Small Schiff Base Molecules—A Possible Strategy to Combat Biofilm-Related Infections
Abstract
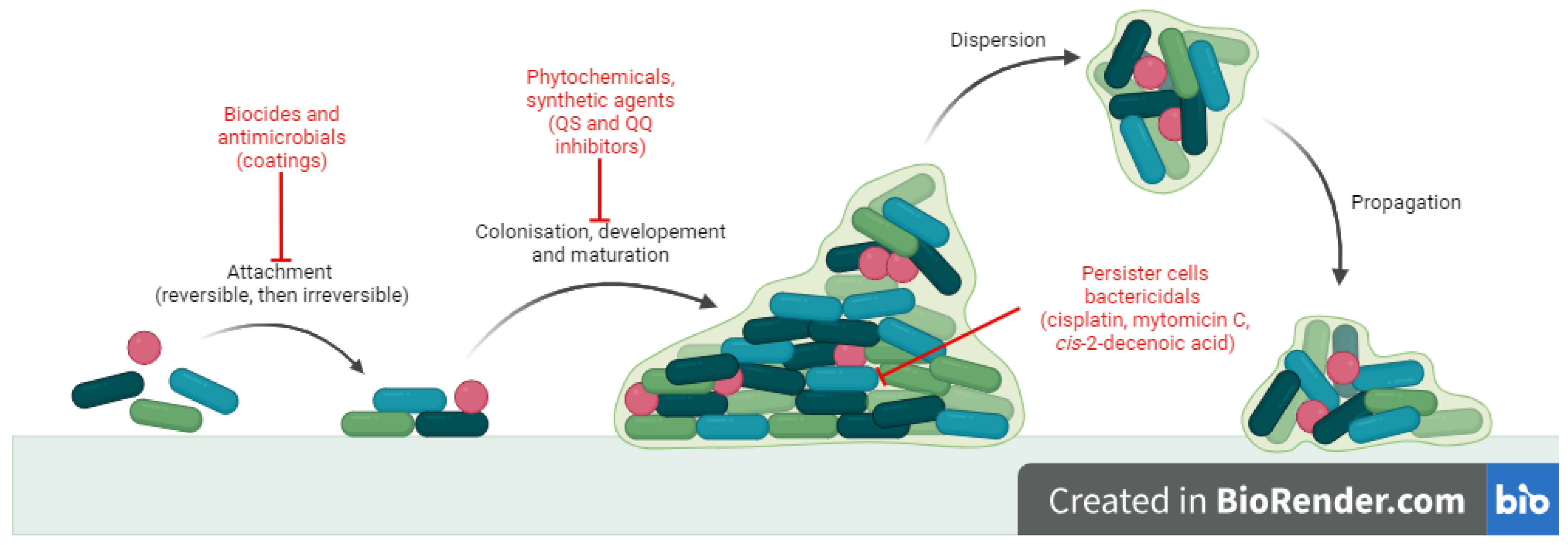
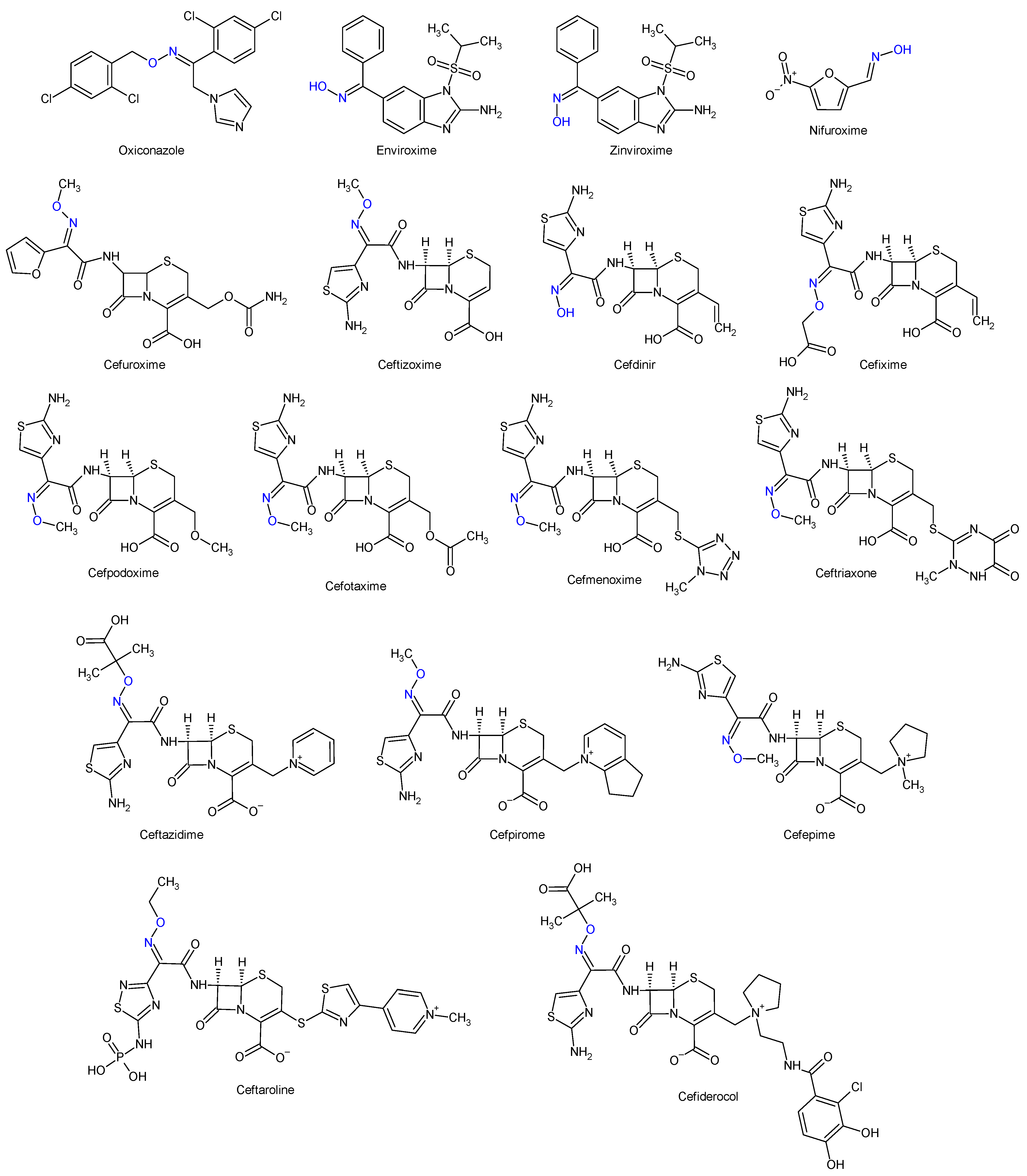
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AB | Acinetobacter baumannii |
| AF | Aspergillus flavus |
| AN | Aspergillus niger |
| ATCC | American Type Culture Collection |
| BC | Bacillus subtilis |
| BI | biofilm inhibitory percentage |
| BI50 | concentration required for 50% inhibition of biofilm formation |
| BEC50 | concentration required for 50% eradication of mature biofilm |
| CA | Candida albicans |
| CK | Candida krusei |
| CL | Candida lusitaniae |
| CP | Candida parapsilosis |
| CT | Candida tropicalis |
| CFU | colony-forming unit |
| DABA | 3,5-diaminobenzoic acid |
| EC | Escherichia coli |
| EF | Enterococcus faecalis |
| EPS | extracellular polymeric substances |
| IAM | Japan Collection of Microorganisms |
| IC50 | half maximal inhibitory concentration |
| IR | inhibition ratio |
| KP | Klebsiella pneumoniae |
| MABA | m-aminobenzoic acid |
| MBC | minimum bactericidal concentration |
| MBEC | minimum biofilm eradication concentration |
| MBIC | minimum biofilm inhibition concentration |
| MDR-TB | multidrug-resistant tuberculosis |
| MIC | minimum inhibitory concentration |
| MIC50 | minimum concentration inhibiting growth of 50% tested strains |
| MIC90 | minimum concentration inhibiting growth of 90% tested strains |
| MRSA | methicillin-resistant Staphylococcus aureus |
| MS | Mycobacterium smegmatis |
| NCTC | National Collection of Type Cultures |
| NRRL | Agriculture Research Culture Collection |
| OD630 | optical density at 630 nm |
| PA | Pseudomonas aeruginosa |
| PABA | p-aminobenzoic acid |
| QS | Quorum sensing |
| SA | Staphylococcus aureus |
| SE | Staphylococcus epidermidis |
| SM | Streptococcus mutans |
| ST | Salmonella typhymurium |
| TI | Trichophytoninterdigitale |
| VRSA | vancomycin-resistant Staphylococcus aureus |
References
- Hall-Stoodley, L.; Stoodley, P.; Kathju, S.; Høiby, N.; Moser, C.; William Costerton, J.; Moter, A.; Bjarnsholt, T. Towards Diagnostic Guidelines for Biofilm-Associated Infections. FEMS Immunol. Med. Microbiol. 2012, 65, 127–145. [Google Scholar] [CrossRef] [PubMed]
- Høiby, N. A Personal History of Research on Microbial Biofilms and Biofilm Infections. Pathog. Dis. 2014, 70, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Lebeaux, D.; Chauhan, A.; Rendueles, O.; Beloin, C. From in Vitro to in Vivo Models of Bacterial Biofilm-Related Infections. Pathogens 2013, 2, 288–356. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Alam, A.; Rani, M.; Ehtesham, N.Z.; Hasnain, S.E. Biofilms: Survival and Defense Strategy for Pathogens. Int. J. Med. Microbiol. 2017, 307, 481–489. [Google Scholar] [CrossRef]
- Madigan, M.; Aiyer, J.; Buckley, D.; Sattley, W.; Stahl, D. Brock Biology of Microorganisms, 16th ed.; Pearson Education Limited: London, UK, 2021; ISBN 1292404795. [Google Scholar]
- Rather, M.A.; Gupta, K.; Mandal, M. Microbial Biofilm: Formation, Architecture, Antibiotic Resistance, and Control Strategies. Braz. J. Microbiol. 2021, 52, 1701–1718. [Google Scholar] [CrossRef]
- Zhao, X.; Zhao, F.; Wang, J.; Zhong, N. Biofilm Formation and Control Strategies of Foodborne Pathogens: Food Safety Perspectives. RSC Adv. 2017, 7, 36670–36683. [Google Scholar] [CrossRef]
- Høiby, N.; Bjarnsholt, T.; Moser, C.; Bassi, G.L.; Coenye, T.; Donelli, G.; Hall-Stoodley, L.; Holá, V.; Imbert, C.; Kirketerp-Møller, K.; et al. ESCMID* Guideline for the Diagnosis and Treatment of Biofilm Infections 2014. Clin. Microbiol. Infect. 2015, 21, S1–S25. [Google Scholar] [CrossRef]
- Stewart, P.S. Prospects for Anti-Biofilm Pharmaceuticals. Pharmaceuticals 2015, 8, 504–511. [Google Scholar] [CrossRef]
- Stewart, P.S.; Parker, A.E. Measuring Antimicrobial Efficacy against Biofilms: A Meta-Analysis. Antimicrob. Agents Chemother. 2019, 63, e00020-19. [Google Scholar] [CrossRef]
- Gómez-Junyent, J.; Benavent, E.; Sierra, Y.; El Haj, C.; Soldevila, L.; Torrejón, B.; Rigo-Bonnin, R.; Tubau, F.; Ariza, J.; Murillo, O. Efficacy of Ceftolozane/Tazobactam, Alone and in Combination with Colistin, against Multidrug-Resistant Pseudomonas Aeruginosa in an in Vitro Biofilm Pharmacodynamic Model. Int. J. Antimicrob. Agents 2019, 53, 612–619. [Google Scholar] [CrossRef]
- Herrmann, G.; Yang, L.; Wu, H.; Song, Z.; Wang, H.; Høiby, N.; Ulrich, M.; Molin, S.; Riethmüller, J.; Döring, G. Colistin-Tobramycin Combinations Are Superior to Monotherapy Concerning the Killing of Biofilm Pseudomonas Aeruginosa. J. Infect. Dis. 2010, 202, 1585–1592. [Google Scholar] [CrossRef]
- ClinicalTrials.gov. Phase 3 Study of Aztreonam for Inhalation Solution (AZLI) in a Continuous Alternating Therapy Regimen for the Treatment of Chronic Pseudomonas Aeruginosa Infection in Patients with CF (AZLI CAT), Identifier NCT01641822. Available online: https://clinicaltrials.gov/study/NCT01641822 (accessed on 10 January 2024).
