Innovative Techniques for Infection Control and Surveillance in Hospital Settings and Long-Term Care Facilities: A Scoping Review
Abstract
1. Introduction
2. Methods
2.1. Search Strategy
2.2. Eligibility Criteria
2.3. Study Selection
2.4. Data Extraction and Quality Assessment
2.5. Data Synthesis and Statistical Analysis
3. Results
3.1. Search Results
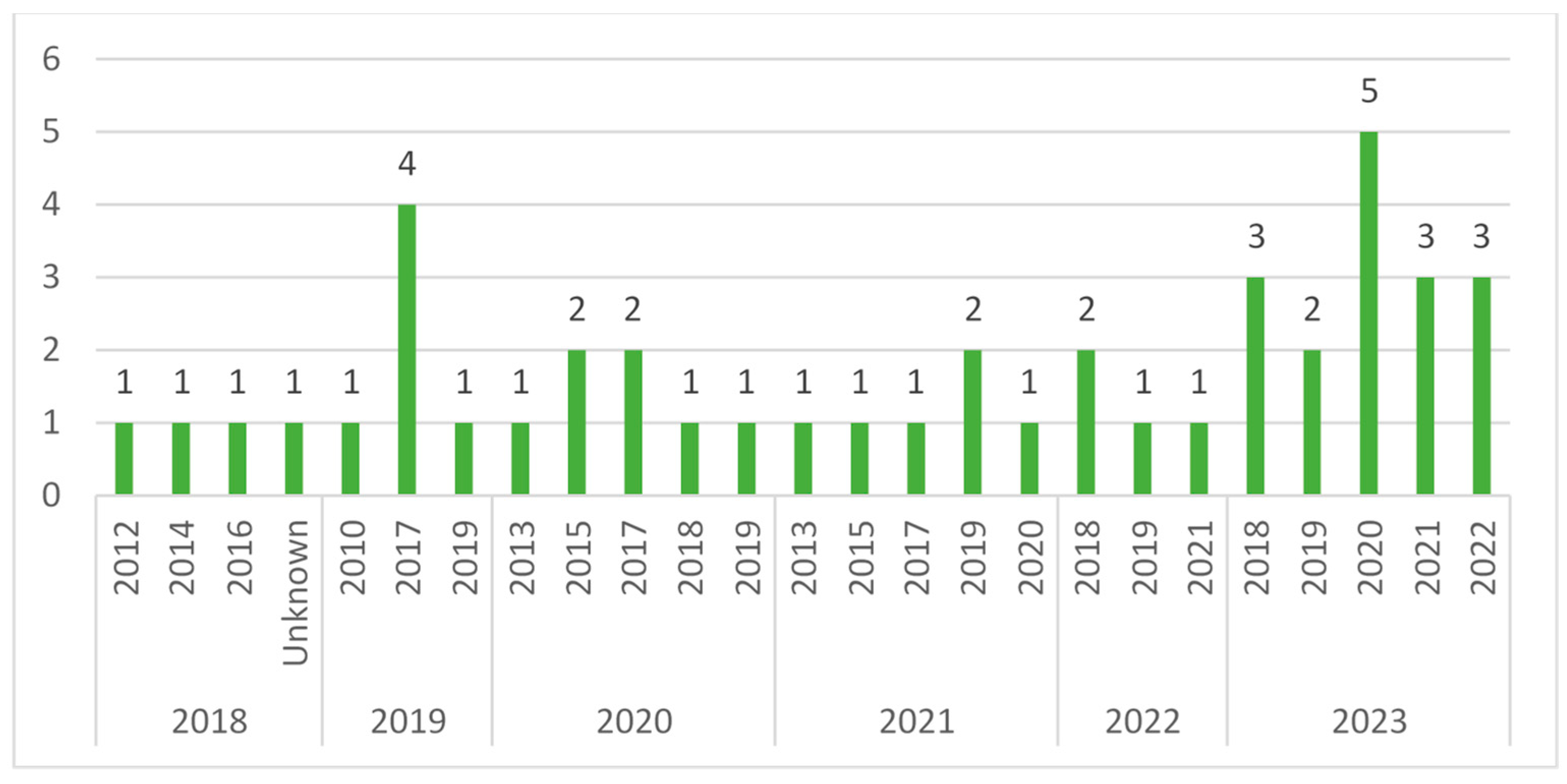
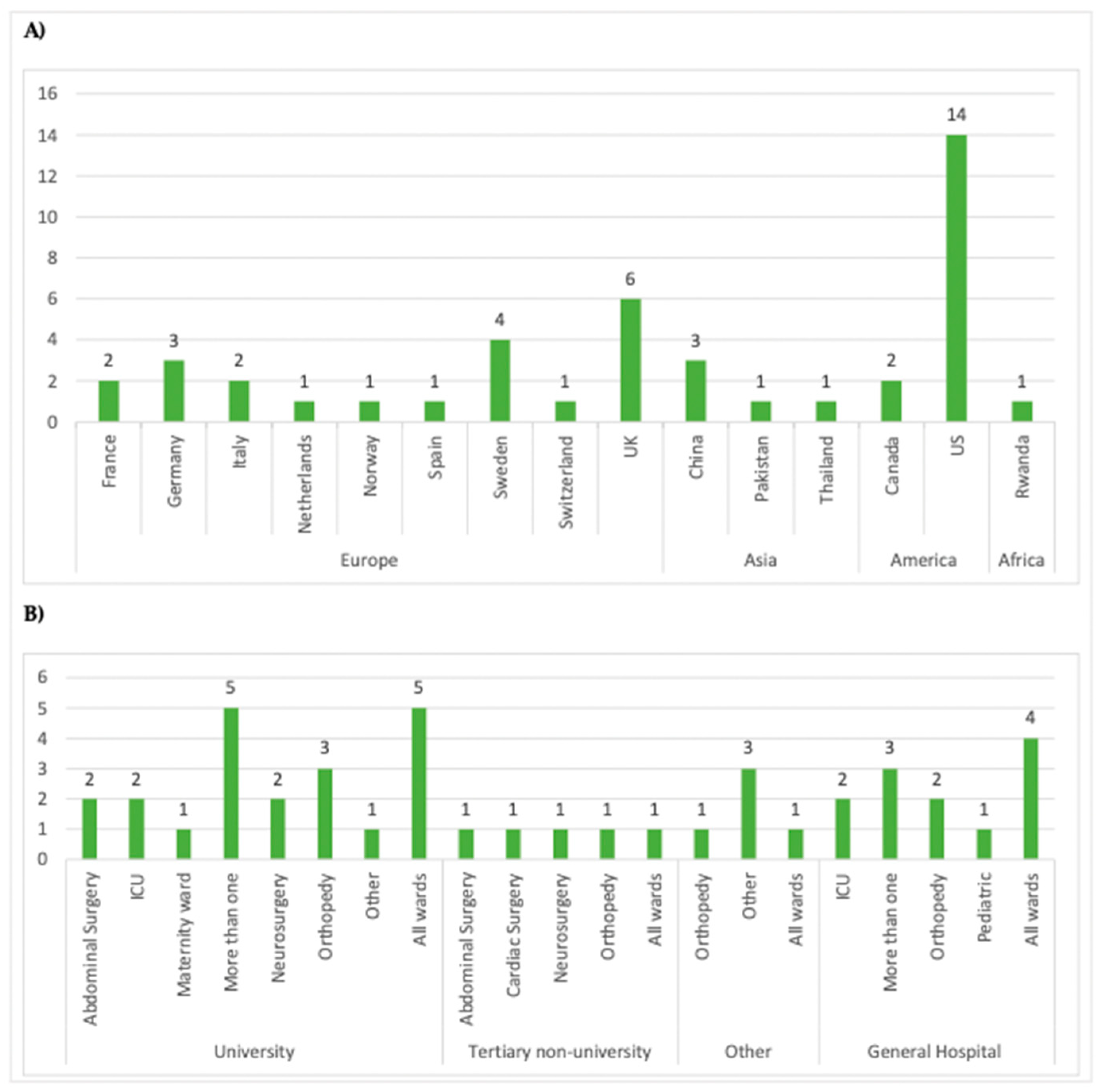
3.2. Description of Studies
Quality Assessment
3.3. Description of Interventions
3.3.1. Healthcare-Associated Infections
3.3.2. Innovations
3.3.3. Thematic Analysis
- Patient empowerment [29,30,33,56,57]: Smartphone and tablet computing devices with e-health and m-health technologies are implemented, especially in postsurgical settings, to improve patients’ management, fostering their empowerment. These outcomes are also measured in the same studies, with patient-reported experience measures (PREMS) and patient-reported outcomes measures (PROMS).
