Profiling Antibiotic Susceptibility among Distinct Enterococcus faecalis Isolates from Dental Root Canals
Abstract
:1. Introduction
2. Results
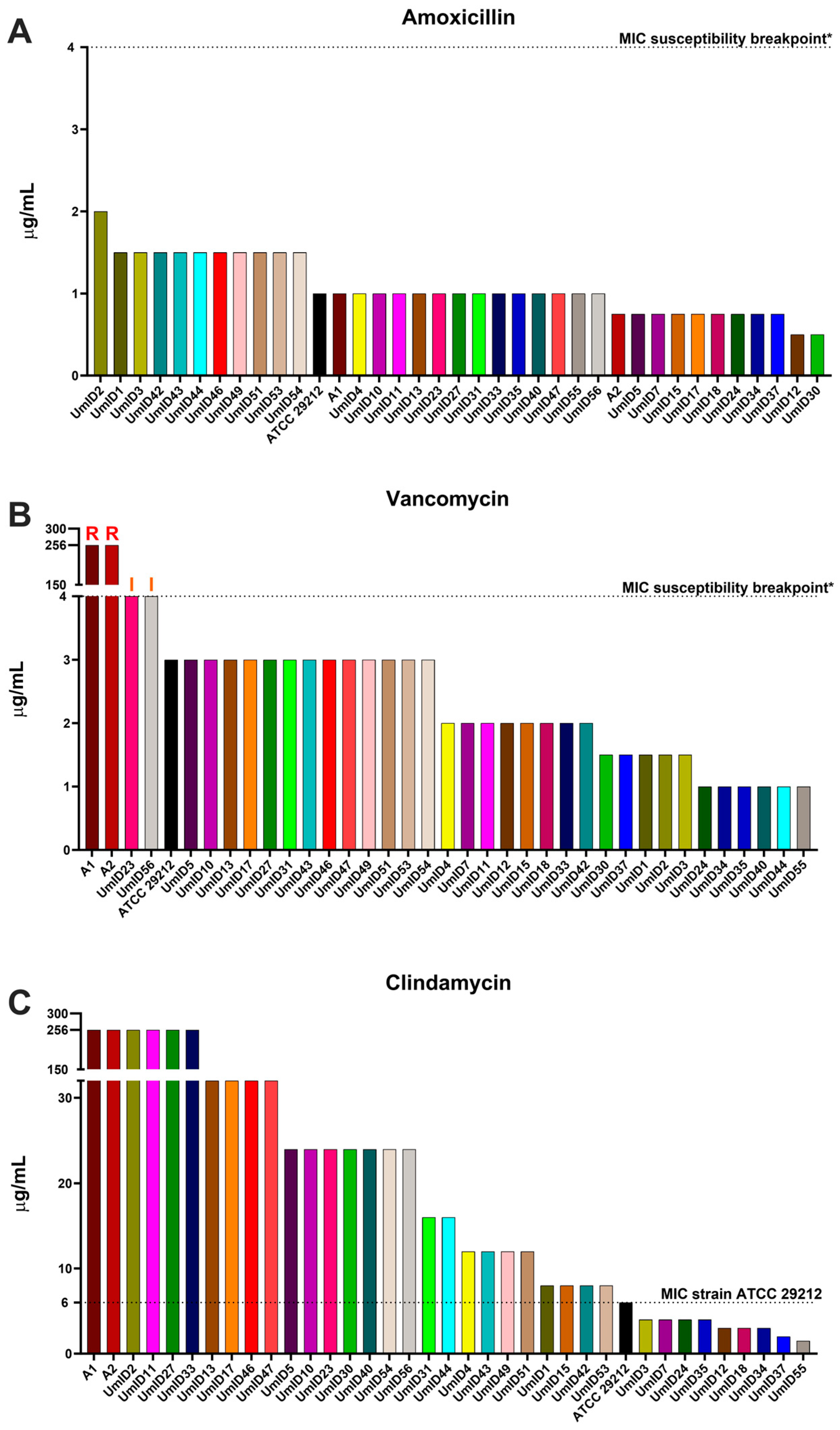
2.1. AST Outcomes for Amoxicillin, Vancomycin, and Clindamycin
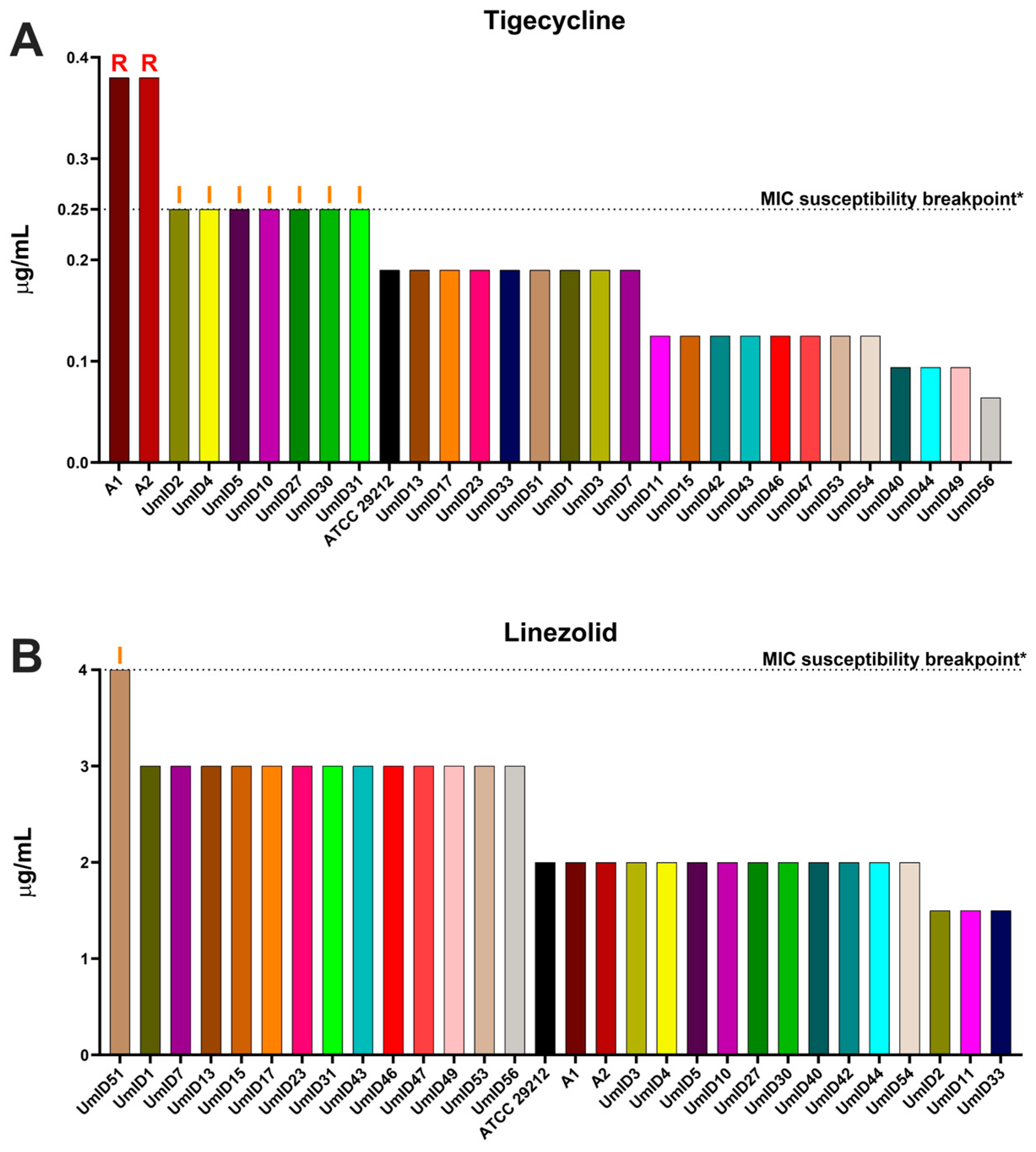
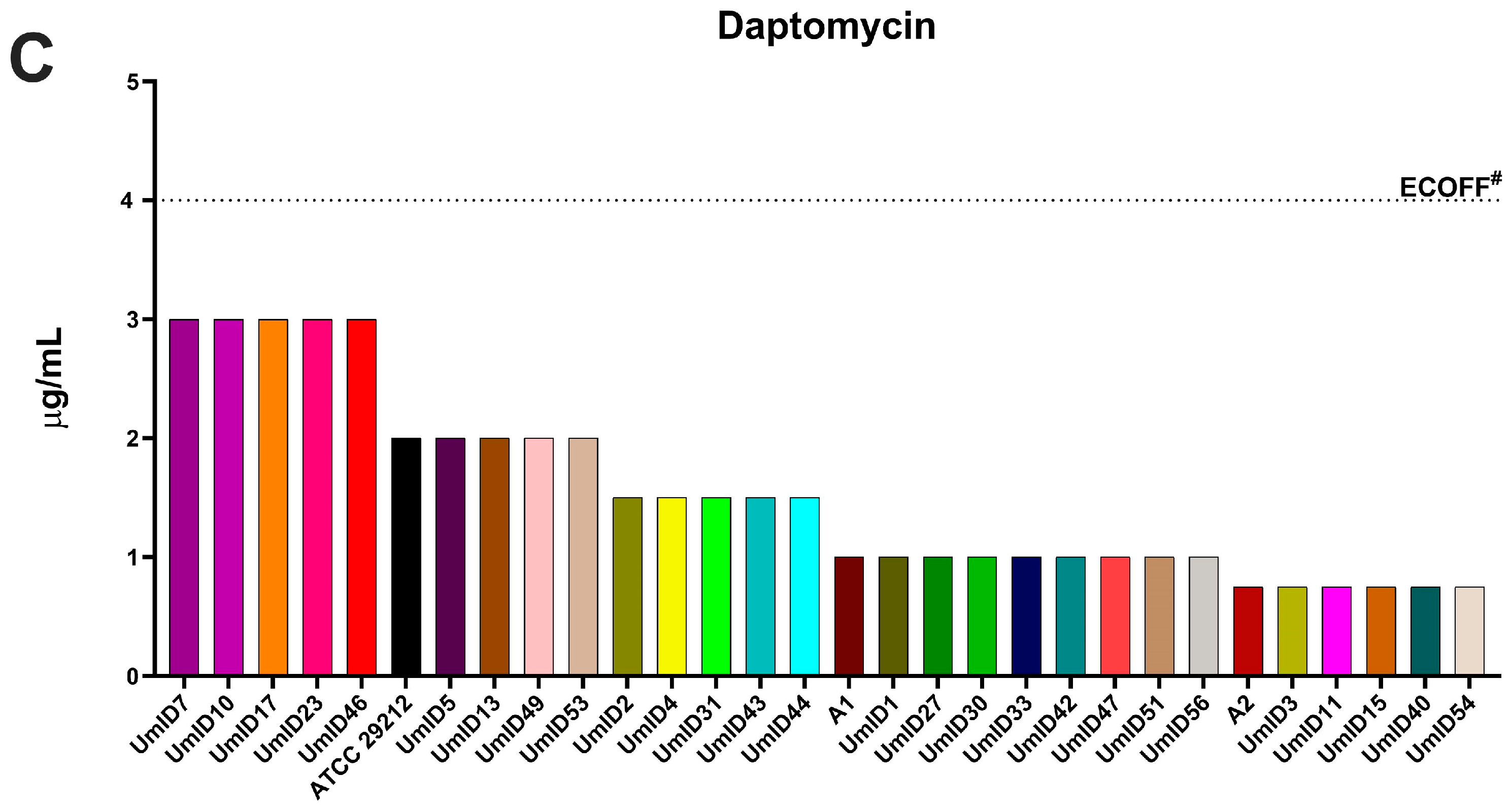
2.2. AST Outcomes in E. faecalis Isolates with High Clindamycin Intrinsic Resistance
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Isolates
4.2. In Vitro Antimicrobial Susceptibility Testing
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ch’ng, J.H.; Chong, K.K.L.; Lam, L.N.; Wong, J.J.; Kline, K.A. Biofilm-associated infection by enterococci. Nat. Rev. Microbiol. 2019, 17, 82–94. [Google Scholar] [CrossRef] [PubMed]
- Ran, S.; Liu, B.; Jiang, W.; Sun, Z.; Liang, J. Transcriptome analysis of Enterococcus faecalis in response to alkaline stress. Front. Microbiol. 2015, 6, 795. [Google Scholar] [CrossRef] [PubMed]
- Manoil, D.; Bouillaguet, S. Oxidative stress in bacteria measured by flow cytometry. Adv. Biotechnol. Microbiol. 2018, 8, 555726. [Google Scholar] [CrossRef]
- Paria, P.; Chakraborty, H.J.; Behera, B.K. Identification of novel salt tolerance-associated proteins from the secretome of Enterococcus faecalis. World J. Microbiol. Biotechnol. 2022, 38, 177. [Google Scholar] [CrossRef] [PubMed]
- Parga, A.; Manoil, D.; Brundin, M.; Otero, A.; Belibasakis, G.N. Gram-negative quorum sensing signalling enhances biofilm formation and virulence traits in gram-positive pathogen Enterococcus faecalis. J. Oral Microbiol. 2023, 15, 2208901. [Google Scholar] [CrossRef] [PubMed]
- Arias, C.A.; Murray, B.E. Emergence and management of drug-resistant enterococcal infections. Expert Rev. Anti-Infect. Ther. 2008, 6, 637–655. [Google Scholar] [CrossRef] [PubMed]
- Brinkwirth, S.; Ayobami, O.; Eckmanns, T.; Markwart, R. Hospital-acquired infections caused by enterococci: A systematic review and meta-analysis, WHO European Region, 1 January 2010 to 4 February 2020. Eurosurveill 2021, 26, 2001628. [Google Scholar] [CrossRef]
- Magill, S.S.; O’Leary, E.; Janelle, S.J.; Thompson, D.L.; Dumyati, G.; Nadle, J.; Wilson, L.E.; Kainer, M.A.; Lynfield, R.; Greissman, S.; et al. Changes in Prevalence of Health Care-Associated Infections in U.S. Hospitals. N. Engl. J. Med. 2018, 379, 1732–1744. [Google Scholar] [CrossRef]
- Aas, J.A.; Paster, B.J.; Stokes, L.N.; Olsen, I.; Dewhirst, F.E. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 2005, 43, 5721–5732. [Google Scholar] [CrossRef]