- Hympanova, M.; Terlep, S.; Markova, A.; Prchal, L.; Dogsa, I.; Pulkrabkova, L.; Benkova, M.; Marek, J.; Stopar, D. The Antibacterial Effects of New N-Alkylpyridinium Salts on Planktonic and Biofilm Bacteria. Front. Microbiol. 2020, 11, 573951. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Chen, J.; Sun, X.; Xu, G.; Li, P.; Deng, Q.; Yu, Z.; Chen, Z.; Zheng, J. The Antibacterial and Antibiofilm Activity of Telithromycin Against Enterococcus spp. Isolated From Patients in China. Front. Microbiol. 2021, 11, 616797. [Google Scholar] [CrossRef]
- Ramos, E.R.; Reitzel, R.; Jiang, Y.; Hachem, R.Y.; Chaftari, A.M.; Chemaly, R.F.; Hackett, B.; Pravinkumar, S.E.; Nates, J.; Tarrand, J.J.; et al. Clinical Effectiveness and Risk of Emerging Resistance Associated with Prolonged Use of Antibiotic-Impregnated Catheters: More than 0.5 Million Catheter Days and 7 Years of Clinical Experience. Crit. Care Med. 2011, 39, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Boban, T.; Nadar, S.; Tauro, S. Breaking down Bacterial Communication: A Review of Quorum Quenching Agents. Future J. Pharm. Sci. 2023, 9, 77. [Google Scholar] [CrossRef]
- Chowdhury, N.; Wood, T.L.; Martínez-Vázquez, M.; García-Contreras, R.; Wood, T.K. DNA-Crosslinker Cisplatin Eradicates Bacterial Persister Cells. Biotechnol. Bioeng. 2016, 113, 1984–1992. [Google Scholar] [CrossRef] [PubMed]
- Chung, E.S.; Ko, K.S. Eradication of Persister Cells of Acinetobacter Baumannii through Combination of Colistin and Amikacin Antibiotics. J. Antimicrob. Chemother. 2019, 74, 1277–1283. [Google Scholar] [CrossRef]
- Baek, M.S.; Chung, E.S.; Jung, D.S.; Ko, K.S. Effect of Colistin-Based Antibiotic Combinations on the Eradication of Persister Cells in Pseudomonas Aeruginosa. J. Antimicrob. Chemother. 2020, 75, 917–924. [Google Scholar] [CrossRef]
- Marques, C.N.H.; Morozov, A.; Planzos, P.; Zelaya, H.M. The Fatty Acid Signaling Molecule Cis-2-Decenoic Acid Increases Metabolic Activity and Reverts Persister Cells to an Antimicrobial-Susceptible State. Appl. Environ. Microbiol. 2014, 80, 6976. [Google Scholar] [CrossRef]
- Moss, G.P.; Smith, P.A.S.; Tavernier, D. Glossary of Class Names of Organic Compounds and Reactivity Intermediates Based on Structure (IUPAC Recommendations 1995). Pure Appl. Chem. 1995, 67, 1307–1375. [Google Scholar] [CrossRef]
- Fabbrizzi, L. Beauty in Chemistry: Making Artistic Molecules with Schiff Bases. J. Org. Chem. 2020, 85, 12212–12226. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S. Recent Advances in the Schiff Bases and N-Heterocyclic Carbenes as Ligands in the Cross-Coupling Reactions: A Comprehensive Review. J. Heterocycl. Chem. 2019, 56, 1168–1230. [Google Scholar] [CrossRef]
- Oiye, É.N.; Ribeiro, M.F.M.; Katayama, J.M.T.; Tadini, M.C.; Balbino, M.A.; Eleotério, I.C.; Magalhães, J.; Castro, A.S.; Silva, R.S.M.; da Cruz Júnior, J.W.; et al. Electrochemical Sensors Containing Schiff Bases and Their Transition Metal Complexes to Detect Analytes of Forensic, Pharmaceutical and Environmental Interest. A Review. Crit. Rev. Anal. Chem. 2019, 49, 488–509. [Google Scholar] [CrossRef] [PubMed]
- Durgun, M.; Turkmen, H.; Ceruso, M.; Supuran, C.T. Synthesis of 4-Sulfamoylphenyl-Benzylamine Derivatives with Inhibitory Activity against Human Carbonic Anhydrase Isoforms I, II, IX and XII. Bioorg. Med. Chem. 2016, 24, 982–988. [Google Scholar] [CrossRef]
- Elie, J.; Vercouillie, J.; Arlicot, N.; Lemaire, L.; Bidault, R.; Bodard, S.; Hosselet, C.; Deloye, J.B.; Chalon, S.; Emond, P.; et al. Design of Selective COX-2 Inhibitors in the (Aza)Indazole Series. Chemistry, in Vitro Studies, Radiochemistry and Evaluations in Rats of a [18F] PET Tracer. J. Enzyme Inhib. Med. Chem. 2019, 34, 1–7. [Google Scholar] [CrossRef]
- Bordei (Telehoiu), A.T.; Nuță, D.C.; Muşat, G.C.; Missir, A.V.; Căproiu, M.T.; Dumitraşcu, F.; Zarafu, I.; Ioniță, P.; Bădiceanu, C.D.; Limban, C.; et al. Microwave Assisted Synthesis and Spectroscopic Characterization of Some Novel Schiff Bases of Carprofen Hydrazide. Farmacia 2019, 67, 955–962. [Google Scholar] [CrossRef]
- Krátký, M.; Konečná, K.; Šimková, A.; Jand’ourek, O.; Maixnerová, J.; Stolaříková, J.; Vejsová, M.; Voxová, B.; Trejtnar, F.; Vinšová, J. Improving the Antimicrobial Activity of Old Antibacterial Drug Mafenide: Schiff Bases and Their Bioactivity Targeting Resistant Pathogens. Future Med. Chem. 2023, 15, 255–274. [Google Scholar] [CrossRef]
- Bendre, R.S.; Patil, R.D.; Patil, P.N.; Patel, H.M.; Sancheti, R.S. Synthesis and Characterization of New Schiff-Bases as Methicillin Resistant Staphylococcus Aureus (MRSA) Inhibitors. J. Mol. Struct. 2022, 1252, 132152. [Google Scholar] [CrossRef]
- Prasad, H.S.N.; Ananda, A.P.; Lohith, T.N.; Prabhuprasad, P.; Jayanth, H.S.; Krishnamurthy, N.B.; Sridhar, M.A.; Mallesha, L.; Mallu, P. Design, Synthesis, Molecular Docking and DFT Computational Insight on the Structure of Piperazine Sulfynol Derivatives as a New Antibacterial Contender against Superbugs MRSA. J. Mol. Struct. 2022, 1247, 131333. [Google Scholar] [CrossRef]
- Li, B.; Zhang, Z.; Zhang, J.F.; Liu, J.; Zuo, X.Y.; Chen, F.; Zhang, G.Y.; Fang, H.Q.; Jin, Z.; Tang, Y.Z. Design, Synthesis and Biological Evaluation of Pleuromutilin-Schiff Base Hybrids as Potent Anti-MRSA Agents in Vitro and in Vivo. Eur. J. Med. Chem. 2021, 223, 113624. [Google Scholar] [CrossRef]
- Das Mahapatra, A.; Patra, C.; Sepay, N.; Sinha, C.; Chattopadhyay, D. Comparative Study on Antibacterial Efficacy of a Series of Chromone Sulfonamide Derivatives against Drug-Resistant and MDR-Isolates. Braz. J. Microbiol. 2023, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Ding, M.; Shi, J.; Luo, N.; Wang, Y.; Lin, D.; Bao, X. Design, Synthesis, X-ray Crystal Structure, and Antimicrobial Evaluation of Novel Quinazolinone Derivatives Containing the 1,2,4-Triazole Schiff Base Moiety and an Isopropanol Linker. Mol. Divers. 2023, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Ergüden, B.; Lüleci, H.B.; Ünver, Y. Chalcone Schiff Bases Disrupt Cell Membrane Integrity of Saccharomyces Cerevisiae and Candida Albicans Cells. Arch. Microbiol. 2023, 205, 246. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Chen, K.; Han, X.; Lou, Y.; Gu, S.; Gao, Y.; Shang, S.; Song, Z.; Song, J.; Li, J. Design and Synthesis of Antifungal Candidates Containing Triazole Scaffold from Natural Rosin against Valsa Mali for Crop Protection. J. Agric. Food Chem. 2023, 71, 9718–9727. [Google Scholar] [CrossRef]
- Mahmood, W.; Ahmad, I.; Khan, M.A.; Ali Shah, S.A.; Ashraf, M.; Shahzad, M.I.; Pervaiz, I.; Sajid-ur-Rehman, M.; Khurshid, U. Synthesis, Characterization, Molecular Docking and Biological Evaluation of Schiff Base Derivatives of Cefpodoxime. Heliyon 2022, 8, e11332. [Google Scholar] [CrossRef]
- Omar, A.Z.; Hamdy, E.; Hamed, E.A.; Hafez, E.; Abdelkhalek, A. The Curative Activity of Some Arylidene Dihydropyrimidine Hydrazone against Tobacco Mosaic Virus Infestation. J. Saudi Chem. Soc. 2022, 26, 101504. [Google Scholar] [CrossRef]
- Tople, M.S.; Patel, N.B.; Patel, P.P.; Purohit, A.C.; Ahmad, I.; Patel, H. An in Silico-in Vitro Antimalarial and Antimicrobial Investigation of Newer 7-Chloroquinoline Based Schiff-Bases. J. Mol. Struct. 2023, 1271, 134016. [Google Scholar] [CrossRef]
- Ibezim, A.; Ofokansi, M.N.; Ndukwe, X.; Chiama, C.S.; Obi, B.C.; Isiogugu, O.N.; Ikechukwu, P.E.; Onwuka, A.M.; Ihim, S.A.; Asegbeloyin, J.N.; et al. Evaluation of Anti-Malarial Potency of New Pyrazole-Hydrazine Coupled to Schiff Base Derivatives. Malar. J. 2022, 21, 243. [Google Scholar] [CrossRef]
- Lalavani, N.H.; Gandhi, H.R.; Bhensdadia, K.A.; Patel, R.K.; Baluja, S.H. Synthesis, Pharmacokinetic and Molecular Docking Studies of New Benzohydrazide Derivatives Possessing Anti-Tubercular Activity against Mycobacterium Tuberculosis H37Rv. J. Mol. Struct. 2022, 1250, 131884. [Google Scholar] [CrossRef]
- Alcaraz, M.; Sharma, B.; Roquet-Banères, F.; Conde, C.; Cochard, T.; Biet, F.; Kumar, V.; Kremer, L. Designing Quinoline-Isoniazid Hybrids as Potent Anti-Tubercular Agents Inhibiting Mycolic Acid Biosynthesis. Eur. J. Med. Chem. 2022, 239, 114531. [Google Scholar] [CrossRef]
- Desale, V.J.; Mali, S.N.; Thorat, B.R.; Yamgar, R.S. Synthesis, AdmetSAR Predictions, DPPH Radical Scavenging Activity, and Potent Anti-Mycobacterial Studies of Hydrazones of Substituted 4-(Anilino Methyl) Benzohydrazides (Part 2). Curr. Comput. Aided Drug Des. 2020, 17, 493–503. [Google Scholar] [CrossRef] [PubMed]
- Pattanayak, P.; Mishra, G.P. Docking and PASS-Assisted Evaluation of Furaldehyde Substituted Benzimidazoles as Anthelmintic Agents. Russ. J. Bioorg. Chem. 2023, 49, 403–411. [Google Scholar] [CrossRef]
- Pattanayak, P.; Kaliyaperumal, S. Design, Synthesis, Characterization and IN VITRO Antimicrobial and Anthelmintic Evaluation of Metronidazole Derivatives Modified at Position 1. Pharm. Chem. J. 2022, 56, 191–196. [Google Scholar] [CrossRef]
- Tirmazi, S.A.A.S.; Qadir, M.A.; Ahmed, M.; Imran, M.; Hussain, R.; Sharif, M.; Yousaf, M.; Muddassar, M. Levofloxacin and Sulfa Drugs Linked via Schiff Bases: Exploring Their Urease Inhibition, Enzyme Kinetics and in Silico Studies. J. Mol. Struct. 2021, 1235, 130226. [Google Scholar] [CrossRef]
- Channar, P.A.; Saeed, A.; Albericio, F.; Larik, F.A.; Abbas, Q.; Hassan, M.; Raza, H.; Seo, S.-Y.Y. Sulfonamide-Linked Ciprofloxacin, Sulfadiazine and Amantadine Derivatives as a Novel Class of Inhibitors of Jack Bean Urease; Synthesis, Kinetic Mechanism and Molecular Docking. Molecules 2017, 22, 1352. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, M.A.A.; Abuo-Rahma, G.E.D.A.A.; Abdelhafez, E.S.M.N.; Hassan, H.A.; Abd El-Baky, R.M. Design, Synthesis, Molecular Docking, Anti-Proteus Mirabilis and Urease Inhibition of New Fluoroquinolone Carboxylic Acid Derivatives. Bioorg. Chem. 2017, 70, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ashma, A.; Yahya, S.; Subramani, A.; Tamilarasan, R.; Sasikumar, G.; Askar Ali, S.J.; Al-Lohedan, H.A.; Karnan, M. Synthesis of New Nicotinic Acid Hydrazide Metal Complexes: Potential Anti-Cancer Drug, Supramolecular Architecture, Antibacterial Studies and Catalytic Properties. J. Mol. Struct. 2022, 1250, 131860. [Google Scholar] [CrossRef]
- Hamurcu, F. Synthesis, Characterization, and Biological Properties of Novel Schiff Bases Containing Pentafluorophenyl Hydrazine. J. Biochem. Mol. Toxicol. 2023, 37, e23512. [Google Scholar] [CrossRef]
- Saied, S.; Shaldam, M.; Elbadawi, M.M.; Giovannuzzi, S.; Nocentini, A.; Almahli, H.; Salem, R.; Ibrahim, T.M.; Supuran, C.T.; Eldehna, W.M. Discovery of Indolinone-Bearing Benzenesulfonamides as New Dual Carbonic Anhydrase and VEGFR-2 Inhibitors Possessing Anticancer and pro-Apoptotic Properties. Eur. J. Med. Chem. 2023, 259, 115707. [Google Scholar] [CrossRef]
- Sashidhara, K.V.; Rosaiah, J.N.; Bhatia, G.; Saxena, J.K. Novel Keto-Enamine Schiffs Bases from 7-Hydroxy-4-Methyl-2-Oxo-2H-Benzo[h] Chromene-8,10-Dicarbaldehyde as Potential Antidyslipidemic and Antioxidant Agents. Eur. J. Med. Chem. 2008, 43, 2592–2596. [Google Scholar] [CrossRef]
- Alkahtani, H.M.; Almehizia, A.A.; Al-Omar, M.A.; Obaidullah, A.J.; Zen, A.A.; Hassan, A.S.; Aboulthana, W.M. In Vitro Evaluation and Bioinformatics Analysis of Schiff Bases Bearing Pyrazole Scaffold as Bioactive Agents: Antioxidant, Anti-Diabetic, Anti-Alzheimer, and Anti-Arthritic. Molecules 2023, 28, 7125. [Google Scholar] [CrossRef] [PubMed]
- Al-Qadsy, I.; Saeed, W.S.; Al-Odayni, A.B.; Alrabie, A.; Al-Faqeeh, L.A.S.; Al-Adhreai, A.; Al-Owais, A.A.; Semlali, A.; Farooqui, M. Antidiabetic, Antioxidant and Cytotoxicity Activities of Ortho- and Para-Substituted Schiff Bases Derived from Metformin Hydrochloride: Validation by Molecular Docking and in Silico ADME Studies. Open Chem. 2023, 21, 20230125. [Google Scholar] [CrossRef]
- Mesripour, A.; Jafari, E.; Hajibeiki, M.R.; Hassanzadeh, F. Design, Synthesis, Docking, and Antidepressant Activity Evaluation of Isatin Derivatives Bearing Schiff Bases. Iran. J. Basic Med. Sci. 2023, 26, 438–444. [Google Scholar] [CrossRef]
- Goleij, M.; Youseftabar-Miri, L.; Montazeri, M.; Khakpai, F. Induction of Anxiolytic, Antidepressant and Analgesic Effects by Shiff Base of (E)-3-(1H-Imidazol-4-Yl)-2-((2-Oxoindolin-3-Ylidene)Amino)Propanoic Acid Derivatives in Diabetic Rats. J. Diabetes Metab. Disord. 2021, 20, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, S.; Tripathi, R.K.P.; Ayyannan, S.R. Scaffold Hopping-Guided Design of Some Isatin Based Rigid Analogs as Fatty Acid Amide Hydrolase Inhibitors: Synthesis and Evaluation. Biomed. Pharmacother. 2018, 107, 1611–1623. [Google Scholar] [CrossRef] [PubMed]
- Tchekalarova, J.; Todorov, P.; Rangelov, M.; Stoyanova, T.; Todorova, N. Additive Anticonvulsant Profile and Molecular Docking Analysis of 5,5′-Diphenylhydantoin Schiff Bases and Phenytoin. Biomedicines 2023, 11, 2912. [Google Scholar] [CrossRef]
- Tchekalarova, J.; Todorov, P.; Stoyanova, T.; Atanasova, M. Comparative Analysis of Anticonvulsant Activity of Trans and Cis 5,5′-Diphenylhydantoin Schiff Bases. Int. J. Mol. Sci. 2023, 24, 16071. [Google Scholar] [CrossRef] [PubMed]
- Avram, S.; Udrea, A.M.; Nuta, D.C.; Limban, C.; Balea, A.C.; Caproiu, M.T.; Dumitrascu, F.; Buiu, C.; Bordei, A.T. Synthesis and Bioinformatic Characterization of New Schiff Bases with Possible Applicability in Brain Disorders. Molecules 2021, 26, 4160. [Google Scholar] [CrossRef]
- Bilen, E.; Özdemir Özmen, Ü.; Çete, S.; Alyar, S.; Yaşar, A. Bioactive Sulfonyl Hydrazones with Alkyl Derivative: Characterization, ADME Properties, Molecular Docking Studies and Investigation of Inhibition on Choline Esterase Enzymes for the Diagnosis of Alzheimer’s Disease. Chem. Biol. Interact. 2022, 360, 109956. [Google Scholar] [CrossRef]
- Alam, A.; Ali, M.; Latif, A.; Rehman, N.U.; Shah, A.J.; Khan, I.A.; Ayaz, M.; Rahman, S.U.; Al-Harrasi, A.; Ahmad, M. Discovery of (S)-Flurbiprofen-Based Novel Azine Derivatives as Prostaglandin Endoperoxide Synthase-II Inhibitors: Synthesis, in-Vivo Analgesic, Anti-Inflammatory Activities, and Their Molecular Docking. Bioorg. Chem. 2023, 141, 106847. [Google Scholar] [CrossRef]
- World Health Organization. WHO Consolidated Guidelines on Tuberculosis. Module 4: Treatment—Drug-Resistant Tuberculosis Treatment, 2022 Update. Available online: https://www.who.int/publications/i/item/9789240063129 (accessed on 5 January 2024).
