigments, antioxidants and as nectar guides [10]. Although some natural aurones such as cephalocerone [11,12] and hispidol [13] have demonstrated antimicrobial activity, aurones exhibiting natural substitution patterns do not generally lead to the most effective antimicrobial agents. On the other hand, a series of synthetical aurone derivatives showed a strong effect against Gram-positive bacteria [14]. Structurally, aurones are characterized by a benzofuran moiety bearing in position 2 a benzylidene type substituent (rings A, B, and C) (Figure 1). Overall, research on scaffold modifications has mainly focused on the substitution at the B-ring scaffold, e.g., with the introduction of either ferrocene [15], 5-nitroimidazole [16] or quinoline [17] groups, with a global tendency to retain naturally present hydroxy groups at the A-ring.
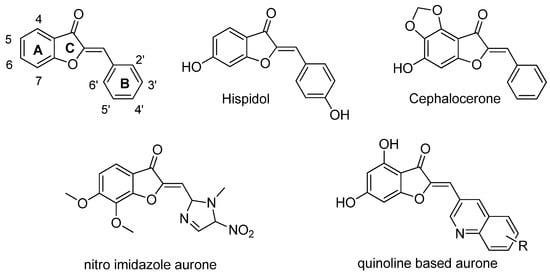
Figure 1.
Structure of the aurone scaffold (top left), natural antimicrobial aurones, and examples of synthetic, heavily modified analogues.
In the present study, modifications of the A-ring were performed by substitution with an amino group combined with substitutions of the B-ring (Figure 1). The 31 new aurone derivatives obtained (Table 1) were tested in terms of antibacterial and antifungal activities.

Table 1.
List of the synthetized aurones and their respective substitution on each position (OBn: Obenzyl, OPh: Ophenyl, OiPr: Oisopropyl).
2. Results
Antimicrobial effect of the aurone derivatives was first evaluated by the determination of their Minimum Inhibitory Concentrations (MICs) on five different bacterial strains representative of Gram-positive bacteria (Bacillus subtilis and Staphylococcus aureus), Gram-negative bacteria (Escherichia coli and Pseudomonas aeruginosa), mycobacteria (Mycobacterium smegmatis), and one fungal strain (Candida albicans) (µM) (Table 2).

Table 2.
Evaluation of the antimicrobial activities of the 31 newly synthetized aurone derivatives. The antimicrobial activities were determined using a MIC assay on species representative of Gram-positive bacteria (B. subtilis, S. aureus), Gram-negative bacteria (E. coli, P. aeruginosa), mycobacteria (M. smegmatis), and fungi (C. albicans). Amphotericin B and gemifloxacin were used as control antimicrobials for fungi and bacteria, respectively. The MIC values are given in µM (n = 2–3).
In this first screening, among the 31 aurone derivatives tested, compounds 10, 12, 15, 16, and 20 were the most active (Figure 2). Compound 10 gave the lowest MIC values on all micro-organisms tested in Table 2 (i.e., Gram-positive and -negative bacteria, mycobacteria and fungi), with MICs ranging from 3.12 to 50 µM. Similarly, compound 20 was active on all tested species (MIC ranging from 12.5 to 50 µM), except P. aeruginosa for which MIC was superior to 100 µM. Compound 16, although active on B. subtilis, S. aureus, E. coli, and M. smegmatis (MIC ranging from 25 to 50 µM), was however inactive on P. aeruginosa and C. albicans. Finally, although compounds 12 and 15 were active on tested Gram-positive bacteria and mycobacteria (MIC ranging from 25 to 100 µM and from 50 to 100 µM, respectively), they were inactive on tested Gram-negative bacteria and C. albicans. Based on MIC values on this first screening, the observed order of antimicrobial activities is as follows: compound 10 > 20 > 12 = 16 > 15.
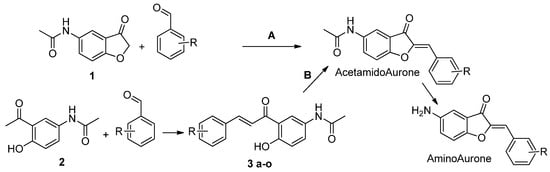
Figure 2.
Synthetic route of the aurone derivatives. A: benzofuranone (1) 1 eq. and various benzaldehydes 1 eq. in Choline chloride/Urea (1/2), 80 °C, 2 h. B: (2) 1 eq. and various benzaldehydes 1 eq. in EtOH, LiOH 3 eq., 90 °C, 2 h. (3 a-o) 1 eq. and Mercuric acetate 1 eq. in pyridine, 110 °C, 1 h. Acetamido Aurone (4–6, 10, 11, 14, 15, 18–20, 24, 25, 29, 31, 32, 34) are converted to their amino analogues (7–9, 12, 13, 16, 17, 21–23, 26–28, 30, 33) in EtOH, 0.5 M HCl, 100 °C, 2 h.
The safety of the five most active derivatives (i.e., compounds 10, 12, 15, 16, and 20) was then evaluated using different human cell types (Figure 3 and Figure 4, Table 3).
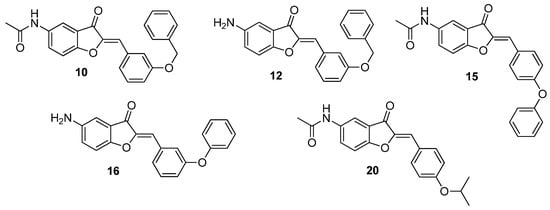
Figure 3.
Structures of the more active aurone derivatives identified during the first screening (compounds 10, 12, 15, 16, and 20).
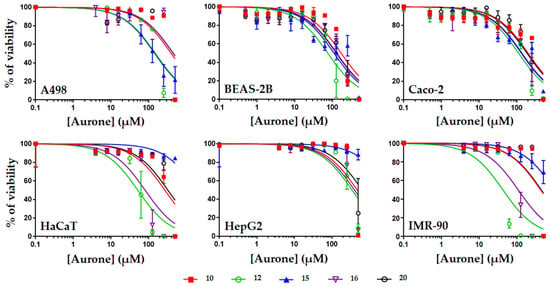
Figure 4.
Evaluation of the toxicity of compounds 10, 12, 15, 16, and 20 on human cells. Human cells corresponding to kidney epithelial cells (A498), lung epithelial cells (BEAS-2B), intestinal epithelial cells (Caco-2), skin cells (HaCaT), liver cells (HepG2), or fibroblasts (IMR-90) were exposed to increasing concentrations of aurones for 48 h before measurement of the cell viability using resazurin. Results are expressed as % of cell viability, DMSO alone being used as negative control giving 100% viability. Data were plotted using GraphPad Prism 7 (means +/− S.D, n = 3).

Table 3.
Determination of the cytotoxic concentrations of compounds 10, 12, 15, 16, and 20 on human cells. The cytotoxic concentrations 50 (CC50, in µM) (i.e., the concentrations of aurones causing 50% reduction of the cell viability after 48 h exposure) were calculated from Figure 3 using GraphPad Prism 7. Results are expressed as means +/− S.D (µM) (n = 3).
Overall, compounds 10, 12, 15, 16, and 20 were found to be safe with most of their CC50 values higher than their MIC ones. Compounds 10 and 20 were the safest molecules with mean CC50 of 321.5 and 305.1 µM (ranging from 169.0 to 472.4 µM and from 125.9 to >500 µM for compounds 10 and 20, respectively). Compounds 12, 15, and 16 were more toxic, with mean CC50 of 129.3, 130.6, and 218.0 µM, respectively. The highest safety of compounds 10 and 20 was further demonstrated when comparing the therapeutic indexes (TI) of the five aurones. Indeed, when calculating the TI of each aurone (by dividing their CC50 on human cells (Table 3) by their MIC on S. aureus (Table 2)), compounds 10 and 20 gave the highest TI values (ranging from 13.5 to 37.7 and from 10.0 to >40, for compounds 10 and 20, respectively) compared to the TI values of compounds 12, 15, and 16 (Table 4) (ranging from 0.4 to 18.0).

Based on antimicrobial activity and toxicity data, the two most active and safest compounds were identified as compounds 10 and 20, which were then tested on a larger panel of bacterial and fungal species in order to evaluate their spectrum of activity (Table 5).

Table 5.
Antimicrobial activities of compounds 10 and 20 on various bacterial and fungal species. The antimicrobial activities were determined using MIC assay as described in Section 4. The MIC values are given in µM (n = 2–3).
The results of this second screening confirmed that compounds 10 and 20 are primarily active on Gram-positive bacteria, with MIC values as low as 0.78 and 3.12 µM for compounds 10 and 20, respectively. Compounds 10 and 20 were particularly active on the various foodborne pathogens. For example, MIC values of 3.12 and 6.25 µM were obtained on Listeria monocytogenes for compounds 10 and 20, respectively. Compounds 10 and 20 gave good activity against Clostridi such as C. difficile (MIC values of 12.5 and 3.12 µM for compounds 10 and 20, respectively) and C. botulinum (MIC values of 0.78 and 3.12 µM for compounds 10 and 20, respectively). In addition, good activities were also observed on the WHO group 3 pathogen Bacillus anthracis, responsible for anthrax disease and used as biological weapon (MIC values of 12.5 and 6.25 µM for compounds 10 and 20, respectively). These MIC values were consistent with the ones obtained on two other Bacillus species, i.e., B. cereus and B. subtilis. On the other hand, C. perfringens, Enterococcus species, P. acnes, and S. pyogenes were found to be weakly sensitive to insensitive to compounds 10 and 20 with MIC values from 50 µM to >100 µM. Overall, compound 10 was more active than compound 20 on most tested Gram-positive bacterial strains, except C. difficile, E. faecium, and B. anthracis, for which compound 20 was more active. S. aureus and methicillin-resistant S. aureus (MRSA) showed good sensitivity with MIC of 12.5–25 µM, showing that resistance to methicillin did not affect the activity of compounds 10 and 20.
Compounds 10 and 20 were also active on Gram-negative bacteria, with A. baumannii, E. coli, and H. pylori being the more sensitive strains (MIC values as low as 12.5 and 25 µM for compounds 10 and 20, respectively). S. enterica, S. flexneri, and V. alginolyticus were also found to be sensitive, with MICs ranging from 25 to 50 µM. Although P. aeruginosa was sensitive to compound 10 (MIC of 25 µM), it was insensitive to compound 20 (MIC > 100 µM). Good activities were also obtained on the WHO group 3 pathogens Brucella melitensis, Francisella tularensis, and Yersinia pestis (MIC values as low as 12.5 µM). E. cloacae and K. pneumoniae were insensitive to compounds 10 and 20 (MIC > 100 µM). Compound 10 was more active than compound 20 on most Gram-negative strains tested, except for B. melitensis and F. tularensis, for which compound 20 gave lower MIC values.