- Segata, N.; Haake, S.K.; Mannon, P.; Lemon, K.P.; Waldron, L.; Gevers, D.; Huttenhower, C.; Izard, J. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 2012, 13, R42. [Google Scholar] [CrossRef]
- Razavi, A.; Gmur, R.; Imfeld, T.; Zehnder, M. Recovery of Enterococcus faecalis from cheese in the oral cavity of healthy subjects. Oral Microbiol. Immunol. 2007, 22, 248–251. [Google Scholar] [CrossRef] [PubMed]
- Zehnder, M.; Guggenheim, B. The mysterious appearance of enterococci in filled root canals. Int. Endod. J. 2009, 42, 277–287. [Google Scholar] [CrossRef] [PubMed]
- Thurnheer, T.; Belibasakis, G.N. Integration of non-oral bacteria into in vitro oral biofilms. Virulence 2015, 6, 258–264. [Google Scholar] [CrossRef] [PubMed]
- Manoil, D.; Al-Manei, K.; Belibasakis, G.N. A Systematic Review of the Root Canal Microbiota Associated with Apical Periodontitis: Lessons from Next-Generation Sequencing. Proteom. Clin. Appl. 2020, 14, e1900060. [Google Scholar] [CrossRef] [PubMed]
- Foschi, F.; Cavrini, F.; Montebugnoli, L.; Stashenko, P.; Sambri, V.; Prati, C. Detection of bacteria in endodontic samples by polymerase chain reaction assays and association with defined clinical signs in Italian patients. Oral Microbiol. Immunol. 2005, 20, 289–295. [Google Scholar] [CrossRef] [PubMed]
- Sedgley, C.; Nagel, A.; Dahlen, G.; Reit, C.; Molander, A. Real-time quantitative polymerase chain reaction and culture analyses of Enterococcus faecalis in root canals. J. Endod. 2006, 32, 173–177. [Google Scholar] [CrossRef] [PubMed]
- Gomes, B.P.; Pinheiro, E.T.; Jacinto, R.C.; Zaia, A.A.; Ferraz, C.C.; Souza-Filho, F.J. Microbial analysis of canals of root-filled teeth with periapical lesions using polymerase chain reaction. J. Endod. 2008, 34, 537–540. [Google Scholar] [CrossRef]
- Siqueira, J.F., Jr.; Antunes, H.S.; Rocas, I.N.; Rachid, C.T.; Alves, F.R. Microbiome in the Apical Root Canal System of Teeth with Post-Treatment Apical Periodontitis. PLoS ONE 2016, 11, e0162887. [Google Scholar] [CrossRef]
- Zandi, H.; Kristoffersen, A.K.; Orstavik, D.; Rocas, I.N.; Siqueira, J.F., Jr.; Enersen, M. Microbial Analysis of Endodontic Infections in Root-filled Teeth with Apical Periodontitis before and after Irrigation Using Pyrosequencing. J. Endod. 2018, 44, 372–378. [Google Scholar] [CrossRef]
- Bouillaguet, S.; Manoil, D.; Girard, M.; Louis, J.; Gaia, N.; Leo, S.; Schrenzel, J.; Lazarevic, V. Root Microbiota in Primary and Secondary Apical Periodontitis. Front. Microbiol. 2018, 9, 2374. [Google Scholar] [CrossRef]