- World Health Organization. WHO Operational Handbook on Tuberculosis. Module 4: Treatment—Drug-Resistant Tuberculosis Treatment, 2022 Update. Web Annexes. Available online: https://iris.who.int/bitstream/handle/10665/365309/9789240065352-eng.pdf (accessed on 5 January 2024).
- Dhuguru, J.; Zviagin, E.; Skouta, R. FDA-Approved Oximes and Their Significance in Medicinal Chemistry. Pharmaceuticals 2022, 15, 66. [Google Scholar] [CrossRef] [PubMed]
- National Center for Biotechnology Information. PubChem Compound Summary for CID 5479537, Cefepime. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/Cefepime (accessed on 30 December 2023).
- National Center for Biotechnology Information. PubChem Compound Summary for CID 9852981, Ceftaroline Fosamil. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/Ceftaroline-Fosamil (accessed on 30 December 2023).
- National Center for Biotechnology Information. PubChem Compound Summary for CID 77843966, Cefiderocol. 2024. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/Cefiderocol. (accessed on 10 January 2024).
- National Center for Biotechnology Information. PubChem Compound Summary for CID 5353853, Oxiconazole. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/Oxiconazole (accessed on 30 December 2023).
- National Center for Biotechnology Information. PubChem Compound Summary for CID 5748733, Enviroxime. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/Enviroxime (accessed on 30 December 2023).
- National Center for Biotechnology Information. PubChem Compound Summary for CID 5361910, Zinviroxime. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/Zinviroxime (accessed on 30 December 2023).
- National Center for Biotechnology Information. PubChem Compound Summary for CID 6478035, 5-Nitro-2-Furaldehyde Oxime. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/5-Nitro-2-furaldehyde-oxime (accessed on 30 December 2023).
- Bailly, C. Toward a Repositioning of the Antibacterial Drug Nifuroxazide for Cancer Treatment. Drug Discov. Today 2019, 24, 1930–1936. [Google Scholar] [CrossRef] [PubMed]
- Mendling, W.; Mailland, F. Microbiological and Pharmacotoxicological Profile of Nifuratel and Its Favourable Risk/Benefit Ratio for the Treatment of Vulvo-Vaginal Infections. Arzneimittelforschung 2002, 52, 8–13. [Google Scholar] [PubMed]
- WHO Collaborating Centre for Drug Statistics Methodology. G01AX06 Furazolidone. Available online: https://www.whocc.no/atc_ddd_index/?code=G01AX06 (accessed on 7 January 2024).
- WHO Collaborating Centre for Drug Statistics Methodology. P01AB51 Metronidazole and Furazolidone. Available online: https://www.whocc.no/atc_ddd_index/?code=P01AB51 (accessed on 7 January 2024).
- WHO Collaborating Centre for Drug Statistics Methodology. A07AX Other Intestinal Antiinfectives. Available online: https://www.whocc.no/atc_ddd_index/?code=A07AX (accessed on 7 January 2024).
- WHO Collaborating Centre for Drug Statistics Methodology. J01XE01 Nitrofurantoin. Available online: https://www.whocc.no/atc_ddd_index/?code=J01XE01 (accessed on 7 January 2024).
- WHO Collaborating Centre for Drug Statistics Methodology. P01CC01 Nifurtimox. Available online: https://www.whocc.no/atc_ddd_index/?code=P01CC01 (accessed on 7 January 2024).
- Domínguez-Asenjo, B.; Gutiérrez-Corbo, C.; Álvarez-Bardón, M.; Pérez-Pertejo, Y.; Balaña-Fouce, R.; Reguera, R.M. Ex Vivo Phenotypic Screening of Two Small Repurposing Drug Collections Identifies Nifuratel as a Potential New Treatment against Visceral and Cutaneous Leishmaniasis. ACS Infect. Dis. 2021, 7, 2390–2401. [Google Scholar] [CrossRef] [PubMed]
- Melcon-Fernandez, E.; Galli, G.; García-Estrada, C.; Balaña-Fouce, R.; Reguera, R.M.; Pérez-Pertejo, Y. Miltefosine and Nifuratel Combination: A Promising Therapy for the Treatment of Leishmania Donovani Visceral Leishmaniasis. Int. J. Mol. Sci. 2023, 24, 1635. [Google Scholar] [CrossRef]
- Yang, L.; Rybtke, M.T.; Jakobsen, T.H.; Hentzer, M.; Bjarnsholt, T.; Givskov, M.; Tolker-Nielsen, T. Computer-Aided Identification of Recognized Drugs as Pseudomonas Aeruginosa Quorum-Sensing Inhibitors. Antimicrob. Agents Chemother. 2009, 53, 2432–2443. [Google Scholar] [CrossRef] [PubMed]
- Roquini, V.; Mengarda, A.C.; Cajas, R.A.; Martins-da-Silva, M.F.; Godoy-Silva, J.; Santos, G.A.; Espírito-Santo, M.C.C.; Pavani, T.F.A.; Melo, V.A.; Salvadori, M.C.; et al. The Existing Drug Nifuroxazide as an Antischistosomal Agent: In Vitro, In Vivo, and In Silico Studies of Macromolecular Targets. Microbiol. Spectr. 2023, 11, e01393-23. [Google Scholar] [CrossRef]
- Nelson, E.A.; Walker, S.R.; Kepich, A.; Gashin, L.B.; Hideshima, T.; Ikeda, H.; Chauhan, D.; Anderson, K.C.; Frank, D.A. Nifuroxazide Inhibits Survival of Multiple Myeloma Cells by Directly Inhibiting STAT3. Blood 2008, 112, 5095–5102. [Google Scholar] [CrossRef]
- Zheng, H.; Chen, Z.; Cai, A.; Lin, X.; Jiang, X.; Zhou, B.; Wang, J.; Yao, Q.; Chen, R.; Kou, L. Nanoparticle Mediated Codelivery of Nifuratel and Doxorubicin for Synergistic Anticancer Therapy through STAT3 Inhibition. Colloids Surf. B Biointerfaces 2020, 193, 111109. [Google Scholar] [CrossRef]
- Sarvi, S.; Crispin, R.; Lu, Y.; Zeng, L.; Hurley, T.D.; Houston, D.R.; von Kriegsheim, A.; Chen, C.H.; Mochly-Rosen, D.; Ranzani, M.; et al. ALDH1 Bio-Activates Nifuroxazide to Eradicate ALDH High Melanoma-Initiating Cells. Cell Chem. Biol. 2018, 25, 1456–1469.e6. [Google Scholar] [CrossRef]
- Yu, J.G.; Ji, C.H.; Shi, M.H. The Anti-Infection Drug Furazolidone Inhibits NF-ΚB Signaling and Induces Cell Apoptosis in Small Cell Lung Cancer. Kaohsiung J. Med. Sci. 2020, 36, 998–1003. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, C.M.; Da Silva, D.L.; Modolo, L.V.; Alves, R.B.; De Resende, M.A.; Martins, C.V.B.; De Fátima, Â. Schiff Bases: A Short Review of Their Antimicrobial Activities. J. Adv. Res. 2011, 2, 1–8. [Google Scholar] [CrossRef]
- Fonkui, T.Y.; Ikhile, M.I.; Ndinteh, D.T.; Njobeh, P.B. Microbial Activity of Some Heterocyclic Schiff Bases and Metal Complexes: A Review. Trop. J. Pharm. Res. 2018, 17, 2507–2518. [Google Scholar] [CrossRef]
- Ceramella, J.; Iacopetta, D.; Catalano, A.; Cirillo, F.; Lappano, R.; Sinicropi, M.S. A Review on the Antimicrobial Activity of Schiff Bases: Data Collection and Recent Studies. Antibiotics 2022, 11, 191. [Google Scholar] [CrossRef] [PubMed]
- Uddin, M.N.; Ahmed, S.S.; Alam, S.M.R. REVIEW: Biomedical Applications of Schiff Base Metal Complexes. J. Coord. Chem. 2020, 73, 3109–3149. [Google Scholar] [CrossRef]
- Abu-Dief, A.M.; Mohamed, I.M.A. A Review on Versatile Applications of Transition Metal Complexes Incorporating Schiff Bases. Beni-Suef Univ. J. Basic Appl. Sci. 2015, 4, 119–133. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Sun, D.; Yang, Y.; Li, M.; Li, H.; Chen, L. Discovery of Metal-Based Complexes as Promising Antimicrobial Agents. Eur. J. Med. Chem. 2021, 224, 113696. [Google Scholar] [CrossRef]