Although compounds 10 and 20 were active on M. smegmatis (Table 2), they were inactive on tested pathogenic mycobacteria, i.e., M. abscessus and M. tuberculosis.
Finally, in term of antifungal effect, compounds 10 and 20 were active on the filamentous fungi F. oxysporum (MIC of 25 µM) but inactive on another important human pathogen A. fumigatus. Antifungal activity was also observed on yeasts, including various Candida species (C. albicans, C. auris, C. glabrata, and C. tropicalis) and Cryptococcus neoformans with MIC values as low as 25 and 12.5 µM for compounds 10 and 20, respectively. In most cases, compounds 10 and 20 gave the same MIC values except for C. auris, for which compound 20 was more active than compound 10 (MIC of 12.5 and 50 µM, respectively). C. tropicalis had a less sensitive MIC value of 100 µM.
The therapeutic indexes (TI) values of compounds 10 and 20 were calculated using the MIC values reported in Table 5 and the CC50 on human cells reported in Table 3 (Table 6 and Table 7).

Table 6.
Safety evaluation for compound 10. Therapeutic indexes (TI) values were calculated by dividing CC50 values by the lowest MIC value (obtained on C. botulinum, i.e., 0.78 µM) for compound 10 (Table 5).

Table 7.
Safety evaluation for compound 20. Therapeutic indexes (TI) values were calculated by dividing CC50 values by the lowest MIC value (obtained on C. botulinum, i.e., 3.12 µM) for compound 20 (Table 5).
TI values ranged from 216.6 to 605.6 and from 40.3 to >160.2 for compounds 10 and 20, respectively, confirming that these two aurone derivatives possess good therapeutic values. Compound 10 was the safest in all cases.
3. Discussion
In the present study, 31 new aurone derivatives were synthetized (Figure 5, see Supplementary Materials) and tested in terms of antimicrobial activity against various bacteria and fungi. These new compounds were obtained by the substitution of the aurone scaffold at position 5 by amino and acetamido groups, and through various substitutions at the 2′, 3′ and 4′ positions. The first screening antimicrobial test performed on representative species of bacteria and fungi allowed us to identify compounds 10, 12, 15, 16, and 20 as the more active aurones. Comparisons between active and inactive structures afforded insightful information to identify the most interesting substitution. Compound 10 can be compared to compound 11, 12 and 13. All these compounds are substituted in position 3′ or 4′ by a benzyloxy group. However, only compound 10 and 12 showed an interesting activity. This suggests that benzyloxy substitution in 3′ may be a key element in the activity of the compound. The same methodology could be used to compare compound 20 with other isopropyloxy compounds such as 18, 19, 21 and 22. These four last mentioned compounds showed a weak or no activity against tested micro-organisms. Similarly to compound 10, this could indicate that the 4′-isopropyl substitution may be a far better alternative to the other position and again that the acetamido group is more effective than the amino group in position 5. From the results, it can be suggested that the acetamido group is important for the antibacterial capacity of the aurones. Thus, compounds 12 and 16 are both 5-aminoaurones; compounds 15 and especially compounds 10 and 20 are 5-acetamidoaurones and showed far better antibacterial activity. Moreover, four of these compounds possess a ring substitution in position 3′ and 4′. Interestingly, benzyloxy-substituted aurones (i.e., 10 and 12) seem to only be active in positions 3′ when phenyl-substituted aurones (i.e., 15 and 16) showed activity when substituted both in 3′ and 4′ position. However, considering the activity of compound 10 compared to 12, 15 and 16, benzyloxy substitution must be considered more promising than phenyl substitution. The other benzyloxy- and phenyl-substituted aurones (i.e., 11, 13, 14 and 17) showed no activity. Isopropyloxy-substituted aurones also are an interesting option as shown by compound 20, which is similar in activity to compound 10. Again only 4′-isopropyl aurones were active; 3′ and 2′ were inactive on the varieties of pathogens tested. Methyl (i.e., 4–9), fluoro (i.e., 24–28), carboxy (i.e., 31), trifluoromethyl (i.e., 29 and 30) and hydroxy (i.e., 32 and 33) substituted aurones showed no activity and thus should not be considered as privileged substitution in the development of antibacterial aurones. Finally, aurone 34 is also inactive and shows that the 5-acetamido substitution is not enough to produce an antibacterial activity to aurones and that a substitution on the B-ring is mandatory. These five aurones were then tested in term of toxicity against various human cell types. The toxicity data demonstrated that compounds 10 and 20 were the safest ones compared to compounds 12, 15, and 16. Again, 5-acetamido aurones seem to be more interesting as they are safer on human cells than 5-aminoaurones. Compounds 10 and 20 were then further tested on a larger panel of pathogens, including Gram-positive and Gram-negative bacteria. In this second screening, these compounds showed an interesting activity as antimicrobials against Gram-positive strains such as S. aureus, methicillin-resistant S. aureus, L. monocytogenes, B. subtilis and C. difficile and Gram-negative strains such as E. coli, A. baumannii and H. pylori. The two selected compounds shared some structural similarities, such as the 5-acetamido substitution. Out of all the compounds, 3′-benzyloxy and 4′-isopropyloxy were the most promising substitutions. Compared to previously described aurones active on various Gram-positive bacteria but only two Gram-negative strains (i.e., H. pylori or V. alginolyticus) [14], compounds 10 and 20 were active on a large number of Gram-positive and -negative bacteria as well as fungi. Structurally, the most active compound synthetized by Olleik et al. [14] was a 5,7-dihydroxyaurone substituted in 4′ by a benzyloxy group and in 3′ by a methoxy group. Again, this shows the promising nature of the benzyloxy substitution and that hydroxy aurones, vastly found in nature, are far less active than synthetic aurones such as amino and acetamidoaurones.
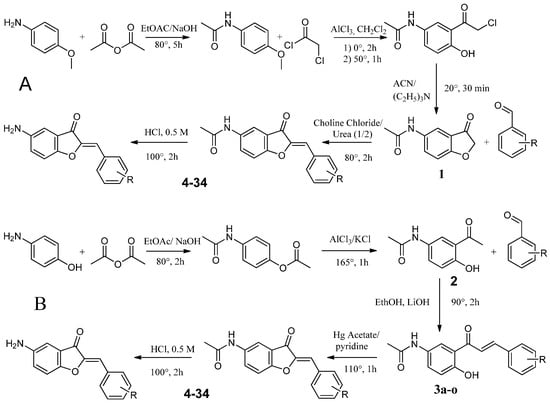
Figure 5.
Route A and B used for the synthesis of 5-amino and 5-acetamido aurones.
4. Materials and Methods
4.1. Biology
4.1.1. Microorganism Strains and Growth Conditions
Bacterial and fungi strains used in this study, except when mentioned, were obtained from either the American Type Culture Collection (ATCC), the German Leibniz Institute (DSMZ), or the French Pasteur Institute (CIP) and correspond to reference strains. They were maintained on agar plates using appropriate media and culture conditions (in terms of temperature and aerobic/microaerobic/anaerobic condition) as previously described [14,18]. Regarding BSL-3 strains, Bacillus anthracis, Francisella tularensis, and Brucella melitensis strains were maintained on Chocolate agar PolyViteX (Biomerieux) agar at 37 °C, and at 26 °C for Yersinia pestis [19,20]. Regarding mycobacteria, M. smegmatis mc2155 (ATCC700084) was grown in Middlebrook 7H9 complete medium containing 0.05% Tween-80 and 0.2% Glycerol (7H9-TG) and M. abscessus (CIP104536T) S and R morphotypes, were cultured in 7H9-TG containing 10% BBL™ Middlebrook OADC Enrichment (7H9-TGOADC) at 37 °C under stirring (200 rpm). M. tuberculosis mc26230, a derivative of H37Rv which contains a deletion of the RD1 region and panCD, resulting in a pan(−) phenotype, was grown in 7H9-TGOADC supplemented with 24 µg/mL D-panthothenate (Sigma-Aldrich, Lyon, France). Cultures were kept at 37 °C without shaking.
4.1.2. Antimicrobial Activity Assay
The antimicrobial activity of aurones on BSL2 bacteria and fungi was evaluated through determination of the Minimum Inhibitory Concentration (MIC) using two-fold serial dilutions in liquid media following the National Committee of Clinical Laboratory Standards (NCCLS, 1997) as previously described [14,18,21,22]. For BSL-3 bacteria, the MIC of aurones was determined following the Clinical and Laboratory Standards Institute (CLSI) recommendations as previously described [23]. For determining the antimycobacterial activity of the different aurones, the microdilution method was used in sterile 96-well flat-bottom Greiner Bio-One polypropylene microplates with lid (Thermo Fisher Scientific) using the resazurin microtiter assay (REMA) as previously described [24,25]. The concentration of aurones leading to 90% inhibition of mycobacteria growth was defined as the MIC. All experiments were performed independently at least three times.
4.1.3. Cytotoxic Assays
The impact of aurones on the viability of human cells were evaluated as previously described [18,22,26]. Human cells used were kidney epithelial cell line A498 (ATCC® HTB-44), normal lung epithelial cells BEAS-2B (ATCC® CRL-9609), intestinal cell line Caco-2 (ATCC® HTB-37), normal epidermal keratinocytes (HaCaT) (from Creative Bioarray, Shirley, NY, USA), liver cell line HepG2 (ATCC® HB-8065), and normal lung fibroblasts IMR-90 (ATCC® CCL186). Cells were cultured in DMEM supplemented with 10% fetal calf serum (FCS), 1% l-glutamine, and 1% antibiotics (all from Invitrogen (Carlsbad, CA, USA). Cells were routinely grown on 25 cm2 flasks and maintained in a 5% CO2 incubator at 37 °C. For toxicity assays, human cells grown on 25 cm2 flasks were detached using trypsin-EDTA solution (from Thermofisher, Waltham, MA, USA), counted using Malassez counting chamber, diluted in appropriate culture media, and seeded into 96-well cell culture plates (Greiner bio-one, Paris, France) at approximately 104 cells per well. The cells were left to grow for 48–72 h at 37 °C in a 5% CO2 incubator until confluence. Media from wells was then aspirated and cells were treated with 100 µL of appropriate culture media containing increasing concentrations of tested aurones (from 0 to 400 µM, 1:2 serial dilutions). Volume of DMSO corresponding to 400 µM of aurones was used as negative control and was found not toxic. The plates were then incubated at 37 °C for 48 h. Resazurin-based in vitro toxicity assay kit (from Sigma-Aldrich, Lyon, France) was then used to assess the viability of the cells following manufacturer’s instructions. Briefly, resazurin stock solution was diluted 1:10 in sterile PBS containing calcium and magnesium (PBS++, pH 7.4). Plates were aspirated and 100 µL of the diluted solution was added per well. After 2 h incubation at 37 °C, fluorescence intensity was measured using a microplate reader (Biotek, Synergy Mx, Colmar, France) (excitation wavelength of 530 nm/emission wavelength of 590 nm). The fluorescence values were normalized by the controls (DMSO treated cells) and expressed as percentage of cell viability. The CC50 values of aurones (i.e., the concentrations causing a reduction of 50% of the cell viability) were calculated using GraphPad® Prism 7 software (San Diego, CA, USA). Experiments were conducted in triplicate (n = 3).