- Molander, A.; Reit, C.; Dahlen, G.; Kvist, T. Microbiological status of root-filled teeth with apical periodontitis. Int. Endod. J. 1998, 31, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Gaeta, C.; Marruganti, C.; Ali, I.A.A.; Fabbro, A.; Pinzauti, D.; Santoro, F.; Neelakantan, P.; Pozzi, G.; Grandini, S. The presence of Enterococcus faecalis in saliva as a risk factor for endodontic infection. Front. Cell. Infect. Microbiol. 2023, 13, 1061645. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, E.T.; Gomes, B.P.; Drucker, D.B.; Zaia, A.A.; Ferraz, C.C.; Souza-Filho, F.J. Antimicrobial susceptibility of Enterococcus faecalis isolated from canals of root filled teeth with periapical lesions. Int. Endod. J. 2004, 37, 756–763. [Google Scholar] [CrossRef] [PubMed]
- Reynaud Af Geijersstam, A.H.; Ellington, M.J.; Warner, M.; Woodford, N.; Haapasalo, M. Antimicrobial susceptibility and molecular analysis of Enterococcus faecalis originating from endodontic infections in Finland and Lithuania. Oral Microbiol. Immunol. 2006, 21, 164–168. [Google Scholar] [CrossRef] [PubMed]
- Barbosa-Ribeiro, M.; De-Jesus-Soares, A.; Zaia, A.A.; Ferraz, C.C.; Almeida, J.F.; Gomes, B.P. Antimicrobial Susceptibility and Characterization of Virulence Genes of Enterococcus faecalis Isolates from Teeth with Failure of the Endodontic Treatment. J. Endod. 2016, 42, 1022–1028. [Google Scholar] [CrossRef] [PubMed]
- Lins, R.X.; Hirata, R.J.; Wilson, M.; MA, O.L.; Fidel, R.A.S.; Williams, D. Comparison of genotypes, antimicrobial resistance and virulence profiles of oral and non oral Enterococcus faecalis from Brazil, Japan and the United Kingdom. J. Dent. 2019, 84, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Solache, M.; Rice, L.B. The Enterococcus: A Model of Adaptability to Its Environment. Clin. Microbiol. Rev. 2019, 32. [Google Scholar] [CrossRef]
- Dadashi, M.; Sharifian, P.; Bostanshirin, N.; Hajikhani, B.; Bostanghadiri, N.; Khosravi-Dehaghi, N.; van Belkum, A.; Darban-Sarokhalil, D. The Global Prevalence of Daptomycin, Tigecycline, and Linezolid-Resistant Enterococcus faecalis and Enterococcus faecium Strains From Human Clinical Samples: A Systematic Review and Meta-Analysis. Front. Med. 2021, 8, 720647. [Google Scholar] [CrossRef]