- Pawariya, V.; De, S.; Dutta, J. Chitosan-Based Schiff Bases: Promising Materials for Biomedical and Industrial Applications. Carbohydr. Polym. 2024, 323, 121395. [Google Scholar] [CrossRef]
- Badea, M.; Uivarosi, V.; Olar, R. Improvement in the Pharmacological Profile of Copper Biological Active Complexes by Their Incorporation into Organic or Inorganic Matrix. Molecules 2020, 25, 5830. [Google Scholar] [CrossRef]
- Olar, R.; Badea, M.; Chifiriuc, M.C. Metal Complexes—A Promising Approach to Target Biofilm Associated Infections. Molecules 2022, 27, 758. [Google Scholar] [CrossRef]
- Yuan, R.; Diao, Y.; Zhang, W.; Lin, Y.; Huang, S.; Zhang, H.; Ma, L. In Vitro Activity of Taurine-5-Bromosalicylaldehyde Schiff Base Against Planktonic and Biofilm Cultures of Methicillin-Resistant Staphylococcus Aureus. J. Microbiol. Biotechnol. 2014, 24, 1059–1064. [Google Scholar] [CrossRef]
- Ding, W.; Zhang, H.; Xu, Y.; Ma, L.; Zhang, W. Proteomic and Morphologic Evidence for Taurine-5-Bromosalicylaldehyde Schiff Base as an Efficient Anti-Mycobacterial Drug. J. Microbiol. Biotechnol. 2019, 29, 1221–1229. [Google Scholar] [CrossRef]
- Krátký, M.; Konečná, K.; Brokešová, K.; Maixnerová, J.; Trejtnar, F.; Vinšová, J. Optimizing the Structure of (Salicylideneamino)Benzoic Acids: Towards Selective Antifungal and Anti-Staphylococcal Agents. Eur. J. Pharm. Sci. 2021, 159, 105732. [Google Scholar] [CrossRef] [PubMed]
- Krátký, M.; Konečná, K.; Janďourek, O.; Diepoltová, A.; Vávrová, P.; Voxová, B.; Vejsová, M.; Bárta, P.; Bősze, S. Insight into the Antibacterial Action of Iodinated Imine, an Analogue of Rafoxanide: A Comprehensive Study of Its Antistaphylococcal Activity. Microbiol. Spectr. 2023, 11, e03064-22. [Google Scholar] [CrossRef]
- Krátký, M.; Konečná, K.; Janoušek, J.; Janďourek, O.; Maixnerová, J.; Kalivodová, S.; Trejtnar, F.; Vinšová, J. Sulfonamide-Salicylaldehyde Imines Active against Methicillin- and Trimethoprim/Sulfonamide-Resistant Staphylococci. Future Med. Chem. 2021, 13, 1945–1962. [Google Scholar] [CrossRef]
- Patil, R.H.; Kalam Khan, F.A.; Jadhav, K.; Damale, M.; Akber Ansari, S.; Alkahtani, H.M.; Ali Khan, A.; Shinde, S.D.; Patil, R.; Sangshetti, J.N. Fungal Biofilm Inhibition by Piperazine-Sulphonamide Linked Schiff Bases: Design, Synthesis, and Biological Evaluation. Arch. Pharm. 2018, 351, 1700354. [Google Scholar] [CrossRef] [PubMed]
- More, P.G.; Karale, N.N.; Lawand, A.S.; Narang, N.; Patil, R.H. Synthesis and Anti-Biofilm Activity of Thiazole Schiff Bases. Med. Chem. Res. 2014, 23, 790–799. [Google Scholar] [CrossRef]
- Arshia; Khan, A.K.; Khan, K.M.; Ahmed, A.; Taha, M.; Perveen, S. Antibiofilm Potential of Synthetic 2-Amino-5-Chlorobenzophenone Schiff Bases and Its Confirmation through Fluorescence Microscopy. Microb. Pathog. 2017, 110, 497–506. [Google Scholar] [CrossRef]
- Aguilar-Llanos, E.; Carrera-Pacheco, S.E.; González-Pastor, R.; Zu, J.; Rodríguez-Pólit, C.; Mayorga-Ramos, A.; Carrillo-Naranjo, O.; Guamán, L.P.; Carlos Romero-Benavides, J.; Cevallos-Morillo, C.; et al. Crystal Structure, Hirshfeld Surface Analysis, and Biological Activities of Schiff-Base Derivatives of 4-Aminoantipyrine. ACS Omega 2023, 8, 42632–42646. [Google Scholar] [CrossRef]
- Sangshetti, J.N.; Khan, F.A.K.; Patil, R.H.; Marathe, S.D.; Gade, W.N.; Shinde, D.B. Biofilm Inhibition of Linezolid-like Schiff Bases: Synthesis, Biological Activity, Molecular Docking and in Silico ADME Prediction. Bioorg. Med. Chem. Lett. 2015, 25, 874–880. [Google Scholar] [CrossRef]
- Khan, F.A.K.; Kaduskar, R.N.; Patil, R.; Patil, R.H.; Ansari, S.A.; Alkahtani, H.M.; Almehizia, A.A.; Shinde, D.B.; Sangshetti, J.N. Synthesis, Biological Evaluations and Computational Studies of N-(3-(-2-(7-Chloroquinolin-2-Yl)Vinyl) Benzylidene)Anilines as Fungal Biofilm Inhibitors. Bioorg. Med. Chem. Lett. 2019, 29, 623–630. [Google Scholar] [CrossRef]
- Damale, M.G.; Chajjed, S.S.; Shelke, S.D.; Patil, R.H.; Sangshetti, J.N. Design, Molecular Modeling, Synthesis and Biological Evaluation of Novel Pyrazole Based Schiff Bases as Fungal Biofilm Inhibitors. J. Med. Pharm. Allied Sci. 2022, 11, 5108–5120. [Google Scholar] [CrossRef]
- Ammar, Y.A.; Ragab, A.; Migahed, M.A.; Al-Sharbasy, S.; Salem, M.A.; Riad, O.K.M.; Selim, H.M.R.M.; Abd-Elmaksoud, G.A.; Abusaif, M.S. Design, Green Synthesis, and Quorum Sensing Quenching Potential of Novel 2-Oxo-Pyridines Containing a Thiophene/Furan Scaffold and Targeting a LasR Gene on P. Aeruginosa. RSC Adv. 2023, 13, 27363–27384. [Google Scholar] [CrossRef] [PubMed]
- Alshammari, N.A.H.; Bakhotmah, D.A. Synthesis, Reactivity, Antimicrobial, and Anti-Biofilm Evaluation of Fluorinated 4-Amino-3-Mercapto-1,2,4-Triazin-5(4H)-One and Their Derivatives. Phosphorus Sulfur Silicon Relat. Elem. 2023, 198, 385–396. [Google Scholar] [CrossRef]
- Alzahrani, A.Y.; Ammar, Y.A.; Salem, M.A.; Abu-Elghait, M.; Ragab, A. Design, Synthesis, Molecular Modeling, and Antimicrobial Potential of Novel 3-[(1H-Pyrazol-3-Yl)Imino]Indolin-2-One Derivatives as DNA Gyrase Inhibitors. Arch. Pharm. 2022, 355, e2100266. [Google Scholar] [CrossRef]
- Alzahrani, A.Y.; Ammar, Y.A.; Abu-Elghait, M.; Salem, M.A.; Assiri, M.A.; Ali, T.E.; Ragab, A. Development of Novel Indolin-2-One Derivative Incorporating Thiazole Moiety as DHFR and Quorum Sensing Inhibitors: Synthesis, Antimicrobial, and Antibiofilm Activities with Molecular Modelling Study. Bioorg. Chem. 2022, 119, 105571. [Google Scholar] [CrossRef]
- Radwan, A.A.; Al-Anazi, F.K.; Al-Agamy, M.; Alghaith, A.F.; Mahrous, G.M.; Alhuzani, M.R.; Alghamdi, A.S.A. Design, Synthesis and Molecular Modeling of Isatin-Aminobenzoic Acid Hybrids as Antibacterial and Antibiofilm Agents. Saudi Pharm. J. 2023, 31, 101781. [Google Scholar] [CrossRef] [PubMed]
- Mohini, Y.; Prasad, R.B.N.; Karuna, M.S.L.; Poornachandra, Y.; Ganesh Kumar, C. Synthesis, Antimicrobial and Anti-Biofilm Activities of Novel Schiff Base Analogues Derived from Methyl-12-Aminooctadec-9-Enoate. Bioorg. Med. Chem. Lett. 2014, 24, 5224–5227. [Google Scholar] [CrossRef]
- Boudiba, S.; Tamfu, A.N.; Hanini, K.; Selatnia, I.; Boudiba, L.; Saouli, I.; Mosset, P.; Ceylan, O.; Egbe, D.A.M.; Sid, A.; et al. Synthesis of a New Diarylhydrazone Derivative and an Evaluation of Its in Vitro Biofilm Inhibition and Quorum Sensing Disruption along with a Molecular Docking Study. J. Chem. Res. 2023, 47, 17475198231184603. [Google Scholar] [CrossRef]
- Noshiranzadeh, N.; Heidari, A.; Haghi, F.; Bikas, R.; Lis, T. Chiral Lactic Hydrazone Derivatives as Potential Bioactive Antibacterial Agents: Synthesis, Spectroscopic, Structural and Molecular Docking Studies. J. Mol. Struct. 2017, 1128, 391–399. [Google Scholar] [CrossRef]
- Halicki, P.C.B.; Radin, V.; Von Groll, A.; Nora, M.V.; Pinheiro, A.C.; Da Silva, P.E.A.; Ramos, D.F. Antibiofilm Potential of Arenecarbaldehyde 2-Pyridinylhydrazone Derivatives Against Acinetobacter Baumannii. Microb. Drug Resist. 2020, 26, 1429–1436. [Google Scholar] [CrossRef] [PubMed]