4.2. Chemistry
1H and 13C NMR spectra were recorded on a Bruker Avance III nanobay—300 MHz instrument (Bruker, Bremen, Germany, 300 MHz for 1H, 75 MHz for 13C). Chemical shifts are reported in ppm relative to the solvent in which the spectrum was recorded [1H: δ (d6-DMSO) = 2.50 ppm, δ (CDCl3) = 7.27 ppm; 13C: δ (d6-DMSO) = 39.52 ppm, δ (CDCl3) = 77.16 ppm], full spectra are presented in Supplementary Materials. Combustion analyses were performed at the analysis facilities of Spectropole (https://fr-chimie.univ-amu.fr/spectropole, accessed on 20 December 2023) with a Thermo Finnigan (San Jose, CA, USA) EA 1112 apparatus; all compounds had purity higher than 95%. Microwave-assisted reactions were performed in a CEM Discover microwave reactor with a focused field (CEM Corporation, Matthews, NC, USA). Silica gel F-254 plates (0.25 mm; Merck, Darmstadt, Germany) were used for thin-layer chromatography (TLC), and silica gel 60 (200–400 mesh; Merck) was used for flash chromatography. Unless otherwise stated, reagents were obtained from commercial sources and were used without further purification.
4.2.1. Synthesis Route A
Synthesis of N-(4-methoxyphenyl)acetamide (1a)
In a solution of 8 g of m-anisidine and 2.5 eq. of NaOH in 50 mL of ethyl acetate, 6.6 g of acetic anhydride (1.5 eq.) was added dropwise. When the addition was completed, the mixture was heated at 80 °C for 5 h. The solution was cooled and filtered. Under pressure, the solvent was removed, the obtained product was dissolved in ethanol and hexane to precipitate, to obtain 5 g of 1a.
Synthesis of 2-Chloro-1-{2-hydroxy-5-[(1-hydroxyethyl)amino]cyclohexyl}ethan-1-one (2a)
A total of 5 g of 1a and 18 g of AlCl3 were dissolved in 30 mL of dichloromethane then at 0 °C, 3.5 eq. of chloroacetyl chloride was added dropwise. When the addition was completed, the solution was heated up to 50 °C for 1 h. The mixture was poured on ice and extracted with ethyl acetate. EtOAc was removed under pressure to obtain 2a.
Synthesis of N-(3-oxo-2,3-dihydro-1-benzofuran-5-yl)acetamide (3a)
In a flask, 2 g of 2a and 1.5 eq. of triethylamine were added to 20 mL of acetonitrile; the solution reacted at 25 °C for 12 h. Solvent was removed under pressure. The left-over mixture was dissolved in EtOAc, washed several times with water then extracted. EtOAc was removed under pressure to obtain 3a.
Synthesis of Substituted 5-Acetamidoaurones
In a flask, 1 mmol of 3a and 1 mmol of the corresponding benzaldehyde were dissolved in 10 mL of choline chloride/urea. Three drops of 50% KOH solution were added. The mixture was heated at 80 °C for 2 h. Water and HCl were added, then the precipitate was filtered and washed several times with ether to obtain acetamido substituted aurones.
Synthesis of Substituted 5-Aminoaurone
A total of 10 mmol of acetamido aurones was added to a mixture of EtOH (20 mL) and 0.5 M HCl (5 mL). The solution was refluxed for 2 h. Upon cooling, the solvent was removed under vacuum and the residue obtained was poured onto iced water (100 mL). The resulting solution was neutralized with NH4OH 16% until pH = 7. The precipitate formed was collected by filtration and washed with excess cold water.
4.2.2. Synthesis Route B
Synthesis of 4-Acetamidophenyl Acetate (1b)
To a solution of 8 g of 4-aminophenol and 2.5 eq. of NaOH in 50 mL of ethyl acetate, 26.5 g of acetic anhydride (3.5 eq.) was added dropwise. When the addition was completed, the solution was heated at 80 °C for 5 h. Upon cooling, the mixture was filtered. Solvent was removed under pressure; the product was recrystallized in ethanol and hexane to obtain 5 g of 1b.
Synthesis of N-(3-acetyl-4-hydroxyphenyl)acetamide (2b)
To a solution of 1b (5 g, 26 mmol), 15 g of AlCl3 (113 mmol, 4 eq.) and 1.9 g of KCl (26 mmol, 1 eq.) were added. The mixture was then heated a 165 °C for 1 h until a brown paste appeared. Upon cooling, ice cold water was added (300 mL) and the mixture was filtered to obtain 2 g of a beige powder (2b).
Synthesis of Substituted of 5-Acetamidochalcones (3b)
In a flask, 193 mg of 2b (0.001 mmol), 1 eq. of chosen benzaldehyde and 188 mg of LiOH (16 eq., 0.016 mmol) were dissolved in ethanol (20 mL). The mixture was heated for 2 h at 90 °C. The solvent was then removed under pressure, cold water and HCl were added, and the precipitate was filtered to obtain the desired chalcone (3b).
Synthesis of Substituted 5-Acetamidoaurones
To a mixture of chosen 3b chalcone in pyridine (20 mL), 1 eq. of mercury acetate was added. The solution was heated for 1 h at 110 °C. Water and HCl were added. The precipitate was filtered and washed several times with ice cold water to get rid of the mercury. Obtention of a powder red-yellow powder depended the substitution.
Synthesis of Substituted 5-Aminoaurone
For synthesis of 5-aminoaurone see Section Synthesis of Substituted 5-Aminoaurone.
Yield: 83%; mp: 234.8 °C; 1H NMR (300 MHz, DMSO-d6): δ 10.15 (s, 1H, NH), 8.20–8.18 (dd, 1H, J = 1.2;7.8 Hz, C-H6′), 8.10 (d, 1H, J = 1.9 Hz, C-H4), 7.83–7.80 (dd, 1H, J = 2.2;8.9 Hz, C-H6), 7.52–7.49 (d, 1H, J = 8.9 Hz, C-H7), 7.46 (dt, 1H, J = 7.1 Hz, C-H4′), 7.19 (s, 1H, C-H10), 7.16–7.09 (m, 2H, C-H3′,5′), 3.91 (s, 3H, OCH3), 2.07 (s, 3H, NHCOCH3). 13C NMR (75 MHz, DMSO-d6): δ 183.53(C-3), 168.41(CO), 161.21(C-8), 158.32(C-2′), 146.75(C-2) 135.53(C-5), 131.99(C-4′), 131.11(C-6′), 128.71(C-6), 120.89(C-1′), 120.68(C-5′), 120.08(C-9), 113.31(C-3′), 113.15(C-7), 111.57(C-10), 105.47(C-4), 55.84(OCH3), 23.83(CH3). Elemental analysis calcd (%) for C18H15NO4: C, 69.89; H, 4.89; N, 4.53; found C, 69.87; H, 4.91; N, 4.52. m/z: 309.1001 (100.0%).
(Z)-N-(2-(3-methoxybenzylidene)-3-oxo-2,3-dihydrobenzofuran-5-yl)acetamide (5):
Yield: 71%; mp: 204.2 °C; 1H NMR (300 MHz, DMSO-d6): δ 10.16 (s, 1H, NH), 8.10 (d, 1H, J = 2 Hz, C-H4), 7.83–7.80 (dd, 1H, J = 2.2;8.9 Hz, C-H6), 7.60–7.55 (m, 2H, C-H2′,4′), 7.54–7.51 (d, 1H, J = 8.9 Hz, C-H7), 7.43 (dt, 1H, J = 8.0 Hz, C-H5′), 7.06–7.03 (dd, 1H, J = 2.6;8.2 Hz, C-H6′), 6.90 (s, 1H, C-H10), 7.16–7.09 (m, 2H, C-H3′,5′), 3.82 (s, 3H, OCH3), 2.07 (s, 3H, NHCOCH3). 13C NMR (75 MHz, DMSO-d6): δ 183.68(C-3), 168.42(CO), 161.33(C-8), 159.42(C-3′), 146.88(C-2), 135.58(C-5), 133.08(C-6′), 130(C-2′), 128.82(C-6), 123.74(C-1′), 120.59(C-9), 116.57(C-6′), 115.76(C-4′), 113.35(C-7), 113.15(C-4), 112.07(C-10), 55.15(OCH3), 23.83(CH3). Elemental analysis calcd (%) for C18H15NO4: C, 69.89; H, 4.89; N, 4.53; found C, 69.84; H, 4.88; N, 4.53. m/z: 309.1001 (100.0%).
(Z)-N-(2-(4-methoxybenzylidene)-3-oxo-2,3-dihydrobenzofuran-5-yl)acetamide (6):
Yield: 91%; mp: 252 °C [1]; 1H NMR (300 MHz, DMSO-d6): δ 10.17 (s, 1H, NH), 8.10 (d, 1H, J = 2 Hz, C-H4), 7.96–7.93 (d, 2H, J = 7.9 Hz, C-H2′,6′), 7.81–7.78 (dd, 1H, J = 2.2;8.9 Hz, C-H6), 7.50–7.47 (d, 1H, J = 8.9 Hz, C-H7), 7.08–7.06 (d, 2H, J = 8.0 Hz, C-H3′,5′), 6.91 (s, 1H, C-H10), 3.83 (s, 3H, OCH3), 2.07 (s, 3H, NHCOCH3). 13C NMR (75 MHz, DMSO-d6): δ 183.37(C-3), 168.46(CO), 161.06(C-8), 161.00(C-4′), 145.75(C-2), 135.47(C-5), 133.40(C-2′,6′), 128.51(C-6), 124.47(C-1′), 120.98(C-9), 114.71(C-3′-5′), 113.30(C-7), 113.07(C-4), 112.74(C-10), 55.40(OCH3), 23.91(CH3). Elemental analysis calcd (%) for C18H15NO4: C, 69.89; H, 4.89; N, 4.53; found C, 69.78; H, 4.87; N, 4.48. m/z: 309.1001 (100.0%).