- Workload reduction and cost reduction [13,19,21,23,26,28,34,35,39,41,42,43,44,45,46,49]: Health informatics, machine learning, and natural language processing are implemented in various settings. Several articles examine the potential of these technologies in reducing the economic burden of infection and prevention control activities and strengthening the workforce, especially in scarcity situations.
3.3.4. Comparative Analysis
4. Discussion
Limitations of this Study
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- WHO. Global Report on Infection Prevention and Control. Available online: https://www.who.int/publications/i/item/9789240051164 (accessed on 30 November 2023).
- Collineau, L.; Godebert, E.; Thibaut, S.; Lemenand, O.; Birgand, G.; Caillon, J.; Bourely, C. Evaluation of the French Surveillance System for Epidemiological Surveillance of Antimicrobial Resistance in the Community and Nursing Homes. JAC Antimicrob. Resist. 2022, 4, dlac078. [Google Scholar] [CrossRef]
- Point Prevalence Survey of Healthcare-Associated Infections and Antimicrobial Use in European Acute Care Hospitals—Protocol Version 6.1. Available online: https://www.ecdc.europa.eu/en/publications-data/point-prevalence-survey-healthcare-associated-infections-and-antimicrobial-use-vs-6-1 (accessed on 30 November 2023).
- Zingg, W.; Holmes, A.; Dettenkofer, M.; Goetting, T.; Secci, F.; Clack, L.; Allegranzi, B.; Magiorakos, A.P.; Pittet, D.; Carmeli, Y.; et al. Hospital Organisation, Management, and Structure for Prevention of Health-Care-Associated Infection: A Systematic Review and Expert Consensus. Lancet Infect. Dis. 2015, 15, 212–224. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Jiang, Y.; Zhi, H.; Dong, Y.; Li, H.; Ma, S.; Wang, Y.; Dong, Q.; Shen, H.; Wang, Y. Artificial Intelligence in Healthcare: Past, Present and Future. Stroke Vasc. Neurol. 2017, 2, 230–243. [Google Scholar] [CrossRef] [PubMed]
- Sips, M.E.; Bonten, M.J.M.; Van Mourik, M.S.M. Automated Surveillance of Healthcare-Associated Infections: State of the Art. Curr. Opin. Infect. Dis. 2017, 30, 425–431. [Google Scholar] [CrossRef]
- Shenoy, E.S.; Branch-Elliman, W. Automating Surveillance for Healthcare-Associated Infections: Rationale and Current Realities (Part I/III). Antimicrob. Steward. Healthc. Epidemiol. 2023, 3, e25. [Google Scholar] [CrossRef] [PubMed]
- Tricco, A.C.; Lillie, E.; Zarin, W.; O’Brien, K.K.; Colquhoun, H.; Levac, D.; Moher, D.; Peters, M.D.J.; Horsley, T.; Weeks, L.; et al. PRISMA Extension for Scoping Reviews (PRISMA-ScR): Checklist and Explanation. Ann. Intern. Med. 2018, 169, 467–473. [Google Scholar] [CrossRef]
- ECDC. Digital Technologies for the Surveillance, Prevention and Control of Infectious Diseases—A Scoping Review of the Research Literature. Available online: https://www.ecdc.europa.eu/en/publications-data/digital-technologies-surveillance-prevention-and-control-infectious-diseases (accessed on 20 November 2023).