- Dhonnar, S.L.; Adole, V.A.; Patil, R.H.; Khairnar, B.B.; Pawar, T.B.; Sadgir, N.V.; Jagdale, B.S. Synthesis, Spectral, Antifungal, in Silico Molecular Docking, ADME and DFT Studies of Some 2-(2-Hydrazineyl)Thiazole Derivatives: Computational and Experimental Perspective. J. Mol. Struct. 2023, 1294, 136411. [Google Scholar] [CrossRef]
- Zhou, X.M.; Hu, Y.Y.; Fang, B.; Zhou, C.H. Benzenesulfonyl Thiazoloimines as Unique Multitargeting Antibacterial Agents towards Enterococcus Faecalis. Eur. J. Med. Chem. 2023, 248, 115088. [Google Scholar] [CrossRef] [PubMed]
- Doğan, Ş.D.; Özcan, E.; Çetinkaya, Y.; Han, M.İ.; Şahin, O.; Bogojevic, S.S.; Nikodinovic-Runic, J.; Gündüz, M.G. Linking Quinoline Ring to 5-Nitrofuran Moiety via Sulfonyl Hydrazone Bridge: Synthesis, Structural Characterization, DFT Studies, and Evaluation of Antibacterial and Antifungal Activity. J. Mol. Struct. 2023, 1292, 136155. [Google Scholar] [CrossRef]
- Aydin, M.; Ozturk, A.; Duran, T.; Ozmen, U.O.; Sumlu, E.; Ayan, E.B.; Korucu, E.N. In Vitro Antifungal and Antibiofilm Activities of Novel Sulfonyl Hydrazone Derivatives against Candida spp. J. Med. Mycol. 2023, 33, 101327. [Google Scholar] [CrossRef] [PubMed]
- Elewa, S.I.; Abdelhamid, A.O.; Hamed, A.A.; Mansour, E. Synthesis, Characterization, Antimicrobial Activities, Anticancer of Some New Pyridines from 2, 3-Dihydro-2-Oxo-4-Phenyl-6-(Thien-2-Yl) Pyridine-3-Carbonitrile. Synth. Commun. 2021, 51, 151–161. [Google Scholar] [CrossRef]
- Bordei, A.T.; Limban, C.; Nuță, D.C.; Zarafu, I.; Denes, E.; Măruțescu, L.; Chifiriuc, M.C.; Popa, M.; Aramă, C. Recent Advances in the Study of Derivatives of (EZ)-N’-Benzylidene-(2RS)-2-(6-Chloro-9H-Carbazol-2-Yl) Propanohydrazide. Farmacia 2022, 70, 589–595. [Google Scholar] [CrossRef]
- Ressler, A.J.; Frate, M.; Hontoria, A.; Ream, A.; Timms, E.; Li, H.; Stettler, L.D.; Bollinger, A.; Poor, J.E.; Parra, M.A.; et al. Synthesis, Anti-Ferroptosis, Anti-Quorum Sensing, Antibacterial and DNA Interaction Studies of Chromene-Hydrazone Derivatives. Bioorg. Med. Chem. 2023, 90, 117369. [Google Scholar] [CrossRef]
- Alnufaie, R.; Kc, H.R.; Alsup, N.; Whitt, J.; Chambers, S.A.; Gilmore, D.; Alam, M.A. Synthesis and Antimicrobial Studies of Coumarin-Substituted Pyrazole Derivatives as Potent Anti-Staphylococcus Aureus Agents. Molecules 2020, 25, 2758. [Google Scholar] [CrossRef]
- Alnufaie, R.; Alsup, N.; Kc, H.R.; Newman, M.; Whitt, J.; Chambers, S.A.; Gilmore, D.; Alam, M.A. Design and Synthesis of 4-[4-Formyl-3-(2-Naphthyl)Pyrazol-1-Yl]Benzoic Acid Derivatives as Potent Growth Inhibitors of Drug-Resistant Staphylococcus Aureus. J. Antibiot. 2020, 73, 818–827. [Google Scholar] [CrossRef]
- Merlani, M.; Nadaraia, N.; Amiranashvili, L.; Petrou, A.; Geronikaki, A.; Ciric, A.; Glamoclija, J.; Carevic, T.; Sokovic, M. Antimicrobial Activity of Some Steroidal Hydrazones. Molecules 2023, 28, 1167. [Google Scholar] [CrossRef] [PubMed]
- Kavaliauskas, P.; Grybaite, B.; Mickevicius, V.; Petraitiene, R.; Grigaleviciute, R.; Planciuniene, R.; Gialanella, P.; Pockevicius, A.; Petraitis, V. Synthesis, ADMET Properties, and in Vitro Antimicrobial and Antibiofilm Activity of 5-Nitro-2-Thiophenecarbaldehyde N-((E)-(5-Nitrothienyl)Methylidene)Hydrazone (KTU-286) against Staphylococcus Aureus with Defined Resistance Mechanisms. Antibiotics 2020, 9, 612. [Google Scholar] [CrossRef]
- Akunuri, R.; Veerareddy, V.; Kaul, G.; Akhir, A.; Unnissa, T.; Parupalli, R.; Madhavi, Y.V.; Chopra, S.; Nanduri, S. Synthesis and Antibacterial Evaluation of (E)-1-(1H-Indol-3-Yl) Ethanone O-Benzyl Oxime Derivatives against MRSA and VRSA Strains. Bioorg. Chem. 2021, 116, 105288. [Google Scholar] [CrossRef]
- Yang, S.-C.; Tang, K.-W.; Lin, C.-H.; Alalaiwe, A.; Tseng, C.-H.; Fang, J.-Y. Discovery of Furanoquinone Derivatives as a Novel Class of DNA Polymerase and Gyrase Inhibitors for MRSA Eradication in Cutaneous Infection. Front. Microbiol. 2019, 10, 1197. [Google Scholar] [CrossRef] [PubMed]
- Stecoza, C.; Majekova, M.; Majek, P.; Caproiu, M.; Marutescu, L. Novel Dibenzothiepins with Antibiofilm Activity Demonstrated by Microbiological Assays and Molecular Modeling. Curr. Org. Chem. 2013, 17, 113–124. [Google Scholar] [CrossRef]
- Ansari, M.F.; Tan, Y.-M.; Sun, H.; Li, S.; Zhou, C.-H. Unique Iminotetrahydroberberine-Corbelled Metronidazoles as Potential Membrane Active Broad-Spectrum Antibacterial Agents. Bioorg. Med. Chem. Lett. 2022, 76, 129012. [Google Scholar] [CrossRef]
- Dawadi, S.; Kordus, S.L.; Baughn, A.D.; Aldrich, C.C. Synthesis and Analysis of Bacterial Folate Metabolism Intermediates and Antifolates. Org. Lett. 2017, 19, 5220–5223. [Google Scholar] [CrossRef]
- Thiede, J.M.; Kordus, S.L.; Turman, B.J.; Buonomo, J.A.; Aldrich, C.C.; Minato, Y.; Baughn, A.D. Targeting Intracellular P-Aminobenzoic Acid Production Potentiates the Anti-Tubercular Action of Antifolates. Sci. Rep. 2016, 6, 38083. [Google Scholar] [CrossRef]
- Kluczyk, A.; Popek, T.; Kiyota, T.; de Macedo, P.; Stefanowicz, P.; Lazar, C.; Konishi, Y. Drug Evolution: P-Aminobenzoic Acid as a Building Block. Curr. Med. Chem. 2012, 9, 1871–1892. [Google Scholar] [CrossRef]
- Joo, M.Y.; Shin, J.H.; Jang, H.-C.; Song, E.S.; Kee, S.J.; Shin, M.G.; Suh, S.P.; Ryang, D.W. Expression of SAP5 and SAP9 in Candida Albicans Biofilms: Comparison of Bloodstream Isolates with Isolates from Other Sources. Med. Mycol. 2013, 51, 892–896. [Google Scholar] [CrossRef]
- More, P.G.; Karale, N.N.; Lawand, A.S.; Rajmane, S.V.; Pawar, S.V.; Patil, R.H. A 4-(o-Methoxyphenyl)-2-Aminothiazole: An Anti-Quorum Sensing Compound. Med. Chem. Res. 2013, 22, 4183–4191. [Google Scholar] [CrossRef]
- Heath, R.J.; Rock, C.O. Enoyl-Acyl Carrier Protein Reductase (FabI) Plays a Determinant Role in Completing Cycles of Fatty Acid Elongation in Escherichia Coli. J. Biol. Chem. 1995, 270, 26538–26542. [Google Scholar] [CrossRef] [PubMed]
- Heath, R.J.; Yu, Y.T.; Shapiro, M.A.; Olson, E.; Rock, C.O. Broad Spectrum Antimicrobial Biocides Target the FabI Component of Fatty Acid Synthesis. J. Biol. Chem. 1998, 273, 30316–30320. [Google Scholar] [CrossRef]
- Lee, J.Y.; Jeong, K.W.; Shin, S.; Lee, J.U.; Kim, Y. Discovery of Novel Selective Inhibitors of Staphylococcus Aureus β-Ketoacyl Acyl Carrier Protein Synthase III. Eur. J. Med. Chem. 2012, 47, 261–269. [Google Scholar] [CrossRef]
- Lai, C.Y.; Cronan, J.E. β-Ketoacyl-Acyl Carrier Protein Synthase III (FabH) Is Essential for Bacterial Fatty Acid Synthesis. J. Biol. Chem. 2003, 278, 51494–51503. [Google Scholar] [CrossRef] [PubMed]
- Weidel, E.; De Jong, J.C.; Brengel, C.; Storz, M.P.; Braunshausen, A.; Negri, M.; Plaza, A.; Steinbach, A.; Müller, R.; Hartmann, R.W. Structure Optimization of 2-Benzamidobenzoic Acids as PqsD Inhibitors for Pseudomonas Aeruginosa Infections and Elucidation of Binding Mode by SPR, STD NMR, and Molecular Docking. J. Med. Chem. 2013, 56, 6146–6155. [Google Scholar] [CrossRef]
- Kiratisin, P.; Tucker, K.D.; Passador, L. LasR, a Transcriptional Activator of Pseudomonas Aeruginosa Virulence Genes, Functions as a Multimer. J. Bacteriol. 2002, 184, 4912–4919. [Google Scholar] [CrossRef] [PubMed]