(Z)-5-amino-2-(2-methoxybenzylidene)benzofuran-3(2H)-one (7):
Yield: 22%; mp: 189.3 °C; 1H NMR (300 MHz, DMSO-d6): δ 8.18–8.16 (dd, 1H, J = 1.6;7.8 Hz, C-H6′), 7.43 (dt, 1H, J = 1.5;8.5 Hz, C-H4′), 7.26–7.23 (d, 1H, J = 8.8 Hz, C-H7), 7.14–7.11 (m, 2H, C-H3′,5′), 7.10 (s, 1H, C-H10), 7.06–7.03 (dd, 1H, J = 2.5;8.8 Hz, C-H6), 6.84 (d, 1H, J = 2.4 Hz, C-H4), 5.23 (bs, 2H, NH2), 3.90 (s, 3H, OCH3). 13C NMR (75 MHz, DMSO-d6): δ 184.09(C-3), 158.13(C-2′), 158.00(C-8), 147.05(C-2), 145.56(C-5), 131.55(C-4′), 130.98(C-6′), 124.55(C-6), 121.01(C-9, 120.84(C-5′), 120.41(C-1′), 113.14(C-7), 111.47(C-10), 105.44(C-3′), 104.18(C-4), 55.79(OCH3). Elemental analysis calcd (%) for C16H13NO3: C, 71.90; H, 4.90; N, 5.24; found C, 71.85; H, 4.95; N, 5.21. m/z: 267.0895 (100.0%).
(Z)-5-amino-2-(3-methoxybenzylidene)benzofuran-3(2H)-one (8):
Yield: 50%; mp: 190 °C; 1H NMR (300 MHz, DMSO-d6): δ 7.59–7.57 (d, 1H, J = 7.7 Hz, C-H6′), 7.54 (d, 1H, J = 2.4 Hz, C-H4), 7.49–7.46 (d, 1H, J = 8.8 Hz, C-H7), 7.42 (dt, 1H, J = 8.2 Hz, C-H5′), 7.38–7.35 (dd, 1H, J = 2.2;8.8 Hz, C-H6), 7.24 (d, 1H, J = 2.11 Hz, C-H2′), 7.06–7.03 (dd, 1H, J = 1.9;8.1 Hz, C-H4′), 6.88 (s, 1H, C-H10), 3.82 (s, 3H, OCH3). 13C NMR (75 MHz, DMSO-d6): δ 183.6 (C-3), 160.79 (C-8), 159.42 (C-3′), 146.96 (C-2), 137.91 (C-5′), 133.13 (C-1′), 129.99 (C-5′), 127.97(C-6), 123.71 (C-7), 121.18 (C-9), 116.54 (C-6′), 115.72 (C-4′), 113.87 (C-4), 111.88 (C-2′), 111.18 (C-10), 55.16 (OCH3). Elemental analysis calcd (%) for C16H13NO3: C, 71.90; H, 4.90; N, 5.24; found C, 71.92; H, 4.92; N, 5.28. m/z: 267.0895 (100.0%).
(Z)-5-amino-2-(4-methoxybenzylidene)benzofuran-3(2H)-one (9):
Yield: 86%; mp: 110.4 °C; 1H NMR (300 MHz, DMSO-d6): δ 7.95–7.92 (d, 2H, J = 8.1 Hz, C-H2′), 7.31–7.28 (d, 1H, J = 8.8 Hz, C-H7), 7.13–7.10 (dd, 1H, J = 2.2;8.8 Hz, C-H6), 7.09–7.06 (d, 2H, J = 8.1 Hz, C-H3′), 6.93 (d, 1H, J = 2.4 Hz, C-H4), 6.83 (s, 1H, C-H10), 3.82 (s, 3H, OCH3). 13C NMR (75 MHz, DMSO-d6): δ 183.77 (C-4), 160.61 (C-4′), 158.51 (C-8), 145.94 (C-5), 143.60 (C-2), 133.16 (C-2′), 125.22 (C-1′), 124.69 (C-6), 121.35 (C-9), 114.65 (C-3′), 113.30 (C-4), 111.68 (C-10), 106.77 (C-7), 55.37 (OCH3). Elemental analysis calcd (%) for C16H13NO3: C, 71.90; H, 4.90; N, 5.24; found C, 71.88; H, 4.97; N, 5.30. m/z: 267.0895 (100.0%).
(Z)-N-(2-(3-(benzyloxy)benzylidene)-3-oxo-2,3-dihydrobenzofuran-5-yl)acetamide (10):
Yield: 80%; mp: 204.5 °C; 1H NMR (300 MHz, DMSO-d6): δ 10.16 (s, 1H, NH), 8.10 (d, 1H, J = 2 Hz, C-H4), 7.84–7.80 (dd, 1H, J = 2.2;8.9 Hz, C-H6), 7.63 (bs, 1H, C-H2′), 7.58–7.32 (m, 8H,), 7.14–7.11 (dd, 1H, J = 7.9 Hz, C-H4′), 6.89 (s, 1H, C-H10), 5.18 (m, 2H, CH2), 2.07 (s, 3H, NHCOCH3). 13C NMR (75 MHz, DMSO-d6): δ 183.74 (C-4), 168.51 (CO), 161.36 (C-8), 158.53 (C-3′), 146.92 (C-2), 136.87 (C-1bn), 135.64 (C-5), 133.14 (C-5′), 130.1 (C-6′), 128.87 (C-6), 128.48 (C-3bn), 127.94 (C-4bn), 127.83 (C-2bn), 124.14 (C-1′), 120.62 (C-9), 117.27 (C-4′), 116.75 (C-2′), 113.45 (C-7), 113.17 (C-4), 112.09 (C-10), 69.34 (CH2), 23.91 (CH3). Elemental analysis calcd (%) for C24H19NO4: C, 74.79; H, 4.97; N, 3.63; found C, 74.74; H, 5.01; N, 3.60. m/z: 385.13141 (100.0%).
(Z)-N-(2-(4-(benzyloxy)benzylidene)-3-oxo-2,3-dihydrobenzofuran-5-yl)acetamide (11):
Yield: 72%; mp: 212.8 °C; 1H NMR (300 MHz, DMSO-d6): δ 10.14 (s, 1H, NH), 8.09 (d, 1H, J = 2.02 Hz, C-H4), 7.96–7.93 (d, 2H, J = 8.8 Hz, C-H2′), 7.82–7.79 (dd, 1H, J = 2.2;8.9 Hz, C-H6), 7.50–7.34 (m, 6H, C-H7, bn), 7.16–7.13 (d, 2H, J = 8.8 Hz, C-H3′), 6.90 (s, 1H, C-H10), 5.18 (m, 2H, CH2), 2.07 (s, 3H, NHCOCH3).
13C NMR (75 MHz, DMSO-d6): δ 183.37 (C-3), 168.48 (CO), 161.06 (C-8), 159.93 (C-4′), 145.75 (C-2), 136.58 (C-1bn), 135.45 (C-5), 133.39 (C-2′), 128.56 (C-1′), 128.49 (C-3bn), 128 (C-4bn), 127.84 (C-2bn), 124.64 (C-6), 120.95 (C-9), 115.52 (C-3′), 113.33 (C-7), 113.08 (C-4), 112.68 (C-10), 69.44 (CH2), 23.9 (CH3). Elemental analysis calcd (%) for C24H19NO4: C, 74.79; H, 4.97; N, 3.63; found C, 74.77; H, 4.96; N, 3.61. m/z: 385.13141 (100.0%).
(Z)-5-amino-2-(3-(benzyloxy)benzylidene)benzofuran-3(2H)-one (12):
Yield: 80%; mp: 132.6 °C; 1H NMR (300 MHz, DMSO-d6): δ 7.61 (bs, 1H, C-H2′), 7.55–7.53 (d, 1H, J = 8.8 Hz, C-H7), 7.50–7.48 (d, 2H, C-H2bn), 7.41 (dt, 2H, C-H3bn), 7.36–7.33 (m, 2H, C-H5′,4bn), 7.11–7.09 (dd, 1H, J = 7.9 Hz, C-H6), 7.06–7.04 (dd, 1H, J = 7.9 Hz, C-H4′), 6.83 (d, 1H, J = 2 Hz, C-H4), 6.77 (s, 1H, C-H10), 5.26 (bs, 2H, NH2), 5.17 (m, 2H, CH2). 13C NMR (75 MHz, DMSO-d6): δ 184.24 (C-3), 158.48 (C-3′), 158.14 (C-8), 147.21 (C-2), 145.63 (C-5), 136.87 (C-1bn), 133.43 (C-1′), 129.97 (C-5′), 128.42 (C-3bn), 127.86 (C-4bn), 127.75 (C-2bn), 124.71 (C-6), 123.89 (C-7), 120.91 (C-9), 117.04 (C-4′), 116.4 (C-2′), 113.21 (C-6′), 110.73 (C-10), 105.45 (C-4), 69.31 (CH2). Elemental analysis calcd (%) for C22H17NO3: C, 76.95; H, 4.99; N, 4.08; found C, 76.88; H, 5.01; N, 4.04. m/z: 343.12084 (100.0%).
(Z)-5-amino-2-(4-(benzyloxy)benzylidene)benzofuran-3(2H)-one (13):
Yield: 80%; mp: 141.6 °C; 1H NMR (300 MHz, DMSO-d6): δ 7.97–7.95 (d, 2H, C-H2′-6′ J = 8.8 Hz), 7.49–7.47 (m, 3H, C-H), 7.41–7.39 (m, 4H, C-Hbn), 7.28 (s, 1H, C-H4), 7.17–7.15 (d, 2H, C-H3′-5, J = 8.8 Hz), 6.92 (s, 1H, C-H10), 5.20 (s, 2H, CH2). 13C NMR (75 MHz, DMSO-d6): δ 183.16 (C-3), 160.82 (C-4′), 159.91 (C-8), 145.76 (C-2), 136, 55 (C-5), 133.36 (C-1′), 128.46 (C-3bn, C-5bn), 128.20 (C-1bn), 127.92 (C-4bn), 127.81 (C-2bn, C-6bn), 124.64 (C-6), 121.55 (C-9), 115.5 (C-2′, C-6′), 113.90 (C-7),112.58 (C-4), 111.79 (C-10), 69.42 (CH2). (CH3). Elemental analysis calcd (%) for C22H17NO3: C, 76.95; H, 4.99; N, 4.08; found C, 76.77; H, 4.98; N, 4.06. m/z: 343.13141 (100.0%).