- Sundermann, A.J.; Chen, J.; Miller, J.K.; Saul, M.I.; Shutt, K.A.; Griffith, M.P.; Mustapha, M.M.; Ezeonwuka, C.; Waggle, K.; Srinivasa, V.; et al. Outbreak of Pseudomonas Aeruginosa Infections from a Contaminated Gastroscope Detected by Whole Genome Sequencing Surveillance. Clin. Infect. Dis. 2021, 73, E638–E642. [Google Scholar] [CrossRef]
- Atkinson, A.; Ellenberger, B.; Piezzi, V.; Kaspar, T.; Salazar-Vizcaya, L.; Endrich, O.; Leichtle, A.B.; Marschall, J. Extending Outbreak Investigation with Machine Learning and Graph Theory: Benefits of New Tools with Application to a Nosocomial Outbreak of a Multidrug-Resistant Organism. Infect. Control Hosp. Epidemiol. 2023, 44, 246–252. [Google Scholar] [CrossRef]
- Valik, J.K.; Ward, L.; Tanushi, H.; Müllersdorf, K.; Ternhag, A.; Aufwerber, E.; Färnert, A.; Johansson, A.F.; Mogensen, M.L.; Pickering, B.; et al. Validation of Automated Sepsis Surveillance Based on the Sepsis-3 Clinical Criteria against Physician Record Review in a General Hospital Population: Observational Study Using Electronic Health Records Data. BMJ Qual. Saf. 2020, 29, 735–745. [Google Scholar] [CrossRef]
- Rabhi, S.; Jakubowicz, J.; Metzger, M.H. Deep Learning versus Conventional Machine Learning for Detection of Healthcare-Associated Infections in French Clinical Narratives. Methods Inf. Med. 2019, 58, 31–41. [Google Scholar] [CrossRef]
- Mull, H.J.; Stolzmann, K.L.; Shin, M.H.; Kalver, E.; Schweizer, M.L.; Branch-Elliman, W. Novel Method to Flag Cardiac Implantable Device Infections by Integrating Text Mining with Structured Data in the Veterans Health Administration’s Electronic Medical Record. JAMA Netw. Open 2020, 3, E2012264. [Google Scholar] [CrossRef]
- van der Werff, S.D.; Verberk, J.D.M.; Buchli, C.; van Mourik, M.S.M.; Nauclér, P. External Validation of Semi-Automated Surveillance Algorithms for Deep Surgical Site Infections after Colorectal Surgery in an Independent Country. Antimicrob. Resist. Infect. Control 2023, 12, 96. [Google Scholar] [CrossRef] [PubMed]
- Beeler, C.; Dbeibo, L.; Kelley, K.; Thatcher, L.; Webb, D.; Bah, A.; Monahan, P.; Fowler, N.R.; Nicol, S.; Judy-Malcolm, A.; et al. Assessing Patient Risk of Central Line-Associated Bacteremia via Machine Learning. Am. J. Infect. Control 2018, 46, 986–991. [Google Scholar] [CrossRef] [PubMed]
- Bauer, J.M.; Welling, S.E.; Bettinger, B. Can We Automate Spine Fusion Surgical Site Infection Data Capture? Spine Deform. 2023, 11, 329–333. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Liu, C.; Zhang, Z.; Liang, T.; Zhu, J.; Zhou, C.; Wu, S.; Yao, Y.; Huang, C.; Zhang, B.; et al. Using Machine Learning to Predict Surgical Site Infection after Lumbar Spine Surgery. Infect. Drug Resist. 2023, 16, 5197–5207. [Google Scholar] [CrossRef] [PubMed]
- Hebert, C.; Flaherty, J.; Smyer, J.; Ding, J.; Mangino, J.E. Development and Validation of an Automated Ventilator-Associated Event Electronic Surveillance System: A Report of a Successful Implementation. Am. J. Infect. Control 2018, 46, 316–321. [Google Scholar] [CrossRef] [PubMed]
- Colborn, K.L.; Zhuang, Y.; Dyas, A.R.; Henderson, W.G.; Madsen, H.J.; Bronsert, M.R.; Matheny, M.E.; Lambert-Kerzner, A.; Myers, Q.W.O.; Meguid, R.A. Development and Validation of Models for Detection of Postoperative Infections Using Structured Electronic Health Records Data and Machine Learning. Surgery 2023, 173, 464–471. [Google Scholar] [CrossRef]
- Verberk, J.D.M.; van der Werff, S.D.; Weegar, R.; Henriksson, A.; Richir, M.C.; Buchli, C.; van Mourik, M.S.M.; Nauclér, P. The Augmented Value of Using Clinical Notes in Semi-Automated Surveillance of Deep Surgical Site Infections after Colorectal Surgery. Antimicrob. Resist. Infect. Control 2023, 12, 117. [Google Scholar] [CrossRef]