- Skindersoe, M.E.; Alhede, M.; Phipps, R.; Yang, L.; Jensen, P.O.; Rasmussen, T.B.; Bjarnsholt, T.; Tolker-Nielsen, T.; Høiby, N.; Givskov, M. Effects of Antibiotics on Quorum Sensing in Pseudomonas Aeruginosa. Antimicrob. Agents Chemother. 2008, 52, 3648–3663. [Google Scholar] [CrossRef]
- Da Silva, J.F.M.; Garden, S.J.; Pinto, A.C. The Chemistry of Isatins: A Review from 1975 to 1999. J. Braz. Chem. Soc. 2001, 12, 273–324. [Google Scholar] [CrossRef]
- Martin, P.K.; Li, T.; Sun, D.; Biek, D.P.; Schmid, M.B. Role in Cell Permeability of an Essential Two-Component System in Staphylococcus Aureus. J. Bacteriol. 1999, 181, 3666–3673. [Google Scholar] [CrossRef]
- Levison, M.E. Pharmacodynamics of Antimicrobial Drugs. Infect. Dis. Clin. N. Am. 2004, 18, 451–465. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.Y.; Wang, J.; Li, T.J.; Yadav Bheemanaboina, R.R.; Ansari, M.F.; Cheng, Y.; Zhou, C.H. An Unexpected Discovery toward Novel Membrane Active Sulfonyl Thiazoles as Potential MRSA DNA Intercalators. Future Med. Chem. 2020, 12, 1709–1727. [Google Scholar] [CrossRef] [PubMed]
149. Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |

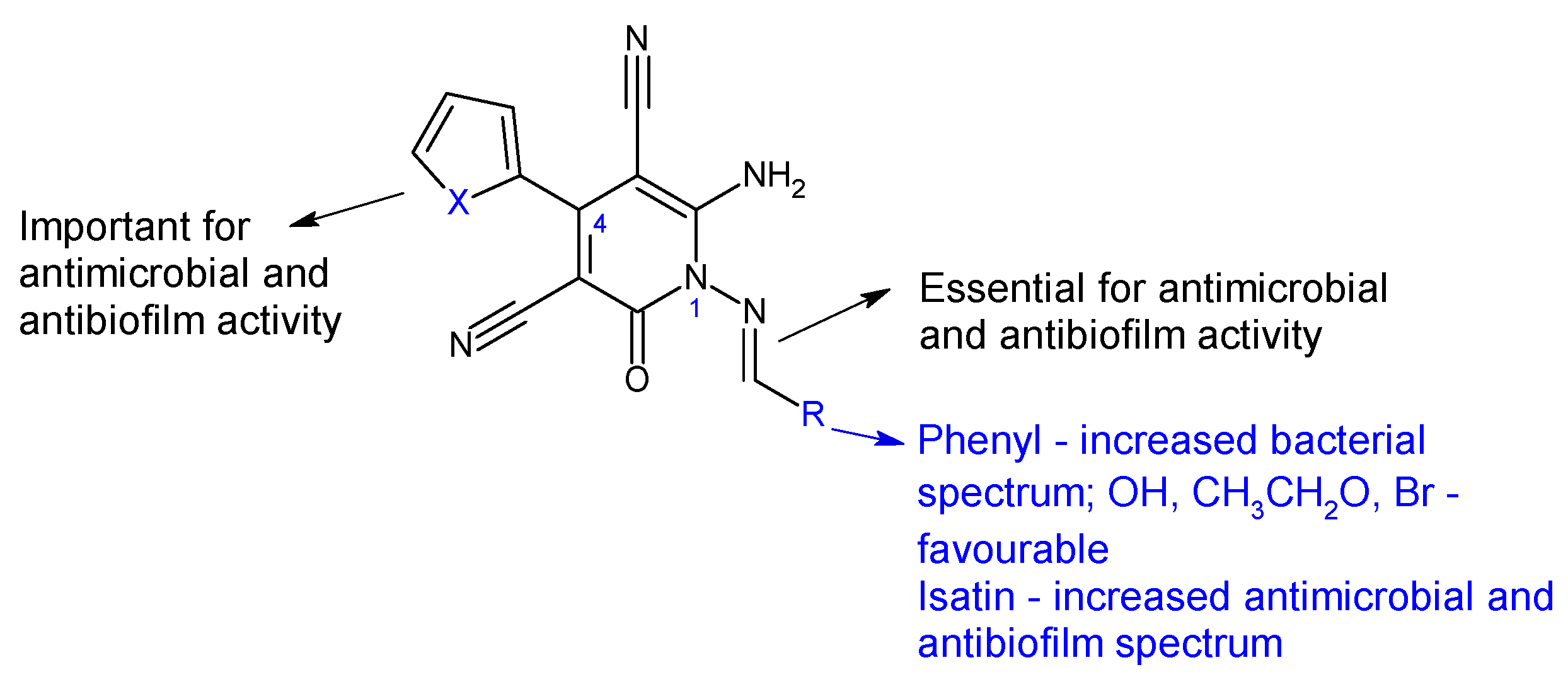
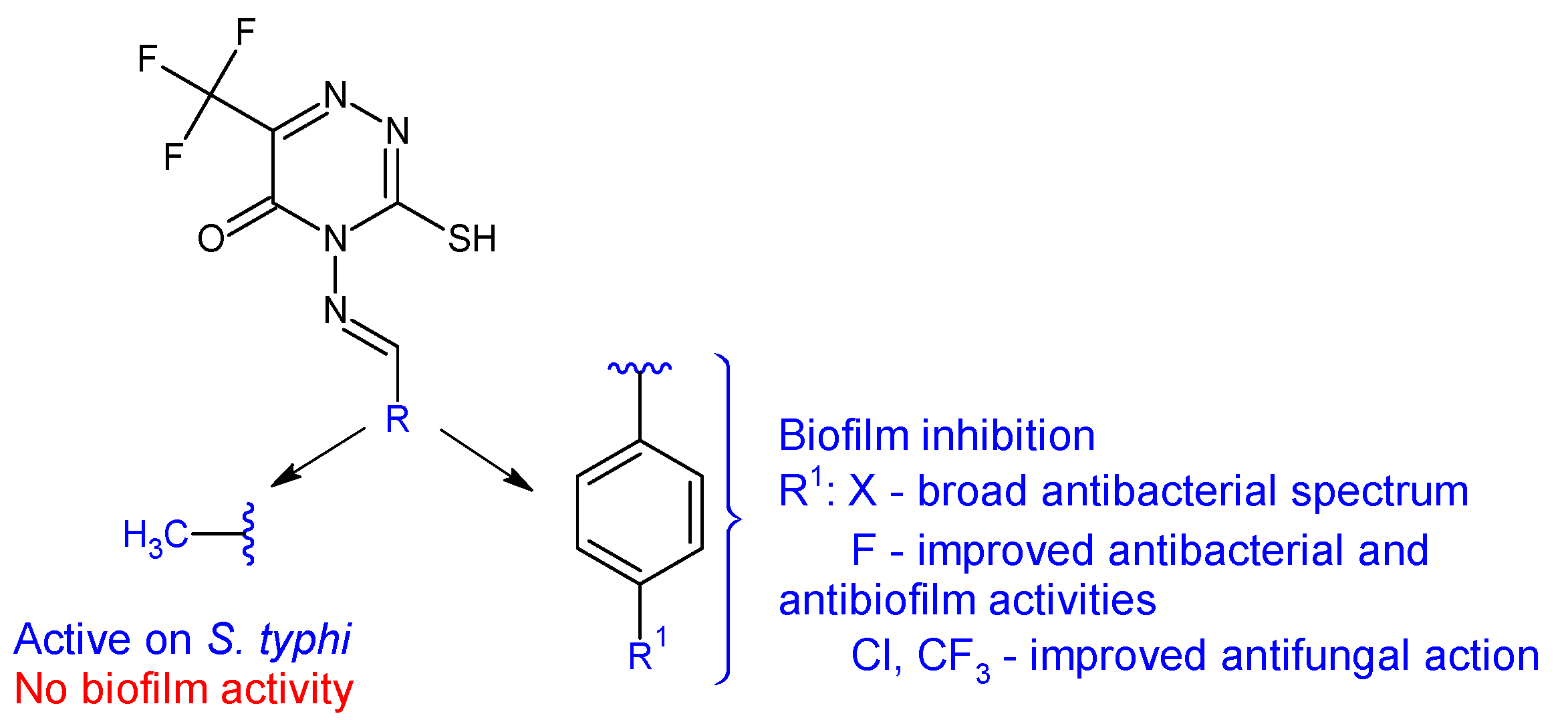
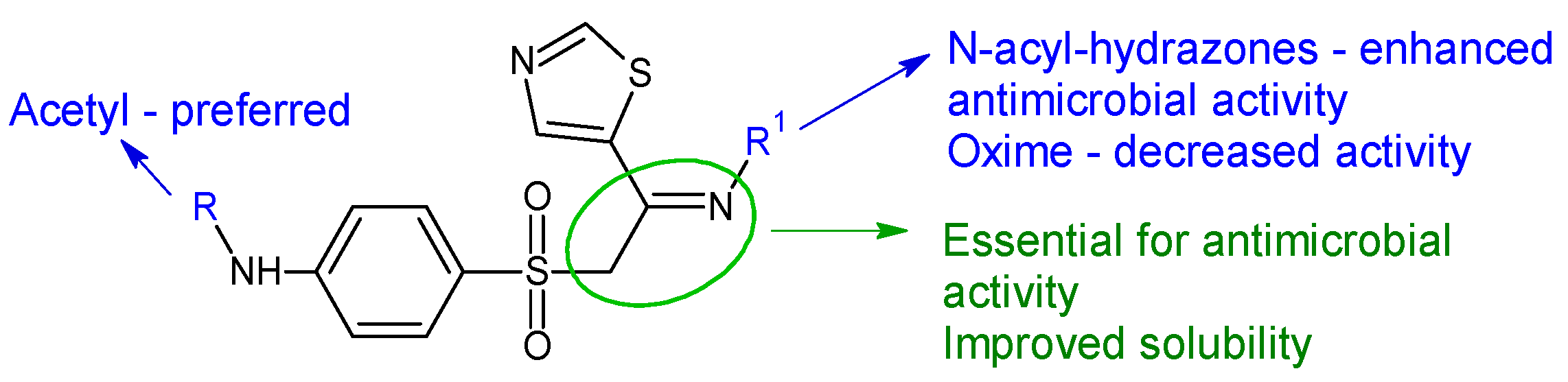
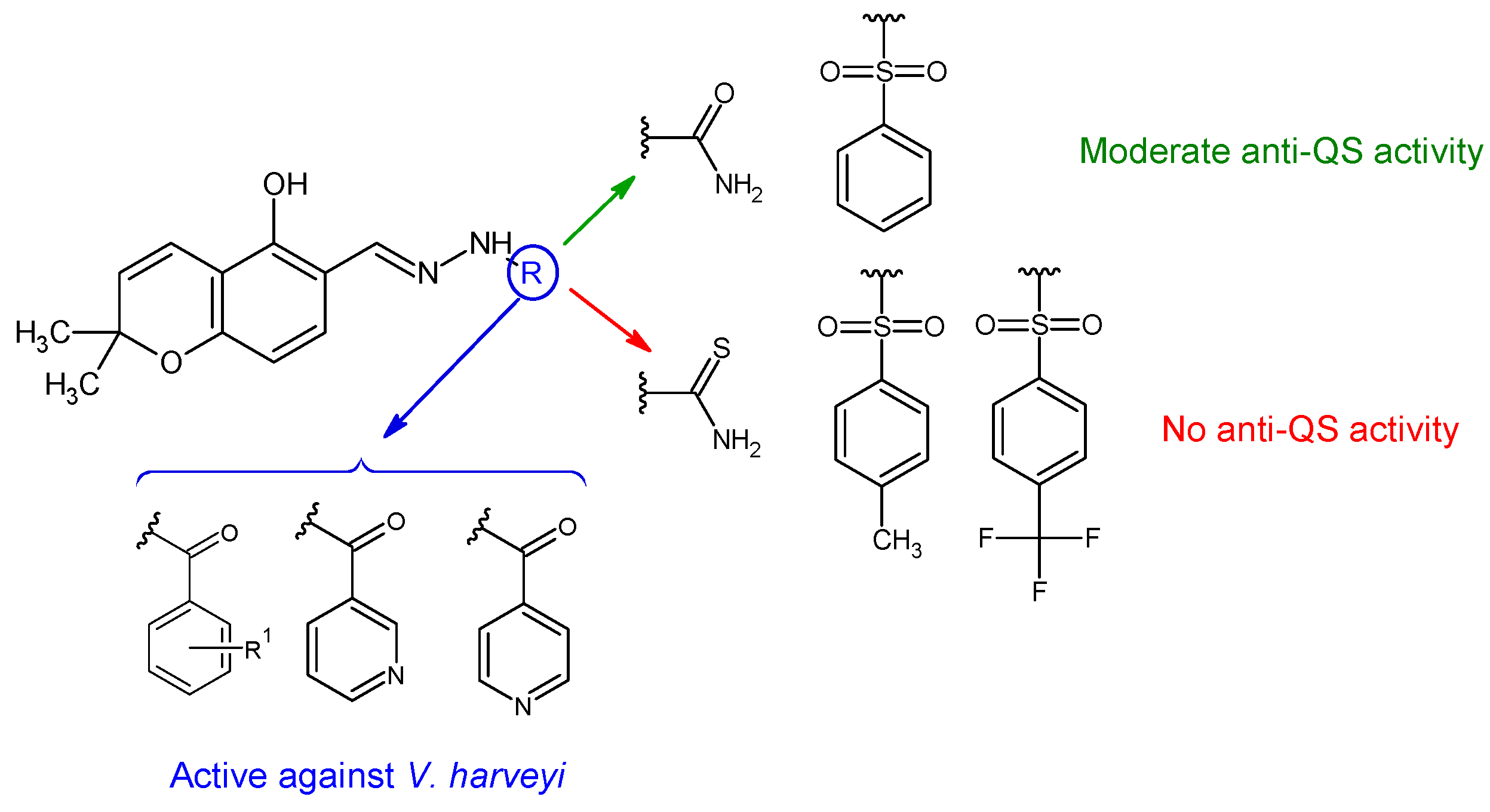
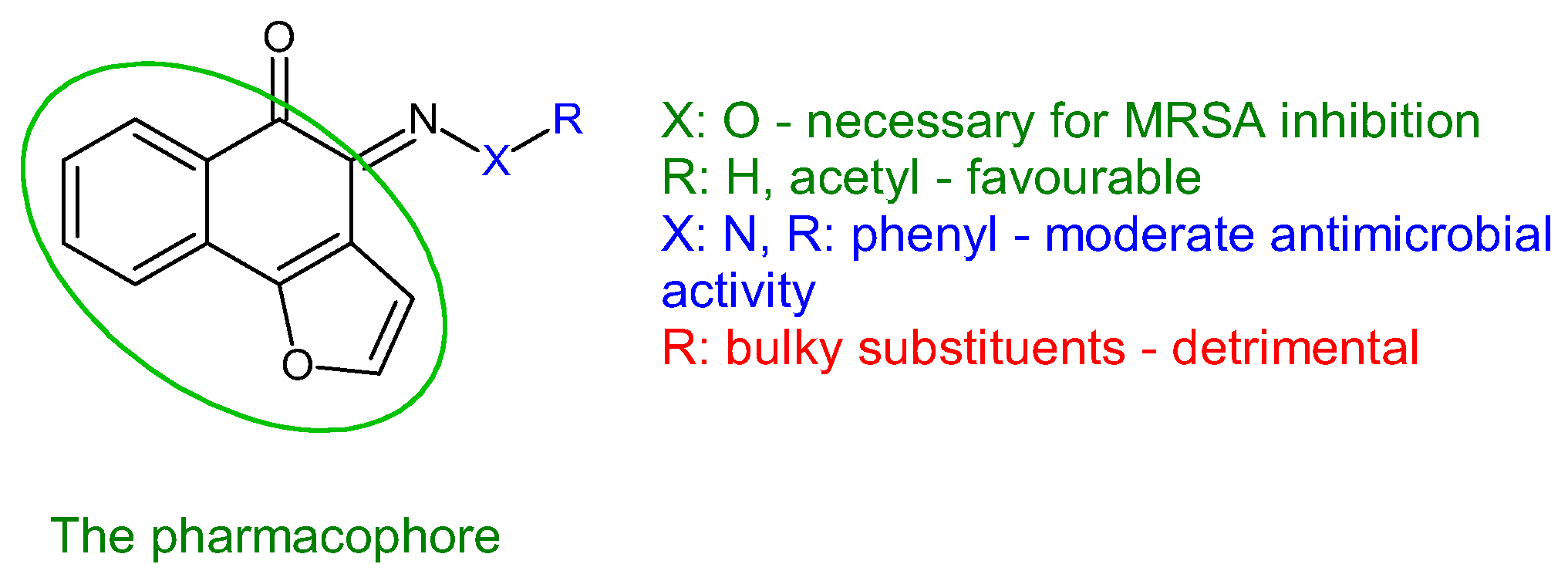
| Compounds | Biological Assay/Microorganism | Observations | Ref. |
|---|---|---|---|
| Classical Schiff Bases | |||
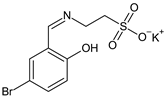 Taurine-5-Bromosalicylaldehyde Schiff base | Antibacterial screening: Staphylococcus aureus ATCC 43300, Mycobacterium smegmatis mc2155 | SA: MIC 32 μg/mL MS: MIC > 60 μg/mL | [97,98] |
| Antibiofilm screening: S. aureus ATCC 43300, M. smegmatis mc2155 | Biofilm inhibition SA: MBIC 8 μg/mL | ||
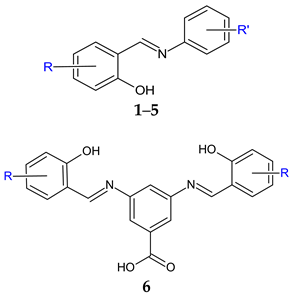 R’: 4-COOH (1); 3-COOH (2); 4-COOCH3 (3); 4-COOCH2CH3 (4); 4-CONHC6H5 (5) R: 3-I-5-Cl (a); 3,5-diI (b) | Antibacterial screening: S. aureus ATCC 29213, methicillin-resistant S. aureus ATCC 43300, S. epidermidis, clinical isolate 143-2016, Enterococcus faecalis ATCC 29212, Escherichia coli ATCC 25922, Klebsiella pneumoniae ATCC 10031, Serratia marcescens, clinical isolate 62-2016, Pseudomonas aeruginosa ATCC 27853. | Gram-positive bacteria—susceptible MIC ≥ 7.81 μM (SA, MRSA) MIC ≥ 15.62 μM (SE, EF) | [99] |
| Antimycobacterial screening: Mycobacterium tuberculosis 331/88 (H37Rv), M. avium 330/88, M. kansasii 6509/96 | No activity | ||
| Antifungal screening: Candida albicans ATCC 24433, Candida krusei ATCC 6258, Candida parapsilosis ATCC 22019, Candida tropicalis ATCC 750, Aspergillus fumigatus ATCC 204305, Aspergillus flavus CCM 8363; Lichtheimia corymbifera CCM 8077, Trichophyton interdigitale ATCC 9533 | CA, TI—susceptible MIC ≥ 3.90 μM (TI) MIC ≥ 7.81 μM (CA) | ||
| Antibiofilm screening: methicillin-resistant S. aureus ATCC 43300, S. epidermidis ATCC 1228 | 3b—MBIC 781.25-1562.5 μg/mL, MBEC 1562.5–3125.0 μg/mL (MRSA) MBIC 781.25–1562.5 μg/mL, MBEC > 1562.5 μg/mL (SE) | ||
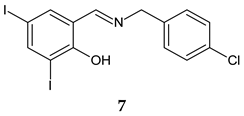 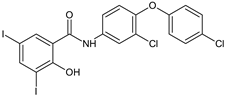 Rafoxanide | Antibacterial screening: methicillin-resistant S. aureus ATCC 43300, and clinical isolates 131/16, 138/16, 153/16; methicillin-sensitive S. aureus 136/16, 141/16, 154/16; vancomycin-resistant S. aureus 203/19 NIPH, CCM 1767; S. epidermidis ATCC 1228; vancomycin-resistant E. faecium 198/16 | Bactericidal MIC 15.625–62.5 μM (SA) MIC 62.5–125 μM (EF) | [100] |
| Antibiofilm screening: methicillin-resistant S. aureus ATCC 43300; S. epidermidis ATCC 12228 | MBIC 62.216–124.432 μg/mL, MBEC 124.432–248.863 μg/mL (MRSA), MBIC 31.108–62.216 μg/mL, MBEC 124.432–248.863 μg/mL (SE) | ||
 R2: 3,5-diCl (a), 3-Br-5-Cl (b), 3-I-5-Cl (c), 3,5-diI (d), 6-Cl (e), H (f) | Antibacterial screening: S. aureus ATCC 2913, CCM 4223; methicillin-resistant S. aureus ATCC 43300, CCM 4750; S. epidermidis H 6966/08; S. epidermidis H2232, S. epidermidis D7944 (clinical isolate), S. epidermidis H2232 (clinical isolate); S. hominis H2202 (clinical isolate); E. faecalis ATCC 29212, CCM 4224; E. coli ATCC 25922, CCM 3954; K. pneumoniae D 11750/08; ESBL-positive K. pneumoniae J 14368/08; P. aeruginosa ATCC 27853, CCM 3955. | Gram-positive bacteria—susceptible Bactericidal 8e, 8f, 9d: MIC 31.25 μM (EF) 9d, 10d: MIC 15.62 μM (SA, MRSA, SE), 10d: MIC 3.91 μM (SE H2202) | [101] |
| Antibiofilm screening: methicillin-resistant S. aureus ATCC 43300, S. epidermidis ATCC 1228 | No biofilm disruption 10a: MBIC 390.6–781.25 μM, MBEC > 3462 μM (MRSA, SE) | ||
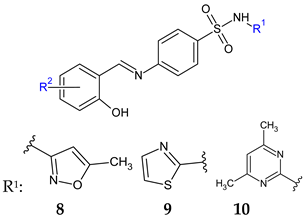
 R: H (a), 2-CF3 (b), 3-CF3 (c), 2-OH (d), 4-OH (e), 4-OCH3 (f) | Antibacterial screening: B. subtilis NCIM-2063, S. aureus NCIM-2901, E. coli NCIM-2256, P. aeruginosa NCIM-2036 | 11b—MIC 35.7 μg/mL (SA) 11c—MIC 84.0 μg/mL (EC) 11f, 11a—MIC 39.0, 40.0 μg/mL (PA) | [102] |
| Antifungal screening: C. albicans NCIM-3471 | 11d, 11c, 11e—MIC 39.6, 45.0, 47.2 μg/mL | ||
| Antibiofilm screening: C. albicans NCIM-3471 | 11d, 11a, 11c, 11e—IC50 31.4, 32.1, 37.2, 39.5 μM | ||
 R1: H (a), 3-CH3 (b), 4-CH3 (c), 5-CH3 (d), 3-OCH3 (e), 5-Br (f) | Antibacterial screening: B. subtilis NCIM 2063; E. coli NCIM 2931 | 12f, 12g—MIC 25μg/mL, MBC 50 μg/mL (BS) 12g—MIC = MBC 100 μg/mL (EC) | [103] |
| Antibiofilm screening: P. aeruginosa | QS mediated mechanism | ||
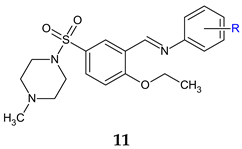
 R1: 2-OH-4-OCH3 (a), 2-NO2-5-OH (b), 3-Br-4-F (c), 2-OH (d), 2-OH-5-F (e), 2-OH-5-Br (f), 2-OH-3-Br-5-Cl (g), 3-OH-4-OH (h), 3-Br-4-OH (i), 3-Cl-4-OH (j), 3-Br-4-OCH3 (k) | Antibacterial screening: S. mutans ATCC 25175, S. aureus ATCC 43300, Proteus mirabilis ATCC 12453, K. pneumoniae ATCC 13882 | 13g—MIC 20 μg/mL (SM), 36.22 μg/mL (SA), 144.9 μg/mL (PM) 13l—MIC > 58.1 μg/mL (PA) 13f—MIC 79.45 μg/mL (KP) | [104] |
| Antibiofilm screening: S. mutans ATCC 25175, S. aureus ATCC 43300, P. mirabilis ATCC 12453, K. pneumoniae ATCC 13882 | MBIC < 100 μg/mL 13i, 13k, 13g: disruption of SA biofilm 13i: Disruption of preformed biofilm (PM) | ||
 R1: Br, R2: H (a), R1: H, R2: 4-NO2 (b), R1: H, R2: 3-OCH3-4-OCOCH3 (c) | Antibacterial screening: S. aureus ATCC 25923, E. faecalis ATCC 29212, Salmonella enterica ATCC 14028, Klebsiella ozaenae (clinical isolate), Enterobacter gergoviae (clinical isolate) P. aeruginosa ATCC 27853 | 14a: MIC 15.60 μM (E. gergoviae), 31.25 μM (S. enterica), 62.5 μM (K. ozonae, SA), 125 μM (EF) 14b: 250 μM (EF, CT) c: 250 μM (EF) | [105] |
| Antifungal screening: C. albicans (clinical isolate), C. krusei (clinical isolate), C. tropicalis (clinical isolate), C. glabrata (clinical isolate) | 14a: MIC 15.60–62.50 μM 14b: 250 μM | ||
| Antibiofilm screening: S. aureus ATCC 25923, E. faecalis ATCC 29212, C. tropicalis (clinical isolate) | 14a: BI 82.77% (SA), 75.69% (EF), 90.41% (CT) 14b: BI 76.63% (EF) | ||