(Z)-N-(3-oxo-2-(3-phenoxybenzylidene)-2,3-dihydrobenzofuran-5-yl)acetamide (14):
Yield: 96%; mp: 196.4 °C; 1H NMR (300 MHz, DMSO-d6): δ 10.16 (s, 1H, NH), 8.10 (d, 1H, J = 2 Hz, C-H4), 7.83–7.79 (dd, 1H, J = 2.2;8.9 Hz, C-H6), 7.75–7.72 (d, 1H, J = 7.9 Hz, C-H6′), 7.67 (bs, 1H, C-H2′), 7.51 (dt, 1H, J = 7.4 Hz, C-H5′), 7.44–7.41 (m, 3H, C-H4′,8′), 7.20 (t, 1H, J = 7.4 Hz, C-H10′), 7.10–7.07 (d, 3H, C-H9′, 7), 6.93 (s, 1H, C-H10), 2.07 (s, 3H, NHCOCH3). 13C NMR (75 MHz, DMSO-d6): δ 183.72 (C-3), 168.52 (CO), 161.30 (C-8), 157.14 (C-7′), 156.19 (C-3′), 147.11 (C-2), 135.68 (C-5), 133.78 (C-1′), 130.60 (C-6), 130.17 (C-9′), 128.92 (C-5′), 126.54 (C-9), 123.88 (C-4), 120.62 (C-6′), 120.57 (C-10′), 119.97 (C-4′), 118.99 (C-8′), 113.30 (C-2′), 113.21 (C-7), 111.41 (C-10), 23.91 (CH3). Elemental analysis calcd (%) for C23H17NO4: C, 74.38; H, 4.61; N, 3.77; found C, 74.33; H, 4.63; N, 3.71. m/z: 371.11576 (100.0%).
(Z)-N-(3-oxo-2-(4-phenoxybenzylidene)-2,3-dihydrobenzofuran-5-yl)acetamide (15):
Yield: 67%; mp: 213.5 °C; 1H NMR (300 MHz, DMSO-d6): δ 10.15 (s, 1H, NH), 8.11 (d, 1H, J = 2 Hz, C-H4), 8.03–8.00 (d, 2H, J = 7.9 Hz, C-H2′), 7.83–7.79 (dd, 1H, J = 2.2;8.9 Hz, C-H6), 7.50–7.42 (m, 3H, C-H7,7′), 7.22 (t, 1H, J = 7.4 Hz, C-H8′), 7.12–7.09 (m, 4H, C-H3′,6′), 6.94 (s, 1H, C-H10), 2.07 (s, 3H, NHCOCH3). 13C NMR (75 MHz, DMSO-d6): δ 183.43 (C-3), 168.38 (CO), 161.14 (C-8), 158.41 (C-1″), 155.43 (C-4′), 146.18 (C-2), 135.5 (C-5), 133.43 (C-3″), 130.15 (C-2′), 128.67 (C-6), 126.79 (C-1′), 124.27 (C-4″), 120.76 (C-9), 119.41 (C-3′), 118.28 (C-2″), 113.24 (C-7), 113.11 (C-4), 111.83 (C-10), 23.82 (CH3). Elemental analysis calcd (%) for C23H17NO4: C, 74.38; H, 4.61; N, 3.77; found C, 74.35; H, 4.67; N, 3.73. m/z: 371.11576 (100.0%).
(Z)-5-amino-2-(3-phenoxybenzylidene)benzofuran-3(2H)-one (16):
Yield: 51%; mp: 145.6 °C; 1H NMR (300 MHz, DMSO-d6): δ 7.71–7.69 (d, 1H, J = 7.9 Hz, C-H6′), 7.64 (bs, 1H, C-H2′), 7.51–7.49 (d, 1H, J = 7.9 Hz, C-H6), 7.44 (dt, 2H, C-H9′), 7.22–7.15 (m, 2H, C-H4,5′), 7.09–7.04 (m, 4H, C-H7,9′,10′), 6.83 (d, 1H, J = 2.02 Hz, C-H4), 6.77 (s, 1H, C-H10), 5.41 (bs, 2H, NH2). 13C NMR (75 MHz, DMSO-d6): δ 184.24(C-3), 158.13(C-7′), 157.1(C-3′), 156.21(C-8), 147.41(C-2), 145.56(C-5), 134.09(C-1′), 130.52(C-5′), 130.15(C-9′), 126.34(C-6), 124.82(C-9), 123.81(C-6′), 120.88(C-10′), 120.43(C-4′), 119.65(C-4), 118.95(C-8′), 113.13(C-8), 110.13(C-10), 105.61(C-7). Elemental analysis calcd (%) for C21H15NO3: C, 76.58; H, 4.59; N, 4.25; found C, 76.56; H, 4.61; N, 4.22. m/z: 329.10519 (100.0%).
(Z)-5-amino-2-(4-phenoxybenzylidene)benzofuran-3(2H)-one (17):
Yield: 76%; mp: 170.6 °C; 1H NMR (300 MHz, DMSO-d6): δ 7.99–7.96 (d, 2H, J = 8.6 Hz, C-H3′), 7.44 (dt, 2H, J = 7.7 Hz, C-H7′), 7.24–7.21 (m, 2H, C-H7,8′), 7.10–7.02 (m, 5H, C-H6,2′,6′), 6.83 (bs, 2H, C-H4,10), 5.25 (bs, 2H, NH2). 13C NMR (75 MHz, DMSO-d6): δ 184.11 (C-3), 158.16 (C-5′), 158.02 (C-8), 155.58 (C-4′), 146.56 (C-2), 145.6 (C-5), 133.27 (C-7′), 130.24 (C-2′), 127.21 (C-1′), 124.62 (C-6), 124.28 (C-8′), 121.15 (C-9), 119.41 (C-3′), 118.39 (C-6′), 113.17 (C-4), 110.62 (C-10), 105.45 (C-7). Elemental analysis calcd (%) for C21H15NO3: C, 76.58; H, 4.59; N, 4.25; found C, 76.43; H, 4.64; N, 4.54. m/z: 329.10519 (100.0%).
(Z)-N-(2-(2-isopropoxybenzylidene)-3-oxo-2,3-dihydrobenzofuran-5-yl)acetamide (18):
Yield: 93%; mp: 230 °C; 1H NMR (300 MHz, DMSO-d6): δ 10.15 (s, 1H, NH), 8.21–8.19 (d, 1H, J = 1.2;7.8 Hz, C-H6′), 8.12 (bs, 1H, C-H4), 7.81–7.78 (d, 1H, J = 8.7 Hz, C-H6), 7.52–7.49 (d, 1H, J = 8.9 Hz, C-H7), 7.42 (dt, 1H, J = 7.7 Hz, C-H4′), 7.20 (s, 1H, C-H10), 7.16–7.13 (d, 1H, J = 8.3 Hz, C-H3′), 7.08 (dt, 1H, J = 7.6 Hz, C-H5′), 4.75 (q, 1H, J = 5.9;11.9 Hz, C-Hisop), 2.07 (s, 3H, NHCOCH3), 1.35–1.33 (d, 6H, J = 5.8 Hz, C-H3isop). 13C NMR (75 MHz, DMSO-d6): δ 183.52 (C-3), 168.39 (CO), 161.15 (C-8), 156.76 (C-2′), 146.69 (C-2), 135.5 (C-5), 131.85 (C-6′), 131.38 (C-4′), 128.64 (C-6), 121.03 (C-9), 120.73 (C-1′), 120.69 (C-5′), 113.85 (C-7), 113.28 (C-4), 113.11 (C-10), 105.92 (C-3′), 70.49 (CHiPr), 23.83 (CH3), 21.74 (CH3iPr). Elemental analysis calcd (%) for C20H19NO4: C, 71.20; H, 5.68; N, 4.15; found C, 71.14; H, 5.67; N, 4.12. m/z: 337.13 (100.0%).
(Z)-N-(2-(3-isopropoxybenzylidene)-3-oxo-2,3-dihydrobenzofuran-5-yl)acetamide (19):
Yield: 91%; mp: 167 °C; 1H NMR (300 MHz, DMSO-d6): δ 10.19 (s, 1H, NH), 8.12 (d, 1H, J = 2 Hz, C-H4), 7.83–7.80 (dd, 1H, J = 2.2; 8.9 Hz, C-H6), 7.57–7.52 (m, 3H, C-H2′,4′,7), 7.40 (dt, 1H, J = 8.0 Hz, C-H5′), 7.04–7.01 (dd, 1H, J = 2.6;8.2 Hz, C-H6′), 6.91 (s, 1H, C-H10), 4.68 (q, 1H, J = 5.9;11.9 Hz, C-Hisop), 2.07 (s, 3H, NHCOCH3), 1.31–1.29 (d, 6H, J = 5.8 Hz, C-H3isop). 13C NMR (75 MHz, DMSO-d6): δ 183.73 (C-3), 168.49 (CO), 161.35 (C-8), 157.66 (C-3′), 146.87 (C-2), 135.62 (C-5), 133.17 (C-1′), 130.1 (C-5′), 128.86 (C-6), 123.56 (C-6′), 120.64 (C-9), 118.25 (C-2′), 117.32 (C-3′), 113.41 (C-7), 113.17 (C-4), 112.28 (C-10), 69.35 (CHiPr), 23.88 (CH3), 21.77 (CH3iPr). Elemental analysis calcd (%) for C20H19NO4: C, 71.20; H, 5.68; N, 4.15; found C, 71.18; H, 5.66; N, 4.16. m/z: 337.13 (100.0%).
(Z)-N-(2-(4-isopropoxybenzylidene)-3-oxo-2,3-dihydrobenzofuran-5-yl)acetamide (20):
Yield: 93%; mp: 200.2 °C; 1H NMR (300 MHz, DMSO-d6): δ 10.14 (s, 1H, NH), 8.10 (d, 1H, J = 1.9 Hz, C-H4), 7.94–7.91 (d, 2H, J = 8.8 Hz, C-H2′), 7.82–7.78 (dd, 1H, J = 2.2;8.8 Hz, C-H6), 7.51–7.48 (d, 1H, J = 8.89 Hz, C-H7), 7.06–7.03 (d, 2H, J = 8.8 Hz, C-H3′), 6.90 (s, 1H, C-H10), 4.72 (q, 1H, J = 5.9;11.9 Hz, C-Hisop), 2.07 (s, 3H, NHCOCH3), 1.30–1.28 (d, 6H, J = 5.8 Hz, C-H3isop). 13C NMR (75 MHz, DMSO-d6): δ 183.25 (C-3), 168.39 (CO), 160.98 (C-8), 159.16 (C-4′), 145.6 (C-2), 135.39 (C-5), 133.41 (C-2′), 128.48 (C-6), 124.02 (C-1′), 120.94 (C-9), 115.94 (C-3′), 113.2 (C-7), 113.06 (C-4), 112.77 (C-10), 69.5 (CHiPr), 23.83 (CH3), 21.68 (CH3iPr). Elemental analysis calcd (%) for C20H19NO4: C, 71.20; H, 5.68; N, 4.15; found C, 71.18; H, 5.69; N, 4.12. m/z: 337.13 (100.0%).