- Anderson, A.C.; von Ohle, C.; Frese, C.; Boutin, S.; Bridson, C.; Schoilew, K.; Peikert, S.A.; Hellwig, E.; Pelz, K.; Wittmer, A.; et al. The oral microbiota is a reservoir for antimicrobial resistance: Resistome and phenotypic resistance characteristics of oral biofilm in health, caries, and periodontitis. Ann. Clin. Microbiol. Antimicrob. 2023, 22, 37. [Google Scholar] [CrossRef]
- Arias, C.A.; Murray, B.E. The rise of the Enterococcus: Beyond vancomycin resistance. Nat. Rev. Microbiol. 2012, 10, 266–278. [Google Scholar] [CrossRef]
- Sukumar, S.; Roberts, A.P.; Martin, F.E.; Adler, C.J. Metagenomic Insights into Transferable Antibiotic Resistance in Oral Bacteria. J. Dent. Res. 2016, 95, 969–976. [Google Scholar] [CrossRef]
- Brauner, A.; Fridman, O.; Gefen, O.; Balaban, N.Q. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol. 2016, 14, 320–330. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, I.; Hilpert, K.; Hancock, R.E. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Leclercq, R.; Canton, R.; Brown, D.F.; Giske, C.G.; Heisig, P.; MacGowan, A.P.; Mouton, J.W.; Nordmann, P.; Rodloff, A.C.; Rossolini, G.M.; et al. EUCAST expert rules in antimicrobial susceptibility testing. Clin. Microbiol. Infect. 2013, 19, 141–160. [Google Scholar] [CrossRef] [PubMed]
- Weber, R.E.; Fleige, C.; Layer, F.; Neumann, B.; Kresken, M.; Werner, G. Determination of a Tentative Epidemiological Cut-Off Value (ECOFF) for Dalbavancin and Enterococcus faecium. Antibiotics 2021, 10, 915. [Google Scholar] [CrossRef] [PubMed]
- Turnidge, J.; Kahlmeter, G.; Kronvall, G. Statistical characterisation of bacterial wild-type MIC value distributions and the determination of epidemiological cut-off values. Clin. Microbiol. Infect. 2006, 12, 418–425. [Google Scholar] [CrossRef]
- Kahlmeter, G.; Turnidge, J. How to: ECOFFs-the why, the how, and the don’ts of EUCAST epidemiological cutoff values. Clin. Microbiol. Infect. 2022, 28, 952–954. [Google Scholar] [CrossRef]
- Nabal Diaz, S.G.; Algara Robles, O.; Garcia-Lechuz Moya, J.M. New definitions of susceptibility categories EUCAST 2019: Clinic application. Rev. Esp. Quimioter. 2022, 35 (Suppl. S3), 84–88. [Google Scholar] [CrossRef]
- Giske, C.G.; Turnidge, J.; Canton, R.; Kahlmeter, G.; Committee, E.S. Update from the European Committee on Antimicrobial Susceptibility Testing (EUCAST). J. Clin. Microbiol. 2022, 60, e0027621. [Google Scholar] [CrossRef]
- Hollenbeck, B.L.; Rice, L.B. Intrinsic and acquired resistance mechanisms in enterococcus. Virulence 2012, 3, 421–433. [Google Scholar] [CrossRef]