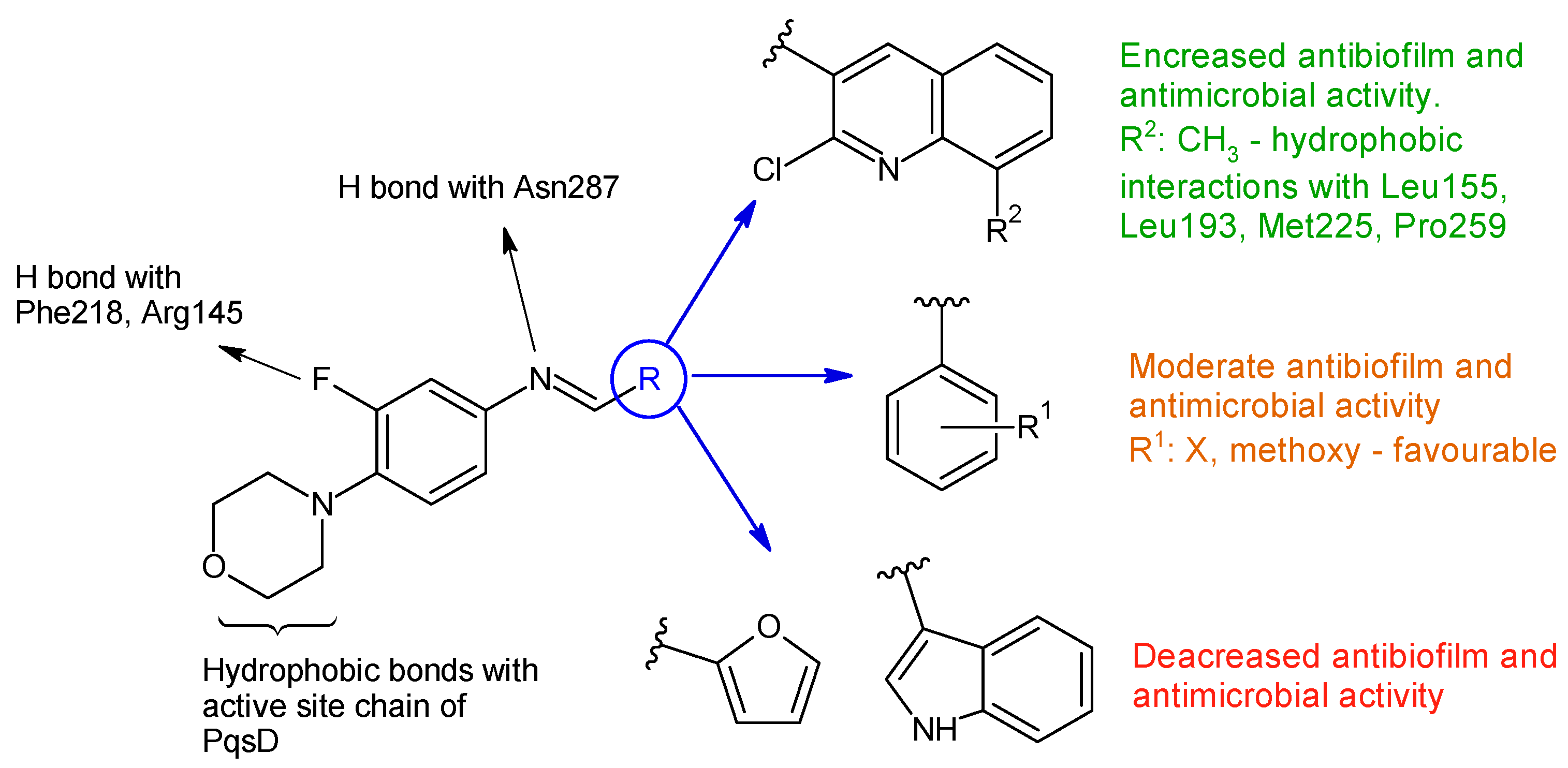
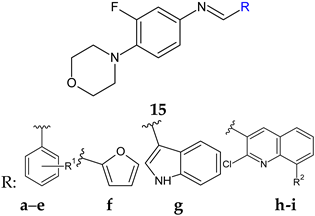 R1: H (a), 4-F (b), 2,6-diCl (c), 3,4-diOH (d), 4-diOCH3 (e) R2: H (h), CH3 (i) 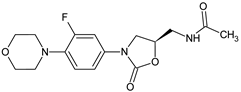 Linezolid | Antibacterial screening: B. subtilis NCIM-2063, E. coli NCIM-2256, P. aeruginosa NCIM-2036 | 15h—MIC 2.5 ± 0.15 15i—MIC 3.5 ± 0.18 μg/mL (PA) | [106] |
| Antibiofilm screening: P. aeruginosa O1 | PqsD inhibition 15h—IC50 12.97 ± 0.33 μM 15i—IC50 15.63 ± 0.20 μM | ||
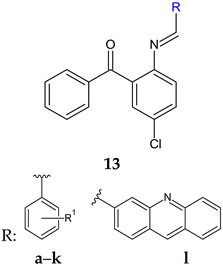
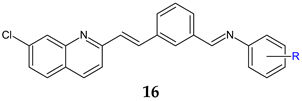 R: H (a), 4-Cl (b), 2-CH3 (c), 4-CH3 (d), 2-CF3 (e), 3-CF3 (f), 4-CF3 (g), 4-OCH3 (h), 3-NO2 (i), 4-NO2 (j) | Antibacterial screening: B. subtilis NCIM-2063, S. aureus NCIM-2901, E. coli NCIM-2256, P. aeruginosa NCIM-2036 | 16b: MIC 45 μg/mL (EC) 16g: MIC 91.5 μg/mL (PA) 16e: MIC 55.3 μg/mL (SA) | [107] |
| Antifungal screening: C. albicans NCIM-3471 | 16b: MIC 94.2 μg/mL 16a: MIC 98.8 μg/mL | ||
| Antibiofilm screening: C. albicans NCIM-3471 | Als-3 adhesin inhibition 16g: IC50 51.2 μM 16b: IC50 66.2 μM | ||
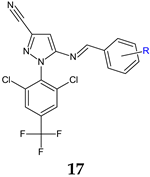 R: 4-NO2 (a), 4-COOH (b), 3,4-diOH (c), 2,4-diOCH3 (d), 2,4-diCl (e), 3,4-diOCH3 (f), 2,5-diOCH3 (g), 3-CN (h), 4-Br (i), 4-Cl (j) | Antifungal screening: C. albicans | 17i: MIC 42.6 μg/mL | [108] |
| Antifungal screening: C. albicans | 17i: IC50 41.5 μM | ||
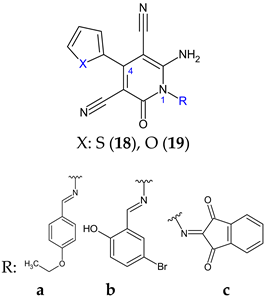 | Antibacterial screening: methicillin sensitive S. aureus ATCC 25923, methicillin resistant S. aureus ATCC 43300, E. coli ATCC-25922, K. pneumoniae ATCC-700603, P. aeruginosa ATCC-2785, Acinetobacter baumannii ATCC-19606 | 19b: MIC 62.5 μg/mL (MRSA) 19a: MIC 125 μg/mL (EC), 15.6 μg/mL (KP) 18c: MIC 62.5 μg/mL (PA) 18a, 19a, 18e: MIC 3.9 μg/mL (AB) | [109] |
| Antifungal screening: C. albicans ATCC-10231 | 18c: MIC 15.6 μg/mL | ||
| Antibiofilm screening: methicillin resistant S. aureus ATCC 43300, E. coli ATCC-25922, P. aeruginosa ATCC-2785, C. albicans ATCC-10231 | Downregulation of LasR 19b: BI 64.7 ± 1.85% (MRSA) 18b, 19c: BI 63.8% (EC) 19c: BI 45.4 ± 1.30% (PA), 75.0 ± 0.51% (CA) | ||
 R1: F (b), Cl (c), Br (d), NO2 (e), CF3 (f) | Antibacterial screening: B. subtilis ATCC 6633, S. aureus NRRL B-767, Salmonella typhi, E. coli ATCC 25955 | 20b: MIC 3.90 μg/mL (SA, EC) 20a: MIC 7.81 μg/mL (ST) | [110] |
| Antifungal screening: A. niger A. flavus | 20c, 20f: MIC 3.90 μg/mL (AF) 20f: MIC 15.62 μg/mL (AN) | ||
| Antibiofilm screening: S. aureus, E. coli | 20b: BI 72.34% (SA), 87.38% (EC) | ||
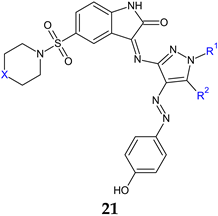 a: X: CH2, R1: H, R2: OH; b: X: CH2, R1: phenyl, R2: NH2; c: X: N(CH3), R1: phenyl, R2: NH2; d: X: CH2, R1: benzoyl, R2: NH2; e: X: N(CH3), R1: benzoyl, R2: NH2; f: X: N(CH3), R1: benzoyl, R2: OH | Antibacterial screening: S. aureus ATCC 6538, E. faecalis ATCC 29212, E. coli ATCC 35218, P. aeruginosa ATCC 27853 | Bactericidal 21b: 56.07 μM (SA, EF, PA), 112.16 μM (EC) 21d: 53.45 μM (EC, EF), 106.91 (SA, PA) | [111] |
| Antifungal screening: C. albicans ATCC 90028 | Fungicidal 21d: 106.91 μM | ||
| Antibiofilm screening: S. aureus ACL51 (MRSA) | 21d: BI 89.9 ± 4.7, 89.7 ± 9, 70.8 ± 2.3% at 0.03, 0.015, 0.007 mg/mL | ||
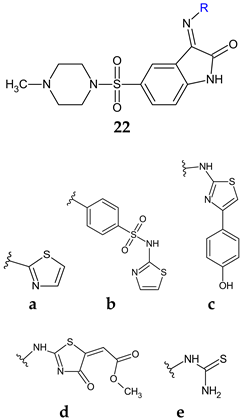 | Antibacterial screening: S. aureus ATCC 25923, B. subtilis ATCC 6051, E. faecalis ATCC 29212, E. coli ATCC 35218, P. aeruginosa ATCC 27853, S. typhimuriumATCC14028 | 22c: MIC 1.9 (EC), 7.8 (ST), 15.6 (SA, PA), 31.2 μg/mL (BS) | [112] |
| Antifungal screening: C. albicans ATCC10213 | 22d: MIC 31.2 μg/mL | ||
| Antibiofilm screening: S. aureus ATCC 29213, P. aeruginosa ATCC 9027 | 22b: BI50 1.95 μg/mL (SA) 22a, 22c, 22d: BI50 15.6 μg/mL (SA) 22c: BI50 7.8 μg/mL (PA) | ||
| Anti-quorum sensing: E. faecalis ATCC 29212 | 22c: 83.9, 73.0 and 64.9% fsr system inhibition at 3.9, 1.9 and 0.9 µg/mL | ||
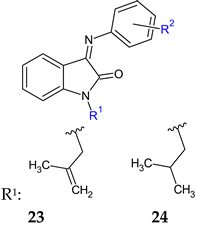 R2: 4-COOH (a), 2-COOH-4-Cl (b) | Antibacterial screening: B. subtilis ATCC10400, S. aureus ATCC29213 | 23a: MIC 0.09 mmol/L (SA, BS) 23b: MIC 0.181 mmol/L (SA, BS) | [113] |
| Antibiofilm screening: S. aureus ATCC29213, methicillin-resistant S. aureus ATCC35501 | 23a, 23b: BI 55% | ||
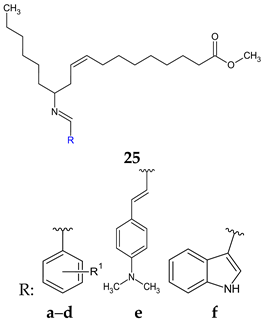 R1: 4-Cl (a), 4-N(CH3) (b), 4-OH-3-OCH3 (c), 4-OH-3,5-(OCH3)2 (d) | Antibacterial screening: Micrococcus luteus MTCC 2470, S. aureus MTCC 96, S. aureus MLS-16 MTCC 2940, B. subtilis MTCC 121, E. coli MTCC 739, P. aeruginosa MTCC 2453, Klebsiella planticola MTCC 2453 | Gram-positive bacteria—susceptible (SA, BS) 25a: MIC 9.0 μM, MBC 9–18 μM 25c: MIC 17.4 μM, MBC 35 μM, 25d: MIC 16.4 μM, MBC 16.4–32.8 μM | [114] |
| Antibiofilm screening: S. aureus MTCC 96, S. aureus MLS-16 MTCC 2940, B. subtilis MTCC 121 | 25a: IC50 4.3–6 μM 25d: IC50 6.5–8.6 μM 25c: IC50 8.0–9.4 μM (SA) 25f: IC50 7.3–9.5 μM (SA) | ||
| Oximes and Hydrazones | |||
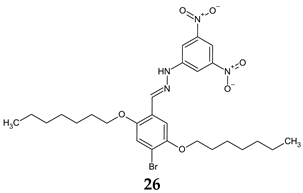 | Antibacterial screening: S. aureus ATCC 25923, E. faecalis ATCC 29212, E. coli ATCC 25922, P. aeruginosa ATCC 27853 | MIC 0.625 mg/mL (EF) MIC 1.25 mg/mL (SA) MIC 2.50 mg/mL (PA, EC) | [115] |
| Antifungal screening: C. albicans ATCC 10239, C. tropicalis ATCC 13803 | MIC 0.625 mg/mL | ||
| Antibiofilm screening: S. aureus ATCC 25923, E. faecalis ATCC 29212, E. coli ATCC 25922, P. aeruginosa ATCC 27853, C. albicans ATCC 10239, C. tropicalis ATCC 13803 | SA: BI 24.30–72.24% (MIC/4–MIC) EF: BI 23.41–49.55% PA: 12.50–28.30% CA: 10.26–25.83% CT: 23.90–40.15% (MIC/2–MIC) | ||
| Violacein inhibition: C. violaceum CV12472 | 5.7–100% (MIC/32–MIC) | ||
| QS inhibition: C. violaceum CV026 | 7.0–10.5 mm (MIC/2–MIC) | ||
| Swarming motility inhibition: P. aeruginosa PA01 | 14.4–45.7% (MIC/4–MIC) | ||
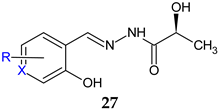 X = CH; R: H (a), 3-OCH3 (b), 5-Br (c), 5-I (d), 5-NO2 (e), 5-OH (f) X = N; R: 3-CH3-6-CH2OH (g) | Antibacterial screening: S. aureus PTCC 1112, S. pneumonia PTCC 1240, E. coli ATCC 25922, P. aeruginosa PAO1 | 27e, 27g: MIC 64 μg/mL (SA, EC) | [116] |
| Antibiofilm screening: P. aeruginosa PAO1 | 27e, 27g: Significantly reduction (1/16 and 1/4 MIC) | ||
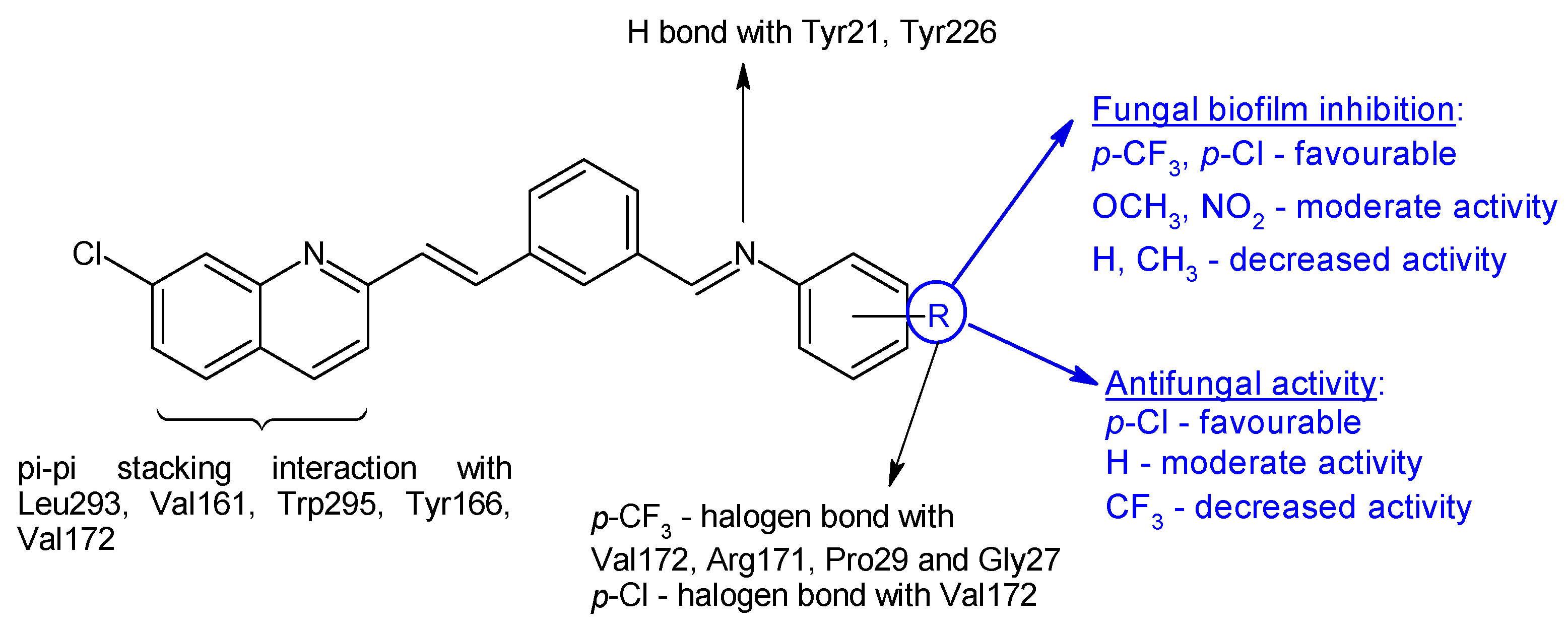
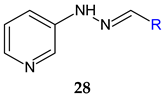 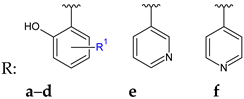 R1: 3-OH (a), 3-OCH3 (b), 4-OCH3 (c), 5-NO2 (d) | Antibacterial screening: A. baumannii ATCC 19606 A. baumannii clinical isolates 1–4 | MIC 25-200 μg/mL 28d: 25 μg/mL | [117] |
| Antibiofilm screening: A. baumannii ATCC 19606 A. baumannii clinical isolates 4 | 28a–f: Biofilm inhibition (MIC-2× MIC) 28a, 28d: Biofilm disruption (12.5 μg/mL) 28a: BEC50 28.2 μg/mL 28d: BEC50 12.8 μg/mL | ||
 R1: H, R2: H (a); R1: H, R2: 4-OCH3 (b); R1: H, R2: 4-CH3 (c); R1: 2,4-diF, R2: 4-NO2 (d); R1: 2,4-diF, R2: 4-CN (e) | Antibiofilm screening: C. albicans, clinical isolate | Upregulation of bcy1, nrg1, tup1 Downregulation of als3, hwp1, ras1 29a–c: 100 μg/mL 29d–e: 50 μg/mL | [118] |