(Z)-5-amino-2-(2-isopropoxybenzylidene)benzofuran-3(2H)-one (21):
Yield: 68%; mp: 126.7 °C; 1H NMR (300 MHz, DMSO-d6): δ 8.19–8.17 (d, 1H, J = 6.9 Hz, C-H6′), 7.39 (t, 1H, J = 7.3 Hz, C-H4′), 7.26–7.24 (d, 1H, J = 8.7 Hz, C-H7), 7.14 (d, 1H, C-H3′), 7.11 (bs, 2H, NH2), 7.08–7.06 (d, 1H, C-H5′), 7.06–7.04 (dd, 1H, J = 2.2;7.7 Hz, C-H6), 6.95 (s, 1H, C-H10), 6.85 (d, 1H, J = 2.2 Hz, C-H4), 4.73 (q, 1H, J = 5.9;11.9 Hz, C-Hisop), 1.34–1.32 (d, 6H, J = 5.8 Hz, C-H3isop). 13C NMR (75 MHz, DMSO-d6): 184.16 (CO), 158.14 (C-8), 156.62 (C-2′), 147.02 (C-2), 145.24 (C-5), 131.55 (C-6′), 131.33 (C-4′), 124.76 (C-6), 121.4 (C-9), 121.11 (C-5′), 120.72 (C-1′), 113.85 (C-7), 113.23 (C-10), 105.77 (C-3′), 104.8 (C-4), 70.46 (CHiPr), 21.82 (CH3iPr). Elemental analysis calcd (%) for C20H19NO4: C, 71.20; H, 5.68; N, 4.15; found C, 71.24; H, 5.74; N, 4.18. m/z: 337.13 (100.0%).
(Z)-5-amino-2-(3-isopropoxybenzylidene)benzofuran-3(2H)-one (22):
Yield: 51%; mp: 198.4 °C; 1H NMR (300 MHz, DMSO-d6): δ 7.57–7.54 (d, 1H, J = 7.8 Hz, C-H6′), 7.51 (bs, 1H, C-H2′), 7.51–7.48 (d, 1H, J = 8.6 Hz, C-H7), 7.43–7.40 (d, 2H, J = 7.8 Hz, C-H4′), 7.41 (dt, 1H, J = 7.8 Hz, C-H5′), 7.27 (d, 1H, J = 1.9 Hz, C-H4), 7.04–7.01 (dd, 1H, J = 2.0;8.0 Hz, C-H6), 6.89 (s, 1H, C-H10), 4.68 (q, 1H, J = 5.9;11.9 Hz, C-Hisop), 1.31–1.29 (d, 6H, J = 5.8 Hz, C-H3isop). 13C NMR (75 MHz, DMSO-d6): δ 183.65 (C-3), 160.93 (C-8), 157.67 (C-3′), 146.95 (C-2), 137.64 (C-5), 133.24 (C-1′), 130.11 (C-5′), 128.17 (C-6), 123.56 (C-6′), 121.25 (C-9), 118.24 (C-2′), 117.28 (C-3′), 113.96 (C-7), 112.14 (C-4), 111.46 (C-10), 69.35 (CHiPr), 21.79 (CH3iPr). Elemental analysis calcd (%) for C10H17NO3: C, 71.20; H, 5.68; N, 4.15; found C, 71.18; H, 5.65; N, 4.12. m/z: 295.12 (100.0%).
(Z)-5-amino-2-(4-isopropoxybenzylidene)benzofuran-3(2H)-one (23):
Yield: 50%; mp: >350 °C; 1H NMR (300 MHz, DMSO-d6): δ 7.97–7.95 (d, 2H, J = 8.8 Hz, C-H2′), 7.66–7.64 (m, 3H, C-H4,6,7), 7.07–7.05 (d, 2H, J = 8.8 Hz, C-H3′), 6.98 (s, 1H, C-H10). 13C NMR (75 MHz, DMSO-d6): δ 183.22 (C-3), 160.22 (C-8), 159.12 (C-4′), 145.7 (C-2), 133.38 (C-2′), 138.33 (C-5), 127.35 (C-7), 124.1 (C-1′), 121.52 (C-9), 115.94 (C-3′), 113.69 (C-6), 112.51 (C-4), 110.58 (C-10), 69.49 (CH2iPr), 21.68 (CH3iPr). Elemental analysis calcd (%) for C10H17NO3: C, 71.20; H, 5.68; N, 4.15; found C, 71.15; H, 5.66; N, 4.18. m/z: 295.12 (100.0%).
(Z)-N-(2-(2-fluorobenzylidene)-3-oxo-2,3-dihydrobenzofuran-5-yl)acetamide (24):
Yield: 68%; mp: 226 °C; 1H NMR (300 MHz, DMSO-d6): δ 10.17 (s, 1h, NH), 8.25 (t, 1H, J = 7.8 Hz, C-H2′), 8.12 (d, 1H, J = 2.2 Hz, C-H4), 7.85–7.82 (dd, 1H, J = 2.5, 8.7 Hz, C-H6′), 7.54–7.51 (m, 2H, C-H4′,7), 7.41–7.39 (d, 1H, C-H3′), 7.35 (dt, 1H, C-H5′), 6.90 (s, 1H, C-H10), 2.08 (s, 3H, CH3). 13C NMR (75 MHz, DMSO-d6): δ 183.5 (C-3), 168.44 (CO), 164.9–161.44 (C-2′, J = 260 Hz), 161.41 (C-8), 147.71 (C-2), 135.78 (C-5), 132.24–132.12 (C-4′, J = 8.8 Hz), 131.31 (C-6′), 129.02 (C-6), 125.11 (C-5′, J = 3.3 Hz), 120.38 (C-9), 119.67–119.52 (C-1′, J = 11 Hz), 115.93–115.64 (C-3′, J = 22 Hz), 113.38 (C-7), 113.25 (C-4), 102.1–102.0 (C-10, J = 7.7 Hz), 23.83 (CH3). Elemental analysis calcd (%) for C17H12FNO3: C, 68.68; H, 4.07; N, 4.71; found C, 68.65; H, 4.01; N, 4.65. m/z: 297.08 (100.0%).
(Z)-N-(2-(3-fluorobenzylidene)-3-oxo-2,3-dihydrobenzofuran-5-yl)acetamide (25):
Yield: 82%; mp: 243.6 °C; 1H NMR (300 MHz, DMSO-d6): δ 10.22 (s, 1h, NH), 8.12 (d, 1H, J = 2.1 Hz, C-H4), 7.83–7.79 (m, 3H, C-H6,2′,6′), 7.59–7.51 (m, 2H, C-H4′,7), 7.31–7.39 (dt, 1H, J = 2.1, 8.4 Hz, C-H5′), 6.59 (s, 1H, C-H10), 2.07 (s, 3H, CH3). 13C NMR (75 MHz, DMSO-d6): δ 183.83(C-3), 168.6(CO), 163.85–160.62 (C-3′, J = 244 Hz), 161.48 (C-8), 147.37 (C-2), 135.77 (C-5), 134.3–134.19 (C-1′, J = 8.25 Hz), 131.08–130.97 (C-5′, J = 8.25 Hz), 129.05 (C-6), 127.62–127.58 (C-6′, J = 2.75 Hz), 120.53 (C-9), 117.45–117.15 (C-4′, J = 22.56 Hz), 117.02–116.73 (C-2′, J = 21.5 Hz), 113.53 (C-7), 113.25 (C-4), 110.69–110.66 (C-10, J = 2.75 Hz), 23.92 (CH3). Elemental analysis calcd (%) for C17H12FNO3: C, 68.68; H, 4.07; N, 4.71; found C, 68.58; H, 4.12; N, 4.73. m/z: 297.08 (100.0%).
(Z)-5-amino-2-(2-fluorobenzylidene)benzofuran-3(2H)-one (26):
Yield: 62%; mp: 161.3 °C; 1H NMR (300 MHz, DMSO-d6): δ 8.23 (dt, 1H, J = 1.65, 7.8 Hz, C-H6′), 7.51–7.47 (m, 1H, C-H4′), 7.37 (t, 1H, C-H3′), 7.34 (dt, 1H, C-H5′), 7.28–7.25 (d, 1H, J = 8.8 Hz, C-H7), 7.09–7.05 (dd, 1H, J = 2.5, 8.7 Hz, C-H6), 6.86 (d, 1H, J = 2.4 Hz, C-H4), 6.81 (s, 1H, C-H10), 5.28 (bs, 2H, NH2). 13C NMR (75 MHz, DMSO-d6): δ 184.05 (C-3), 164.75–161.71 (C-2′, J = 260 Hz), 158.19 (C-8), 148.04 (C-2), 145.83 (C-5), 131.84–131.73 (C-4′, J = 8 Hz), 131.2 (C-6′), 125.11–125.06 (C-5′, J = 3 Hz), 124.84 (C-6), 120.69 (C-9), 119.97–119.81 (C-1′, J = 12 Hz), 115.85–115.56 (C-3′), 113.22 (C-7), 105.54 (C-4), 100.82–100.72 (C-10).
Elemental analysis calcd (%) for C15H10FNO2: C, 70.58; H, 3.95; N, 5.49; found C, 70.44; H, 3.99; N, 5.32. m/z: 255.07 (100.0%).
(Z)-5-amino-2-(3-fluorobenzylidene)benzofuran-3(2H)-one (27):
Yield: 85%; mp: 159.7 °C; 1H NMR (300 MHz, DMSO-d6): δ 7.80–7.78 (m, 2H, C-H2′,4′), 7.57–7.50 (dt, 1H, C-H5′), 7.32–7.27 (m, 2H, C-H7,6′), 7.10–7.07 (dd, 1H, J = 2.5, 8.7 Hz, C-H6), 6.87 (d, 1H, J = 2.4 Hz, C-H4), 6.85 (s, 1H, C-H10). 13C NMR (75 MHz, DMSO-d6): δ 184.26(C-3), 163.81–160.58 (C-3′, J = 244 Hz), 158.49 (C-8), 147.63 (C-2), 145.01 (C-5), 134.58–134.47 (C-1′, J = 8.25 Hz), 130.85 (C-5′), 127.34 (C-6), 125.18 (C-6′), 120.84(C-9), 117.22–116.92 (C-4′, J = 22.5 Hz), 116.64–116.36 (C-2′), 113.38(C-7), 109.49–109.45 (C-10, J = 2.75 Hz), 106.08 (C-4). Elemental analysis calcd (%) for C15H10FNO2: C, 70.58; H, 3.95; N, 5.49; found C, 70.66; H, 4.08; N, 5.31. m/z: 255.07 (100.0%).