- Singh, K.V.; Weinstock, G.M.; Murray, B.E. An Enterococcus faecalis ABC homologue (Lsa) is required for the resistance of this species to clindamycin and quinupristin-dalfopristin. Antimicrob. Agents Chemother. 2002, 46, 1845–1850. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, Y.; Wu, C.; Shen, Z.; Schwarz, S.; Du, X.D.; Dai, L.; Zhang, W.; Zhang, Q.; Shen, J. First report of the multidrug resistance gene cfr in Enterococcus faecalis of animal origin. Antimicrob. Agents Chemother. 2012, 56, 1650–1654. [Google Scholar] [CrossRef] [PubMed]
- Turnidge, J.; Kahlmeter, G.; Canton, R.; MacGowan, A.; Giske, C.G.; European Committee on Antimicrobial Susceptibility, T. Daptomycin in the treatment of enterococcal bloodstream infections and endocarditis: A EUCAST position paper. Clin. Microbiol. Infect. 2020, 26, 1039–1043. [Google Scholar] [CrossRef] [PubMed]
- Sauvage, E.; Kerff, F.; Terrak, M.; Ayala, J.A.; Charlier, P. The penicillin-binding proteins: Structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 2008, 32, 234–258. [Google Scholar] [CrossRef] [PubMed]
- Tran, T.T.; Panesso, D.; Mishra, N.N.; Mileykovskaya, E.; Guan, Z.; Munita, J.M.; Reyes, J.; Diaz, L.; Weinstock, G.M.; Murray, B.E.; et al. Daptomycin-resistant Enterococcus faecalis diverts the antibiotic molecule from the division septum and remodels cell membrane phospholipids. MBio 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Rashid, R.; Cazenave-Gassiot, A.; Gao, I.H.; Nair, Z.J.; Kumar, J.K.; Gao, L.; Kline, K.A.; Wenk, M.R. Comprehensive analysis of phospholipids and glycolipids in the opportunistic pathogen Enterococcus faecalis. PLoS ONE 2017, 12, e0175886. [Google Scholar] [CrossRef]
- Yaghoubi, S.; Zekiy, A.O.; Krutova, M.; Gholami, M.; Kouhsari, E.; Sholeh, M.; Ghafouri, Z.; Maleki, F. Tigecycline antibacterial activity, clinical effectiveness, and mechanisms and epidemiology of resistance: Narrative review. Eur. J. Clin. Microbiol. Infect. Dis. 2022, 41, 1003–1022. [Google Scholar] [CrossRef]
- Hashemian, S.M.R.; Farhadi, T.; Ganjparvar, M. Linezolid: A review of its properties, function, and use in critical care. Drug Des. Devel. Ther. 2018, 12, 1759–1767. [Google Scholar] [CrossRef]
- Dai, X.; Ma, R.; Jiang, W.; Deng, Z.; Chen, L.; Liang, Y.; Shao, L.; Zhao, W. Enterococcus faecalis-Induced Macrophage Necroptosis Promotes Refractory Apical Periodontitis. Microbiol. Spectr. 2022, 10, e0104522. [Google Scholar] [CrossRef]
- Dorn, S.O.; Cheung, G.S.-P. Management of Endodontic Emergencies. In Cohen’s Pathways of the Pulp, 11th ed.; Hargreaves, K.M., Berman, L.H., Eds.; Elsevier: Amsterdam, The Netherlands, 2016; pp. 706–721. [Google Scholar]