- Tunthanathip, T.; Sae-heng, S.; Oearsakul, T.; Sakarunchai, I.; Kaewborisutsakul, A.; Taweesomboonyat, C. Machine Learning Applications for the Prediction of Surgical Site Infection in Neurological Operations. Neurosurg. Focus 2019, 47, E7. [Google Scholar] [CrossRef]
- Thirukumaran, C.P.; Zaman, A.; Rubery, P.T.; Calabria, C.; Li, Y.; Ricciardi, B.F.; Bakhsh, W.R.; Kautz, H. Natural Language Processing for the Identification of Surgical Site Infections in Orthopaedics. J. Bone Jt. Surg. Am. Vol. 2019, 101, 2167–2174. [Google Scholar] [CrossRef]
- Myall, A.; Price, J.R.; Peach, R.L.; Abbas, M.; Mookerjee, S.; Zhu, N.; Ahmad, I.; Ming, D.; Ramzan, F.; Teixeira, D.; et al. Prediction of Hospital-Onset COVID-19 Infections Using Dynamic Networks of Patient Contact: An International Retrospective Cohort Study. Lancet Digit. Health 2022, 4, e573–e583. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Lu, Z.; You, L.; Zhou, L.; Xu, J.; Chen, K. Artificial Intelligence-Based Multimodal Risk Assessment Model for Surgical Site Infection (AMRAMS): Development and Validation Study. JMIR Med. Inform. 2020, 8, e18186. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Cheligeer, C.; Southern, D.A.; Martin, E.A.; Xu, Y.; Leal, J.; Ellison, J.; Bush, K.; Williamson, T.; Quan, H.; et al. Development of Machine Learning Models for the Detection of Surgical Site Infections Following Total Hip and Knee Arthroplasty: A Multicenter Cohort Study. Antimicrob. Resist. Infect. Control 2023, 12, 88. [Google Scholar] [CrossRef] [PubMed]
- McLean, K.A.; Mountain, K.E.; Shaw, C.A.; Drake, T.M.; Pius, R.; Knight, S.R.; Fairfield, C.J.; Sgrò, A.; Bouamrane, M.; Cambridge, W.A.; et al. Remote Diagnosis of Surgical-Site Infection Using a Mobile Digital Intervention: A Randomised Controlled Trial in Emergency Surgery Patients. npj Digit. Med. 2021, 4, 160. [Google Scholar] [CrossRef] [PubMed]
- Schaumburg, T.; Köhler, N.; Breitenstein, Y.; Kolbe-Busch, S.; Hasenclever, D.; Chaberny, I.F. ICU Infection Surveillance Can Be Based on Electronic Routine Data: Results of a Case Study. BMC Infect. Dis. 2023, 23, 126. [Google Scholar] [CrossRef] [PubMed]
- McLean, K.A.; Sgrò, A.; Brown, L.R.; Buijs, L.F.; Daines, L.; Potter, M.A.; Bouamrane, M.-M.; Harrison, E.M. Evaluation of Remote Digital Postoperative Wound Monitoring in Routine Surgical Practice. npj Digit. Med. 2023, 6, 85. [Google Scholar] [CrossRef]
- Rochon, M.; Jawarchan, A.; Fagan, F.; Otter, J.A.; Tanner, J. Image-Based Digital Post-Discharge Surveillance in England: Measuring Patient Enrolment, Engagement, Clinician Response Times, Surgical Site Infection, and Carbon Footprint. J. Hosp. Infect. 2023, 133, 15–22. [Google Scholar] [CrossRef]
- Liu, W.C.; Ying, H.; Liao, W.J.; Li, M.P.; Zhang, Y.; Luo, K.; Sun, B.L.; Liu, Z.L.; Liu, J.M. Using Preoperative and Intraoperative Factors to Predict the Risk of Surgical Site Infections After Lumbar Spinal Surgery: A Machine Learning–Based Study. World Neurosurg. 2022, 162, e553–e560. [Google Scholar] [CrossRef]
- Grammatico-Guillon, L.; Banaei-Bouchareb, L.; Solomiac, A.; Miliani, K.; Astagneau, P.; May-Michelangeli, L. Validation of the First Computerized Indicator for Orthopaedic Surgical Site Infections in France: ISO-ORTHO. Antimicrob. Resist. Infect. Control 2023, 12, 44. [Google Scholar] [CrossRef]