(Z)-5-amino-2-(4-fluorobenzylidene)benzofuran-3(2H)-one (28):
Yield: 82%; mp: 164.4 °C; 1H NMR (300 MHz, DMSO-d6): δ 8.09–8.04 (dd, 2H, J = 7.8 Hz, C-H2′), 7.81–7. 78 (d, 1H, C-H4′), 7.57–7.54 (d, 1H, J = 8.4 Hz, C-H7), 7.36 (t, 2H, C-H3′), 7.33 (d, 1H, C-H4), 6.98 (s, 1H, C-H10). 13C NMR (75 MHz, DMSO-d6): δ 183.5 (C-4), 165.39 (C-8), 164.38–161.07 (C-4′, J = 250 Hz), 145.91 (C-2), 137.62 (C-5), 133.73–133.62 (C-2′, J = 9 Hz), 128.55 (C-1′, J = 3 Hz), 124.24 (C-6), 123.95 (C-7), 120.81 (C-9), 116.26–115.97 (C-3′, J = 22 Hz), 113.14 (C-4), 111.04 (C-10). Elemental analysis calcd (%) for C15H10FNO2: C, 70.58; H, 3.95; N, 5.49; found C, 70.52; H, 4.07; N, 5.23. m/z: 255.06956 (100.0%).
(Z)-N-(3-oxo-2-(3-(trifluoromethyl)benzylidene)-2,3-dihydrobenzofuran-5-yl)acetamide (29):
Yield: 82%; mp: 252.1 °C; 1H NMR (300 MHz, DMSO-d6): δ 10.20 (bs, 1H, NH), 8.30–8.28 (m, 2H, C-H2′,4′), 8.13 (d, 1H, J = 2.1 Hz, C-H4), 7.85–7.81 (dd, 1H, J = 2.3, 8.8 Hz, C-H6), 7.79–7.72 (m, 2H, C-H5′,6′), 7.56–7.53 (d, 1H, J = 8.9 Hz, C-H7), 7.06 (s, 1H, C-H10), 2.07 (s, 3H, CH3). 13C NMR (75 MHz, DMSO-d6): δ 183.81 (C-3), 168.54 (CO), 161.44 (C-8), 147.58 (C-2), 135.82 (C-5), 134.74 (C-1′), 133.09 (C-6), 130.15 (C-6′), 129.94 (C-5′), 129.63–129.03 (C-3′, J = 31.7 Hz), 127.46 (C-4′), 126.17 (C-2′), 125.32–122.61 (CF3, J = 270 Hz), 120.48 (C-9), 113.5 (C-7), 113.23 (C-4), 110.18 (C-10), 23.91 (CH3). Elemental analysis calcd (%) for C18H12F3NO3: C, 62.25; H, 3.48; N, 4.03; found C, 62.09; H, 3.54; N, 3.98. m/z: 347.07693 (100.0%).
(Z)-5-amino-2-(3-(trifluoromethyl)benzylidene)benzofuran-3(2H)-one (30):
Yield: 82%; mp: >350 °C; 1H NMR (300 MHz, DMSO-d6): δ 8.31–8.29 (m, 2H, C-H2′,4′), 7.80–7.73 (m, 2H, C-H-6′,5′), 7.52–7.49 (d, 1H, J = 8.9 Hz, C-H7), 7.40–7.36 (dd, 1H, J = 2.3, 8.9 Hz, C-H7), 7.26 (s, 1H, C-H4), 7.05 (s, 1H, C-H10). 13C NMR (75 MHz, DMSO-d6): δ 183.7 (C-3), 161.09 (C-8), 147.65 (C-2), 134.75 (C-1′), 133.13 (C-5), 130.16 (C-6′), 130.01–129.59 (C-3′, J = 31 Hz), 128.44 (C-7), 127.48 (C-4′), 127.43 (C-2′), 126.11–122.78 (CF3, J = 250 Hz), 121.1 (C-9), 114.05 (C-6), 111.68 (C-4), 110.05 (C-10). Elemental analysis calcd (%) for C16H10F3NO2: C, 62.96; H, 3.30; N, 4.59; found C, 63.11; H, 3.35; N, 4.55. m/z: 305.06636 (100.0%).
(Z)-4-((5-acetamido-3-oxobenzofuran-2(3H)-ylidene)methyl)benzoic acid (31):
Yield: 71%; mp: 165.3 °C; 1H NMR (300 MHz, DMSO-d6): δ 10.28 (s, 1H, NH), 8.14 (d, 1H, J = 1.9 Hz, C-H4), 8.10–8.07 (d, 2H, J = 8.8 Hz, C-H2′), 8.05–8.02 (d, 2H, J = 8.8 Hz, C-H3′), 7.87–7.83 (dd, 1H, J = 2.2;8.8 Hz, C-H6), 7.55–7.53 (d, 1H, J = 8.89 Hz, C-H7), 6.98 (s, 1H, C-H10), 2.08 (s, 3H, NHCOCH3). 13C NMR (75 MHz, DMSO-d6): δ 183.85 (C-3), 168.54 (CO), 166.77 (COOH), 161.47 (C-8), 147.69 (C-2), 136.08 (C-1′), 135.82 (C-5), 131.35 (C-4′), 131.23 (C-3′), 129.74 (C-2′), 129.04 (C-6), 120.47 (C-9), 113.44 (C-7), 113.25 (C-4), 110.61 (C-10), 23.89 (CH3). Elemental analysis calcd (%) for C18H13NO5: C, 66.87; H, 4.05; N, 4.33; found C, 66.85; H, 4.12; N, 4.27. m/z: 323.07937 (100.0%).
(Z)-N-(7-nitro-3-oxo-2-(3-phenoxybenzylidene)-2,3-dihydrobenzofuran-5-yl)acetamide (32):
Yield: 91%; mp: 230.3 °C; 1H NMR (300 MHz, DMSO-d6): δ 10.51 (bs, 1H, NH), 8.71 (d, 1H, J = 2.2 Hz, C-H6), 8.35 (d, 1H, J = 2.2 Hz, C-H4), 7.89–7.86 (d, 2H, J = 7.8 Hz, C-H2″), 7.56 (t, 1H, J = 8.16 Hz, C-H5′), 7.42 (dt, 2H, C-H3″), 7.20–7.17 (d, 1H, J = 7.3 Hz, C-H4′), 7.14 (s, 1H, C-H2′), 7.09–7.06 (m, 2H, C-H10, 6′), 2.11 (s, 3H, CH3). 13C NMR (75 MHz, DMSO-d6): δ 178.66 (C-3), 169.01 (CO), 157.07 (C-1″), 156.36 (C-3′), 153.21 (C-8), 146.13 (C-2), 143.98 (C-7), 135.31 (C-5), 133.21 (C-1′), 130.64 (C-5′), 130.09 (C-3″), 127.01 (C-6), 124.60 (C-9), 123.69 (C-4″), 121.59 (C-6′), 120.86 (C-4), 119.76 (C-4′), 119.35 (C-2′), 118.66 (C-2″), 113.73 (C-10), 23.86 (CH3). Elemental analysis calcd (%) for C23H16N2O6: C, 66.34; H, 3.87; N, 6.73; found C, 66.21; H, 3.74; N, 6.71. m/z: 416.10084 (100.0%).
(Z)-N-(2-(3-hydroxybenzylidene)-3-oxo-2,3-dihydrobenzofuran-5-yl)acetamide (33):
Yield: 75%; mp: >350 °C; 1H NMR (300 MHz, DMSO-d6): δ 10.15 (s, 1H, NH), 9.67 (bs, 1H, OH), 8.11–8.10 (d, 1H, J = 2.2 Hz, C-H4), 7.83–7.80 (dd, 1H, J = 2.2;8.8 Hz, C-H6), 7.50–7.47 (d, 1H, J = 8.9 Hz, C-H7), 7.42 (d, 1H, C-H2′), 7.40–7.38 (d, 1H, J = 7.8 Hz, C-H6′), 7.30 (t, 1H, J = 7.8 Hz, C-H5′), 6.89–6.86 (dd, 1H, C-H4′), 6.82 (s, 1H, C-H10), 2.07 (s, 3H, C-H3′). 13C NMR (75 MHz, DMSO-d6): δ 183.65 (C-3), 168.41 (CO), 161.27 (C-8), 157.58 (C-3′), 146.69 (C-2), 135.54 (C-5), 132.9 (C-1′), 129.89 (C-5′), 128.8 (C-6), 122.64 (C-6′), 120.65 (C-9), 117.53 (C-3′), 117.46 (C-2′), 113.21 (C-7), 113.16 (C-4), 112.47 (C-10), 23.83 (CH3). Elemental analysis calcd (%) for C17H13NO4: C, 69.15; H, 4.44; N, 4.74; found C, 69.01; H, 4.48; N, 4.69. m/z: 295.08 (100.0%).
(Z)-5-amino-2-(3-hydroxybenzylidene)benzofuran-3(2H)-one (34):
Yield: 95%; mp: 225.2 °C; 1H NMR (300 MHz, DMSO-d6): δ 9.63 (bs, 1H, OH), 7.39 (d, 1H, C-H2′), 7.37–7.34 (d, 1H, J = 7.8 Hz, C-H6′), 7.28 (t, 1H, J = 7.8 Hz, C-H5′), 7.25–7.22 (d, J = 8.8 Hz, C-H7), 7.07–7.04 (dd, 1H, J = 2.2;8.5 Hz, C-H4′), 6.86–6.83 (m, 2H, C-H4,6), 6.70 (s, 1H, C-H10), 5.24 (bs, 2H, NH2). 13C NMR (75 MHz, DMSO-d6): δ 184.22 (C-3), 158.08 (C-8), 157.53 (C-3′), 146.99 (C-2), 145.57 (C-5), 133.21 (C-1′), 129.82 (C-5′), 124.66 (C-6), 122.42 (C-6′), 120.98 (C-9), 117.34 (C-3′), 117.1 (C-2′), 113.06 (C-7), 111.17 (C-4), 105.46 (C-10). Elemental analysis calcd (%) for C15H11NO3: C, 71.14; H, 4.38; N, 5.53; found C, 71.21; H, 4.34; N, 5.49. m/z: 253.07 (100.0%).
5. Conclusions
In the present study, 31 new aurone derivative compounds were synthetized. These new compounds were obtained by the substitution of the aurone scaffold at position 5 by amino and acetamido groups, and through various substitutions at the 2′, 3′ and 4′ positions. Antimicrobial testing identified two of these compounds, i.e., 10 and 20, as the most active on both Gram-positive and -negative bacteria with MIC values as low as 0.78 µM. These were also the safest regarding human cells. The two selected compounds shared some structural similarity with the 5-acetamido substitution. The SAR study from this work correlates with the results previously obtained by Olleik et al. [14] showing that benzyloxy and isopropyloxy lead to interesting activities in aurone scaffolds with substitution on the A ring with amino or acetamido groups, improving the activity compared to the natural OH group. Taken together, these results confirm that the aurone scaffold is a promising structure that could be the starting point for the design of new antibacterial agents by diversifying the substitution pattern on A and B rings altogether.