- Zehnder, M.; Belibasakis, G.N. On the dynamics of root canal infections-what we understand and what we don’t. Virulence 2015, 6, 216–222. [Google Scholar] [CrossRef]
- Shah, A.C.; Leong, K.K.; Lee, M.K.; Allareddy, V. Outcomes of hospitalizations attributed to periapical abscess from 2000 to 2008: A longitudinal trend analysis. J. Endod. 2013, 39, 1104–1110. [Google Scholar] [CrossRef] [PubMed]
- Anderson, A.C.; Andisha, H.; Hellwig, E.; Jonas, D.; Vach, K.; Al-Ahmad, A. Antibiotic Resistance Genes and Antibiotic Susceptibility of Oral Enterococcus faecalis Isolates Compared to Isolates from Hospitalized Patients and Food. Adv. Exp. Med. Biol. 2018, 1057, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Belibasakis, G.N.; Lund, B.K.; Kruger Weiner, C.; Johannsen, B.; Baumgartner, D.; Manoil, D.; Hultin, M.; Mitsakakis, K. Healthcare Challenges and Future Solutions in Dental Practice: Assessing Oral Antibiotic Resistances by Contemporary Point-of-Care Approaches. Antibiotics 2020, 9, 810. [Google Scholar] [CrossRef]
- Ahmadi, H.; Ebrahimi, A.; Ahmadi, F. Antibiotic Therapy in Dentistry. Int. J. Dent. 2021, 2021, 6667624. [Google Scholar] [CrossRef] [PubMed]
- Guardabassi, L.; Larsen, J.; Skov, R.; Schonheyder, H.C. Gentamicin-resistant Enterococcus faecalis sequence type 6 with reduced penicillin susceptibility: Diagnostic and therapeutic implications. J. Clin. Microbiol. 2010, 48, 3820–3821. [Google Scholar] [CrossRef] [PubMed]
- Metzidie, E.; Manolis, E.N.; Pournaras, S.; Sofianou, D.; Tsakris, A. Spread of an unusual penicillin- and imipenem-resistant but ampicillin-susceptible phenotype among Enterococcus faecalis clinical isolates. J. Antimicrob. Chemother. 2006, 57, 158–160. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Cormican, M.; Flamm, R.K.; Mendes, R.E.; Jones, R.N. Temporal and Geographic Variation in Antimicrobial Susceptibility and Resistance Patterns of Enterococci: Results From the SENTRY Antimicrobial Surveillance Program, 1997–2016. Open Forum Infect. Dis. 2019, 6, S54–S62. [Google Scholar] [CrossRef]
- Khan, A.; Miller, W.R.; Axell-House, D.; Munita, J.M.; Arias, C.A. Antimicrobial Susceptibility Testing for Enterococci. J. Clin. Microbiol. 2022, 60, e0084321. [Google Scholar] [CrossRef]