- Fletcher, R.R.; Schneider, G.; Hedt-Gauthier, B.; Nkurunziza, T.; Alayande, B.; Riviello, R.; Kateera, F. Use of Convolutional Neural Nets and Transfer Learning for Prediction of Surgical Site Infection from Color Images. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, EMBS, Guadalajara, Mexico, 1–5 November 2021; pp. 5047–5050. [Google Scholar] [CrossRef]
- Sundermann, A.J.; Chen, J.; Kumar, P.; Ayres, A.M.; Cho, S.T.; Ezeonwuka, C.; Griffith, M.P.; Miller, J.K.; Mustapha, M.M.; Pasculle, A.W.; et al. Whole-Genome Sequencing Surveillance and Machine Learning of the Electronic Health Record for Enhanced Healthcare Outbreak Detection. Clin. Infect. Dis. 2022, 75, 476–482. [Google Scholar] [CrossRef]
- Petrosyan, Y.; Thavorn, K.; Smith, G.; Maclure, M.; Preston, R.; van Walravan, C.; Forster, A.J. Predicting Postoperative Surgical Site Infection with Administrative Data: A Random Forests Algorithm. BMC Med. Res. Methodol. 2021, 21, 179. [Google Scholar] [CrossRef] [PubMed]
- Rewley, J.; Koehly, L.; Marcum, C.S.; Reed-Tsochas, F. A Passive Monitoring Tool Using Hospital Administrative Data Enables Earlier Specific Detection of Healthcare-Acquired Infections. J. Hosp. Infect. 2020, 106, 562–569. [Google Scholar] [CrossRef]
- Barchitta, M.; Maugeri, A.; Favara, G.; Riela, P.M.; Gallo, G.; Mura, I.; Agodi, A. A Machine Learning Approach to Predict Healthcare-Associated Infections at Intensive Care Unit Admission: Findings from the SPIN-UTI Project. J. Hosp. Infect. 2021, 112, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Asundi, A.; Stanislawski, M.; Mehta, P.; Mull, H.J.; Schweizer, M.L.; Barón, A.E.; Ho, P.M.; Gupta, K.; Branch-Elliman, W. Development and Validation of a Semi-Automated Surveillance Algorithm for Cardiac Device Infections: Insights from the VA CART Program. Sci. Rep. 2020, 10, 5276. [Google Scholar] [CrossRef] [PubMed]
- Skagseth, H.; Danielsen, A.S.; Kacelnik, O.; Trondsen, U.J.; Berg, T.C.; Sorknes, N.K.; Eriksen-Volle, H.M. Clusters of Healthcare-Associated SARS-CoV-2 Infections in Norwegian Hospitals Detected by a Fully Automatic Register-Based Surveillance System. J. Hosp. Infect. 2023, 135, 50–54. [Google Scholar] [CrossRef]
- Flores-Balado, Á.; Castresana Méndez, C.; Herrero González, A.; Mesón Gutierrez, R.; de las Casas Cámara, G.; Vila Cordero, B.; Arcos, J.; Pfang, B.; Martín-Ríos, M.D. Using Artificial Intelligence to Reduce Orthopedic Surgical Site Infection Surveillance Workload: Algorithm Design, Validation, and Implementation in 4 Spanish Hospitals. Am. J. Infect. Control 2023, 51, 1225–1229. [Google Scholar] [CrossRef]
- Aghdassi, S.J.S.; Kohlmorgen, B.; Schröder, C.; Peña Diaz, L.A.; Thoma, N.; Rohde, A.M.; Piening, B.; Gastmeier, P.; Behnke, M. Implementation of an Automated Cluster Alert System into the Routine Work of Infection Control and Hospital Epidemiology: Experiences from a Tertiary Care University Hospital. BMC Infect. Dis. 2021, 21, 1075. [Google Scholar] [CrossRef]
- Caǧlayan, Ç.; Barnes, S.L.; Pineles, L.L.; Harris, A.D.; Klein, E.Y. A Data-Driven Framework for Identifying Intensive Care Unit Admissions Colonized With Multidrug-Resistant Organisms. Front. Public Health 2022, 10, 853757. [Google Scholar] [CrossRef]
- Atti, M.L.C.D.; Pecoraro, F.; Piga, S.; Luzi, D.; Raponi, M. Developing a Surgical Site Infection Surveillance System Based on Hospital Unstructured Clinical Notes and Text Mining. Surg. Infect 2020, 21, 716–721. [Google Scholar] [CrossRef]
- Schröder, C.; Diaz, L.A.P.; Rohde, A.M.; Piening, B.; Aghdassi, S.J.S.; Pilarski, G.; Thoma, N.; Gastmeier, P.; Leistner, R.; Behnke, M. Lean Back and Wait for the Alarm? Testing an Automated Alarm System for Nosocomial Outbreaks to Provide Support for Infection Control Professionals. PLoS ONE 2020, 15, e0227955. [Google Scholar] [CrossRef]