6. Patents
Aurone derivatives and uses thereof for controlling bacteria and/or fungi. PCT/EP2021/069047. BOLLA Jean Michel., MARESCA Marc, NEULAT-RIPOLL Fabienne, OLLEIK Hamza, PERRIER-VIRET Josette, PIQUE Valérie. ROBIN Maxime.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/antibiotics13040300/s1, File S1: NMR Spectra of compounds 4–34.
Author Contributions
Conceptualization, M.R.; methodology, M.R., J.-M.B. and M.M.; validation, M.R., J.-M.B. and M.M.; formal analysis, A.D.M., H.O., E.C.-D., S.G., F.N.-R., R.H., M.C., J.-F.C., S.C., V.P., Y.C., M.M. and M.R.; investigation, A.D.M., H.O., E.C.-D., S.G., F.N.-R., R.H., M.C., J.-F.C., S.C., V.P., Y.C., M.M. and M.R.; data curation, M.R., S.C., J.-M.B. and M.M.; writing—original draft preparation, A.D.M., M.R. and M.M.; writing—review and editing, A.D.M., H.O., E.C.-D., S.G., F.N.-R., R.H., M.C., J.-F.C., S.C., V.P., Y.C., E.B., A.H., J.P., M.M. and M.R.; supervision, E.B., A.H., J.P., M.R. and M.M.; project administration, M.M. and M.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Région PACA EJD no. 2021-04403 for A.D.M. PhD fellowship.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data generated for this study are contained within the article.
Conflicts of Interest
The authors declare no conflicts of interest. Funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- O’Neill, A. New Antibacterial Agents for Treating Infections Caused by Multi-Drug Resistant Gram-Negative Bacteria. Expert Opin. Investig. Drugs 2008, 17, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Abdeen, S.; Kunkle, T.; Salim, N.; Ray, A.-M.; Mammadova, N.; Summers, C.; Stevens, M.; Ambrose, A.J.; Park, Y.; Schultz, P.G.; et al. Sulfonamido-2-Arylbenzoxazole GroEL/ES Inhibitors as Potent Antibacterials against Methicillin-Resistant Staphylococcus Aureus (MRSA). J. Med. Chem. 2018, 61, 7345–7357. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Wang, L.; Lü, Y.; Yue, C. Development and Research Progress of Anti-Drug Resistant Fungal Drugs. J. Infect. Public Health 2022, 15, 986–1000. [Google Scholar] [CrossRef]
- Bastos, R.W.; Rossato, L.; Goldman, G.H.; Santos, D.A. Fungicide Effects on Human Fungal Pathogens: Cross-Resistance to Medical Drugs and Beyond. PLoS Pathog. 2021, 17, e1010073. [Google Scholar] [CrossRef] [PubMed]
- Sati, H.; Alastruey-Izquierdo, A.; Perfect, J.; Govender, N.P.; Harrison, T.S.; Chiller, T.; Sorrell, T.C.; Bongomin, F.; Oladele, R.; Chakrabarti, A.; et al. HIV and Fungal Priority Pathogens. Lancet HIV 2023, 10, e750–e754. [Google Scholar] [CrossRef] [PubMed]
- Bongomin, F.; Gago, S.; Oladele, R.O.; Denning, D.W. Global and Multi-National Prevalence of Fungal Diseases—Estimate Precision. J. Fungi 2017, 3, 57. [Google Scholar] [CrossRef] [PubMed]
- Haudecoeur, R.; Boumendjel, A. Recent Advances in the Medicinal Chemistry of Aurones. Curr. Med. Chem. 2012, 19, 2861–2875. [Google Scholar] [CrossRef] [PubMed]
- Boumendjel, A. Aurones: A Subclass of Flavones with Promising Biological Potential. Curr. Med. Chem. 2003, 10, 2621–2630. [Google Scholar] [CrossRef]
- Lazinski, L.M.; Royal, G.; Robin, M.; Maresca, M.; Haudecoeur, R. Bioactive Aurones, Indanones, and Other Hemiindigoid Scaffolds: Medicinal Chemistry and Photopharmacology Perspectives. J. Med. Chem. 2022, 65, 12594–12625. [Google Scholar] [CrossRef]
- Boucherle, B.; Peuchmaur, M.; Boumendjel, A.; Haudecoeur, R. Occurrences, Biosynthesis and Properties of Aurones as High-End Evolutionary Products. Phytochemistry 2017, 142, 92–111. [Google Scholar] [CrossRef]
- Pare, P.W.; Dmitrieva, N.; Mabry, T.J. Phytoalexin Aurone Induced in Cephalocereus Senilis Liquid Suspension Culture. Phytochemistry 1991, 30, 1133–1135. [Google Scholar] [CrossRef]
- Paré, P.W.; Mischke, C.F.; Edwards, R.; Dixon, R.A.; Norman, H.A.; Mabry, T.J. Induction of Phenylpropanoid Pathway Enzymes in Elicitor-Treated Cultures ofCephalocereus Senilis. Phytochemistry 1992, 31, 149–153. [Google Scholar] [CrossRef]
- Farag, M.A.; Deavours, B.E.; de Fátima, A.; Naoumkina, M.; Dixon, R.A.; Sumner, L.W. Integrated Metabolite and Transcript Profiling Identify a Biosynthetic Mechanism for Hispidol in Medicago Truncatula Cell Cultures. Plant Physiol. 2009, 151, 1096–1113. [Google Scholar] [CrossRef] [PubMed]
- Olleik, H.; Yahiaoui, S.; Roulier, B.; Courvoisier-Dezord, E.; Perrier, J.; Pérès, B.; Hijazi, A.; Baydoun, E.; Raymond, J.; Boumendjel, A.; et al. Aurone Derivatives as Promising Antibacterial Agents against Resistant Gram-Positive Pathogens. Eur. J. Med. Chem. 2019, 165, 133–141. [Google Scholar] [CrossRef]
- Tiwari, K.N.; Monserrat, J.-P.; Hequet, A.; Ganem-Elbaz, C.; Cresteil, T.; Jaouen, G.; Vessières, A.; Hillard, E.A.; Jolivalt, C. In Vitro Inhibitory Properties of Ferrocene-Substituted Chalcones and Aurones on Bacterial and Human Cell Cultures. Dalton Trans. 2012, 41, 6451–6457. [Google Scholar] [CrossRef] [PubMed]
- Hadj-esfandiari, N.; Navidpour, L.; Shadnia, H.; Amini, M.; Samadi, N.; Faramarzi, M.A.; Shafiee, A. Synthesis, Antibacterial Activity, and Quantitative Structure–Activity Relationships of New (Z)-2-(Nitroimidazolylmethylene)-3(2H)-Benzofuranone Derivatives. Bioorganic Med. Chem. Lett. 2007, 17, 6354–6363. [Google Scholar] [CrossRef] [PubMed]
- Kumar, G.; Lathwal, E.; Saroha, B.; Kumar, S.; Kumar, S.; Chauhan, N.S.; Kumar, T. Synthesis and Biological Evaluation of Quinoline-Based Novel Aurones. ChemistrySelect 2020, 5, 3539–3543. [Google Scholar] [CrossRef]
- Olleik, H.; Nicoletti, C.; Lafond, M.; Courvoisier-Dezord, E.; Xue, P.; Hijazi, A.; Baydoun, E.; Perrier, J.; Maresca, M. Comparative Structure-Activity Analysis of the Antimicrobial Activity, Cytotoxicity, and Mechanism of Action of the Fungal Cyclohexadepsipeptides Enniatins and Beauvericin. Toxins 2019, 11, 514. [Google Scholar] [CrossRef]
- Berthier, M.; Fauchère, J.-L.; Perrin, J.; Grignon, B.; Oriot, D. Fulminant Meningitis Due to Bacillus Anthracis in 11-Year-Old Girl during Ramadan. Lancet 1996, 347, 828. [Google Scholar] [CrossRef]
- Doll, J.M.; Zeitz, P.S.; Ettestad, P.; Bucholtz, A.L.; Davis, T.; Gage, K. Cat-Transmitted Fatal Pneumonic Plague in a Person Who Traveled from Colorado to Arizona. Am. J. Trop. Med. Hyg. 1994, 51, 109–114. [Google Scholar] [CrossRef]
- Olleik, H.; Yacoub, T.; Hoffer, L.; Gnansounou, S.M.; Benhaiem-Henry, K.; Nicoletti, C.; Mekhalfi, M.; Pique, V.; Perrier, J.; Hijazi, A.; et al. Synthesis and Evaluation of the Antibacterial Activities of 13-Substituted Berberine Derivatives. Antibiotics 2020, 9, 381. [Google Scholar] [CrossRef] [PubMed]
- Casanova, M.; Olleik, H.; Hdiouech, S.; Roblin, C.; Cavalier, J.-F.; Point, V.; Jeannot, K.; Caron, B.; Perrier, J.; Charriau, S.; et al. Evaluation of the Efficiency of Random and Diblock Methacrylate-Based Amphiphilic Cationic Polymers against Major Bacterial Pathogens Associated with Cystic Fibrosis. Antibiotics 2023, 12, 120. [Google Scholar] [CrossRef] [PubMed]
- Loupias, P.; Laumaillé, P.; Morandat, S.; Mondange, L.; Guillier, S.; El Kirat, K.; Da Nascimento, S.; Biot, F.; Taudon, N.; Dassonville-Klimpt, A.; et al. Synthesis and Study of New Siderophore Analog-Ciprofloxacin Conjugates with Antibiotic Activities against Pseudomonas aeruginosa and Burkholderia spp. Eur. J. Med. Chem. 2023, 245, 114921. [Google Scholar] [CrossRef]
- Broncano-Lavado, A.; Senhaji-Kacha, A.; Santamaría-Corral, G.; Esteban, J.; García-Quintanilla, M. Alternatives to Antibiotics against Mycobacterium Abscessus. Antibiotics 2022, 11, 1322. [Google Scholar] [CrossRef] [PubMed]
- Madani, A.; Ridenour, J.N.; Martin, B.P.; Paudel, R.R.; Abdul Basir, A.; Le Moigne, V.; Herrmann, J.-L.; Audebert, S.; Camoin, L.; Kremer, L.; et al. Cyclipostins and Cyclophostin Analogues as Multitarget Inhibitors That Impair Growth of Mycobacterium Abscessus. ACS Infect. Dis. 2019, 5, 1597–1608. [Google Scholar] [CrossRef]
- Benkhaled, B.T.; Hadiouch, S.; Olleik, H.; Perrier, J.; Ysacco, C.; Guillaneuf, Y.; Gigmes, D.; Maresca, M.; Lefay, C. Elaboration of Antimicrobial Polymeric Materials by Dispersion of Well-Defined Amphiphilic Methacrylic SG1-Based Copolymers. Polym. Chem. 2018, 9, 3127–3141. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).