- Rice, L.B.; Desbonnet, C.; Tait-Kamradt, A.; Garcia-Solache, M.; Lonks, J.; Moon, T.M.; D’Andrea, E.D.; Page, R.; Peti, W. Structural and Regulatory Changes in PBP4 Trigger Decreased beta-Lactam Susceptibility in Enterococcus faecalis. mBio 2018, 9, 10-1128. [Google Scholar] [CrossRef]
- Manoil, D.; Parga, A.; Hellesen, C.; Khawaji, A.; Brundin, M.; Durual, S.; Ozenci, V.; Fang, H.; Belibasakis, G.N. Photo-oxidative stress response and virulence traits are co-regulated in E. faecalis after antimicrobial photodynamic therapy. J. Photochem. Photobiol. B 2022, 234, 112547. [Google Scholar] [CrossRef]
- Rams, T.E.; Feik, D.; Mortensen, J.E.; Degener, J.E.; van Winkelhoff, A.J. Antibiotic susceptibility of periodontal Enterococcus faecalis. J. Periodontol. 2013, 84, 1026–1033. [Google Scholar] [CrossRef] [PubMed]
- Olson, M.W.; Ruzin, A.; Feyfant, E.; Rush, T.S., 3rd; O’Connell, J.; Bradford, P.A. Functional, biophysical, and structural bases for antibacterial activity of tigecycline. Antimicrob. Agents Chemother. 2006, 50, 2156–2166. [Google Scholar] [CrossRef] [PubMed]
- Kresken, M.; Leitner, E.; Seifert, H.; Peters, G.; von Eiff, C. Susceptibility of clinical isolates of frequently encountered bacterial species to tigecycline one year after the introduction of this new class of antibiotics: Results of the second multicentre surveillance trial in Germany (G-TEST II, 2007). Eur. J. Clin. Microbiol. Infect. Dis. 2009, 28, 1007–1011. [Google Scholar] [CrossRef] [PubMed]
- Werner, G.; Gfrorer, S.; Fleige, C.; Witte, W.; Klare, I. Tigecycline-resistant Enterococcus faecalis strain isolated from a German intensive care unit patient. J. Antimicrob. Chemother. 2008, 61, 1182–1183. [Google Scholar] [CrossRef]
- Cordina, C.; Hill, R.; Deshpande, A.; Hood, J.; Inkster, T. Tigecycline-resistant Enterococcus faecalis associated with omeprazole use in a surgical patient. J. Antimicrob. Chemother. 2012, 67, 1806–1807. [Google Scholar] [CrossRef]
- Arthur, M.; Depardieu, F.; Molinas, C.; Reynolds, P.; Courvalin, P. The vanZ gene of Tn1546 from Enterococcus faecium BM4147 confers resistance to teicoplanin. Gene 1995, 154, 87–92. [Google Scholar] [CrossRef]
- Dabul, A.N.G.; Avaca-Crusca, J.S.; Navais, R.B.; Merlo, T.P.; Van Tyne, D.; Gilmore, M.S.; Camargo, I. Molecular basis for the emergence of a new hospital endemic tigecycline-resistant Enterococcus faecalis ST103 lineage. Infect. Genet. Evol. 2019, 67, 23–32. [Google Scholar] [CrossRef]