- Sanger, P.C.; Granich, M.; Olsen-Scribner, R.; Jain, R.; Lober, W.B.; Stapleton, A.; Pottinger, P.S. Electronic Surveillance For Catheter-Associated Urinary Tract Infection Using Natural Language Processing. AMIA Annu. Symp. Proc. 2017, 2017, 1507–1516. [Google Scholar] [PubMed]
- Kiser, A.C.; Eilbeck, K.; Bucher, B.T. Developing an LSTM Model to Identify Surgical Site Infections Using Electronic Healthcare Records. AMIA Jt. Summits Transl. Sci. Proc. 2023, 2023, 330–339. [Google Scholar]
- Chapman, A.B.; Mowery, D.L.; Swords, D.S.; Chapman, W.W.; Bucher, B.T. Detecting Evidence of Intra-Abdominal Surgical Site Infections from Radiology Reports Using Natural Language Processing. AMIA Annu. Symp. Proc. 2017, 2017, 515–524. [Google Scholar] [PubMed]
- Rafaqat, W.; Fatima, H.S.; Kumar, A.; Khan, S.; Khurram, M. Machine Learning Model for Assessment of Risk Factors and Postoperative Day for Superficial vs Deep/Organ-Space Surgical Site Infections. Surg. Innov. 2023, 30, 455–462. [Google Scholar] [CrossRef] [PubMed]
- van der Werff, S.D.; Thiman, E.; Tanushi, H.; Valik, J.K.; Henriksson, A.; Ul Alam, M.; Dalianis, H.; Ternhag, A.; Nauclér, P. The Accuracy of Fully Automated Algorithms for Surveillance of Healthcare-Associated Urinary Tract Infections in Hospitalized Patients. J. Hosp. Infect. 2021, 110, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Dalcól, C.; Tanner, J.; de Brito Poveda, V. Digital Tools for Post-Discharge Surveillance of Surgical Site Infection. J. Adv. Nurs. 2023, 80, 96–109. [Google Scholar] [CrossRef]
- Januel, J.M.; Lotfinejad, N.; Grant, R.; Tschudin-Sutter, S.; Schreiber, P.W.; Grandbastien, B.; Jent, P.; Lo Priore, E.; Scherrer, A.; Harbarth, S.; et al. Predictive Performance of Automated Surveillance Algorithms for Intravascular Catheter Bloodstream Infections: A Systematic Review and Meta-Analysis. Antimicrob. Resist. Infect. Control 2023, 12, 87. [Google Scholar] [CrossRef]
- Scardoni, A.; Balzarini, F.; Signorelli, C.; Cabitza, F.; Odone, A. Artificial Intelligence-Based Tools to Control Healthcare Associated Infections: A Systematic Review of the Literature. J. Infect. Public Health 2020, 13, 1061–1077. [Google Scholar] [CrossRef]
- Roel Streefkerk, H.A.; Roel Verkooijen, P.A.J.; Wichor Bramer, M.; Henri Verbrugh, A. Electronically Assisted Surveillance Systems of Healthcare-Associated Infections: A Systematic Review. Eurosurveillance 2020, 25, 1900321. [Google Scholar] [CrossRef]
- Bates, D.W.; Levine, D.; Syrowatka, A.; Kuznetsova, M.; Craig, K.J.T.; Rui, A.; Jackson, G.P.; Rhee, K. The Potential of Artificial Intelligence to Improve Patient Safety: A Scoping Review. npj Digit. Med. 2021, 4, 54. [Google Scholar] [CrossRef]
- Zhang, J.; Dushaj, K.; Rasquinha, V.J.; Scuderi, G.R.; Hepinstall, M.S. Monitoring Surgical Incision Sites in Orthopedic Patients Using an Online Physician-Patient Messaging Platform. J. Arthroplast. 2019, 34, 1897–1900. [Google Scholar] [CrossRef] [PubMed]
- Scheper, H.; Derogee, R.; Mahdad, R.; van der Wal, R.J.P.; Nelissen, R.G.H.H.; Visser, L.G.; de Boer, M.G.J. A Mobile App for Postoperative Wound Care after Arthroplasty: Ease of Use and Perceived Usefulness. Int. J. Med. Inform. 2019, 129, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Macefield, R.C.; Blazeby, J.M.; Reeves, B.C.; King, A.; Rees, J.; Pullyblank, A.; Avery, K. Remote Assessment of Surgical Site Infection (SSI) Using Patient-Taken Wound Images: Development and Evaluation of a Method for Research and Routine Practice. J. Tissue Viability 2023, 32, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Junaid, S.B.; Imam, A.A.; Balogun, A.O.; De Silva, L.C.; Surakat, Y.A.; Kumar, G.; Abdulkarim, M.; Shuaibu, A.N.; Garba, A.; Sahalu, Y.; et al. Recent Advancements in Emerging Technologies for Healthcare Management Systems: A Survey. Healthcare 2022, 10, 1940. [Google Scholar] [CrossRef]