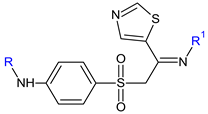 R: H (30), -CO-CH3 (31) R1: OH (a), n-OC6H13 (b), 4-chlorobenzyloxy (c), 2,4-dinitrophenylamino (d), 4-carboxyphenylamino (e), pyridine-2-carboxamido (f), pyridine-3-carboxamido (g), yridine-4-carboxamido (h) | Antibacterial screening: S. aureus ATCC 25923, S. aureus ATCC 29213, methicillin-resistant S. aureus N315, E. faecalis p1-2007226001, E. faecalis p1-2007225053, K. pneumoniae; E. coli ATCC 25922, P. aeruginosa ATCC 27853, A. baumannii | Gram-positive bacteria—susceptible 31f: MIC 1–4 μg/mL (EF) | [119] |
| Antibiofilm screening: E. faecalis | 31f: BI 35% (6× MIC) | ||
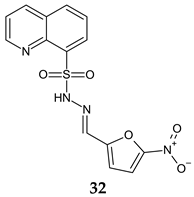 | Antibacterial screening: S. aureus ATCC 25923, S. aureus ATCC 43300, E. coli NCTC 9001, Listeria monocytogenes NCTC 11994 | MIC 125 μg/mL (SA) | [120] |
| Antifungal screening: C. albicans ATCC 10231, C. albicans ATCC 24433, C. parapsilosis ATC 22019, C. krusei ATCC 6258, C. glabrata ATCC 2001, clinical isolates (veterinary samples) | MIC 31.2 μg/mL (CA ATCC 10231, CP) | ||
| Anti-filamentation assay: C. albicans ATCC 10231 | 24.96 μg/mL— inhibition | ||
| Antibiofilm screening: C. albicans ATCC 10231 | 31.2 μg/mL—38% inhibition | ||
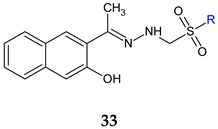 R: C2H5 (a), n-C3H7 (b), n-C4H9 (c) | Antifungal and antibiofilm screening: C. glabrata ATCC 90030 C. krusei ATCC 6258 C. krusei clinical isolates C. albicans ATCC 10231 C. albicans clinical isolates C. parapsilosis ATCC 22019 C. parapsilosis clinical isolates C. tropicalis NRRLY-12968 C. lusitaniae clinical isolates | MIC 32–64 μg/mL BIC 32–64 μg/mL (CA, CT, CK) BIC 64–128 μg/mL (CP, CL) Downregulation of hwp1, als3, ece1 and sap5 genes | [121] |
 R1: H (a), 4-CH3 (b), 4-OCH3 (c), 4-Cl (d) | Antibacterial screening: S. aureus, B. subtilis, E. coli, P. aeruginosa | 34a: IR 64.81% (EC) 34c: IR 64.61% (EC) | [122] |
| Antibiofilm screening: S. aureus, B. subtilis, E. coli, P. aeruginosa | 34a: BI 78.75% (EC) | ||
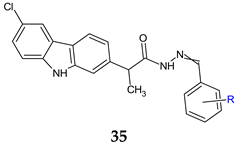 R = 2-OH (a), 2-OH-3-OCH3 (b), 2-OH-5-OCH3 (c), 4-Cl (d), 2,6-diCl (e), 3,5-diCl (f) | Antibacterial screening: S. aureus ATCC 25923, E. faecalis ATCC 29212, E. coli ATCC 25922, P. aeruginosa ATCC 27853 | 35d: MIC 0.15 mg/mL (EF) | [123] |
| Antifungal screening: C. albicans ATCC 10231 | 35a, 35c, 35d: MIC 0.31 mg/mL | ||
| Antibiofilm screening: S. aureus ATCC 25923, E. faecalis ATCC 29212, E. coli ATCC 25922, P. aeruginosa ATCC 27853, C. albicans ATCC 10231 | 35d: MBIC 0.078 mg/mL (EC, SA) 35c: MBIC 0.009 mg/mL (CA) | ||
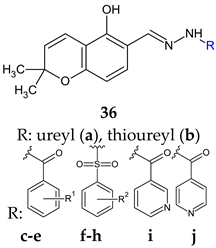 R1: H (c), 4-OH (d), 2,4-di-OH (e) R2: H (f), 4-CH3 (g), 4-CF3 (h) | Antibacterial screening: Vibrio harveyi BB120, S. aureus MW2, E. coli | 36d: MIC 3.9 μg/mL (VH) 36e: MIC 64 μg/mL (SA) | [124] |
| Anti-quorum sensing: V. harveyi BB120 | 36f: IC50 22 μM 36a: IC50 27 μM | ||
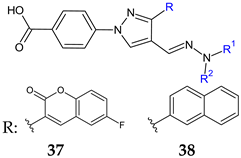 37: R1 = R2: C6H5 (a), R1: H, R2: 3-Cl-C6H4 (b), R1: H, R2: 3-Br-C6H4 (c), R1: H, R2: 4-CF3-C6H4 (d) 38: R1 = R2: C6H5 (a), R1 = R2: CH2C6H5 (b), R1: H, R2: 2,5-F2-C6H3 (c), R1: H, R2: 2-F-3-Cl-C6H3 (d), R1: H, R2: 4-CF3-C6H4 (e) | Antibacterial screening: S. aureus ATCC 25923, S. aureus BAA-2312, S. aureus ATCC 33591, S. aureus ATCC 700699, S. aureus ATCC 33592, S. epidermidis 700296, B. subtilis ATCC 6623; A. baumannii ATCC 19606, A. baumannii ATCC BAA-1605, A. baumannii ATCC 747 | 37a–d: MIC 3.125–12.5 μg/mL (SA, SE, BS) 37b,c: 6.25–25 μg/mL (AB) 38a–e: 0.78–25 μg/mL (SA, SE, BS) | [125,126] |
| Antibiofilm screening: S. aureus ATCC 25923 | Biofilm inhibition 37a,c,d, 38a–c: > 85% (1/2–2× MIC) Biofilm destruction 37a–c: >90%, 38b,e: > 70% (1/2–2× MIC) | ||
 | Antifungal screening: A. fumigatus ATCC 1022, A. niger ATCC 6275, Trichoderma viride IAM 5061, Penicillium funiculosum ATCC 36839, Penicillium verrucosum var. cyclopium (food isolates), C. albicans ATCC 10231 | 39a: 0.37 mg/mL 39b: 0.37–0.75 mg/mL | [127] |
| Antibiofilm screening: C. albicans ATCC 10231 | 39a: BI 33% (MIC), 18% (MIC/2-MIC/4) 39b: BI 16% (MIC/2–MIC), 5% (MIC/4) | ||
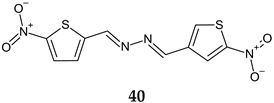 | Antibacterial screening: S. aureus SA-1001, S. aureus ME-311, S. aureus VA13 | MIC50 0.5–4.0 μg/mL MIC90 1–4.0 μg/mL Bactericidal | [128] |
 R1: H, R:4-Cl-C6H4 (a), R1: H, R:4-Br-C6H4 (b), R1: CH3, R:2,4-Cl2-C6H4 (c) | Antibiofilm screening: S. aureus | Altered biofilm integrity (10× MIC) | |
| Antibacterial screening: S. aureus ATCC 29213, MRSA clinical isolates, VRSA clinical isolates | MIC 2–8 μg/mL 41c: 1–4 μg/mL | [129] | |
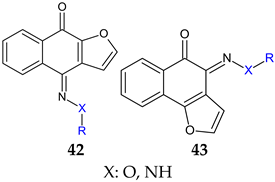 R: H, methyl, benzyl, acetyl, phenyl, 4-fluoro-phenyl, 4-methoxy-phenyl, 4-tolyl, methyl-sulfonyl 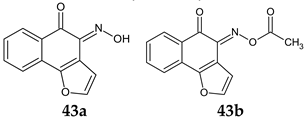 | Antibacterial screening: Methicillin-resistant S. aureus ATCC 33591, S. aureus clinical isolates (KM-1, KM-5), vancomycin-intermediate S. aureus (KV-1, KV-5), E. coli ATCC 8739 | 43a: MIC 9.7–19.5 μg/mL, MBC 3.9–156 μg/mL 43b: MIC 2.4–9.7 μg/mL, MBC 19.5–39 μg/mL | [130] |
| Antibiofilm screening: Methicillin-resistant S. aureus ATCC 33591 | Cell outside—complete inhibition at 100 μg/mL Cell inside— 43a: 4-log CFU reduction at 100 μg/mL | ||
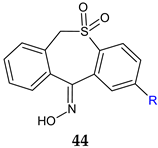 R: H (a), CH3 (b) | Antibacterial screening B. subtilis 12488 A. baumannii 221 P. aeruginosa 207 | 44a: 125 μg/mL (AB), 250 μg/mL (PA) 44b: 250 μg/mL (BS, AB) | [131] |
| Antibiofilm screening: S. aureus IC 13202 B. subtilis 12488 A. baumannii 221 C. albicans 101404 C. albicans IC249 | 44a,b: 125 μg/mL (AB) 44a: 250 μg/mL (BS, SA, CA) 44b: 250 μg/mL (CA) | ||
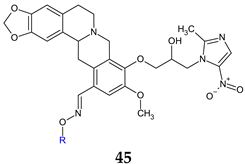 R: H (a), methyl (b), ethyl (c), propyl (d), butyl (e), t-butyl (f), pentyl (g), hexyl (h), allyl (i), benzyl (j) | Antibacterial screening: Methicillin-resistant S. aureus, S. aureus 25923, S. aureus 29213, E. faecalis, K. pneumoniae, E. coli, E. coli 25922, P. aeruginosa, P. aeruginosa 27853, A. baumannii | 45j: MIC 0.024 (PA)-0.199 mM | [132] |
| Antifungal screening: C. albicans, C. albicans ATCC 90023, C. tropicalis, C. parapsilosis 22019, A. fumigatus | 45j: MIC 0.024–0.199 mM (except C. parapsilosis) | ||
| Antibiofilm screening: P. aeruginosa | 45j: 45% inhibition at 8× MIC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Coandă, M.; Limban, C.; Nuță, D.C. Small Schiff Base Molecules—A Possible Strategy to Combat Biofilm-Related Infections. Antibiotics 2024, 13, 75. https://doi.org/10.3390/antibiotics13010075
Coandă M, Limban C, Nuță DC. Small Schiff Base Molecules—A Possible Strategy to Combat Biofilm-Related Infections. Antibiotics. 2024; 13(1):75. https://doi.org/10.3390/antibiotics13010075
Chicago/Turabian StyleCoandă, Maria, Carmen Limban, and Diana Camelia Nuță. 2024. "Small Schiff Base Molecules—A Possible Strategy to Combat Biofilm-Related Infections" Antibiotics 13, no. 1: 75. https://doi.org/10.3390/antibiotics13010075
APA StyleCoandă, M., Limban, C., & Nuță, D. C. (2024). Small Schiff Base Molecules—A Possible Strategy to Combat Biofilm-Related Infections. Antibiotics, 13(1), 75. https://doi.org/10.3390/antibiotics13010075