- Fiedler, S.; Bender, J.K.; Klare, I.; Halbedel, S.; Grohmann, E.; Szewzyk, U.; Werner, G. Tigecycline resistance in clinical isolates of Enterococcus faecium is mediated by an upregulation of plasmid-encoded tetracycline determinants tet(L) and tet(M). J. Antimicrob. Chemother. 2016, 71, 871–881. [Google Scholar] [CrossRef]
- Jepsen, K.; Falk, W.; Brune, F.; Fimmers, R.; Jepsen, S.; Bekeredjian-Ding, I. Prevalence and antibiotic susceptibility trends of periodontal pathogens in the subgingival microbiota of German periodontitis patients: A retrospective surveillance study. J. Clin. Periodontol. 2021, 48, 1216–1227. [Google Scholar] [CrossRef]
- Antimicrobial Resistance, C. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Collaborators, G.B.D.A.R. Global mortality associated with 33 bacterial pathogens in 2019: A systematic analysis for the Global Burden of Disease Study 2019. Lancet 2022, 400, 2221–2248. [Google Scholar] [CrossRef]
- Kohler, P.; Eshaghi, A.; Kim, H.C.; Plevneshi, A.; Green, K.; Willey, B.M.; McGeer, A.; Patel, S.N.; Toronto Invasive Bacterial Diseases, N. Prevalence of vancomycin-variable Enterococcus faecium (VVE) among vanA-positive sterile site isolates and patient factors associated with VVE bacteremia. PLoS ONE 2018, 13, e0193926. [Google Scholar] [CrossRef] [PubMed]
- Sedgley, C.M.; Nagel, A.C.; Shelburne, C.E.; Clewell, D.B.; Appelbe, O.; Molander, A. Quantitative real-time PCR detection of oral Enterococcus faecalis in humans. Arch. Oral Biol. 2005, 50, 575–583. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manoil, D.; Cerit, E.E.; Fang, H.; Durual, S.; Brundin, M.; Belibasakis, G.N. Profiling Antibiotic Susceptibility among Distinct Enterococcus faecalis Isolates from Dental Root Canals. Antibiotics 2024, 13, 18. https://doi.org/10.3390/antibiotics13010018
Manoil D, Cerit EE, Fang H, Durual S, Brundin M, Belibasakis GN. Profiling Antibiotic Susceptibility among Distinct Enterococcus faecalis Isolates from Dental Root Canals. Antibiotics. 2024; 13(1):18. https://doi.org/10.3390/antibiotics13010018
Chicago/Turabian StyleManoil, Daniel, Ender Efe Cerit, Hong Fang, Stéphane Durual, Malin Brundin, and Georgios N. Belibasakis. 2024. "Profiling Antibiotic Susceptibility among Distinct Enterococcus faecalis Isolates from Dental Root Canals" Antibiotics 13, no. 1: 18. https://doi.org/10.3390/antibiotics13010018
APA StyleManoil, D., Cerit, E. E., Fang, H., Durual, S., Brundin, M., & Belibasakis, G. N. (2024). Profiling Antibiotic Susceptibility among Distinct Enterococcus faecalis Isolates from Dental Root Canals. Antibiotics, 13(1), 18. https://doi.org/10.3390/antibiotics13010018