- Van Mourik, M.S.M.; Perencevich, E.N.; Gastmeier, P.; Bonten, M.J.M. Designing Surveillance of Healthcare-Associated Infections in the Era of Automation and Reporting Mandates. Clin. Infect. Dis. 2018, 66, 970–976. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention Is All You Need. Adv. Neural Inf. Process. Syst. 2017, 30, 5999–6009. [Google Scholar]
- Eckmann, C.; Kramer, A.; Assadian, O.; Flessa, S.; Huebner, C.; Michnacs, K.; Muehlendyck, C.; Podolski, K.M.; Wilke, M.; Heinlein, W.; et al. Clinical and Economic Burden of Surgical Site Infections in Inpatient Care in Germany: A Retrospective, Cross-Sectional Analysis from 79 Hospitals. PLoS ONE 2022, 17, e0275970. [Google Scholar] [CrossRef]
- Badia, J.M.; Casey, A.L.; Petrosillo, N.; Hudson, P.M.; Mitchell, S.A.; Crosby, C. Impact of Surgical Site Infection on Healthcare Costs and Patient Outcomes: A Systematic Review in Six European Countries. J. Hosp. Infect. 2017, 96, 1–15. [Google Scholar] [CrossRef]
- Reich, N.G.; Lessler, J.; Varma, J.K.; Vora, N.M. Quantifying the Risk and Cost of Active Monitoring for Infectious Diseases. Sci. Rep. 2018, 8, 1093. [Google Scholar] [CrossRef]
- Anshari, M.; Almunawar, M.N.; Younis, M.Z.; Kisa, A. Modeling Users’ Empowerment in E-Health Systems. Sustainability 2021, 13, 12993. [Google Scholar] [CrossRef]
- Upadhyay, U.; Gradisek, A.; Iqbal, U.; Dhar, E.; Li, Y.-C.; Syed-Abdul, S. Call for the Responsible Artificial Intelligence in the Healthcare. BMJ Health Care Inform. 2023, 30, e100920. [Google Scholar] [CrossRef] [PubMed]
- WHO. Regional Digital Health Action Plan for the WHO European Region 2023–2030 (RC72). Available online: https://www.who.int/europe/publications/i/item/EUR-RC72-5 (accessed on 4 January 2024).


| Variable | Inclusion | Exclusion |
|---|---|---|
| Study design/type |
|
|
| Country |
|
|
| Study subject |
|
|
| Study population |
|
|
| Specific outcomes of interest |
|
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arzilli, G.; De Vita, E.; Pasquale, M.; Carloni, L.M.; Pellegrini, M.; Di Giacomo, M.; Esposito, E.; Porretta, A.D.; Rizzo, C. Innovative Techniques for Infection Control and Surveillance in Hospital Settings and Long-Term Care Facilities: A Scoping Review. Antibiotics 2024, 13, 77. https://doi.org/10.3390/antibiotics13010077
Arzilli G, De Vita E, Pasquale M, Carloni LM, Pellegrini M, Di Giacomo M, Esposito E, Porretta AD, Rizzo C. Innovative Techniques for Infection Control and Surveillance in Hospital Settings and Long-Term Care Facilities: A Scoping Review. Antibiotics. 2024; 13(1):77. https://doi.org/10.3390/antibiotics13010077
Chicago/Turabian StyleArzilli, Guglielmo, Erica De Vita, Milena Pasquale, Luca Marcello Carloni, Marzia Pellegrini, Martina Di Giacomo, Enrica Esposito, Andrea Davide Porretta, and Caterina Rizzo. 2024. "Innovative Techniques for Infection Control and Surveillance in Hospital Settings and Long-Term Care Facilities: A Scoping Review" Antibiotics 13, no. 1: 77. https://doi.org/10.3390/antibiotics13010077
APA StyleArzilli, G., De Vita, E., Pasquale, M., Carloni, L. M., Pellegrini, M., Di Giacomo, M., Esposito, E., Porretta, A. D., & Rizzo, C. (2024). Innovative Techniques for Infection Control and Surveillance in Hospital Settings and Long-Term Care Facilities: A Scoping Review. Antibiotics, 13(1), 77. https://doi.org/10.3390/antibiotics13010077





