Impact and Diversity of ESBL-Producing Klebsiella pneumoniae Recovered from Raw Chicken Meat Samples in Türkiye
Abstract
1. Introduction
2. Results
2.1. MDR in K. pneumoniae from Meat
2.2. Acquired AMR and Single-Nucleotide Variations in the Chromosome
2.3. Virulence Factors of K. pneumoniae
2.4. Transmissibility of the blaCTX-M-15 Determinants
2.5. Phylogenetic Comparison of ESBL-Producing K. pneumoniae from Chicken Meat and Human Infections
3. Discussion
4. Materials and Methods
4.1. Sample Collection and K. pneumoniae Isolation
4.2. Antimicrobial Susceptibility Testing (AST) Using Broth Microdilution
4.3. Pulsed-Field Gel Electrophoresis (PFGE) for Macrorestriction Profiling
4.4. In Vitro Conjugation Assay
4.5. WGS and Bioinformatics Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Central Asian and Eastern European Surveillance of Antimicrobial Resistance: Annual Report 2016; WHO: Geneva, Switzerland, 2016. [Google Scholar]
- WHO. Central Asian and Eastern European Surveillance of Antimicrobial Resistance: Annual Report 2018; WHO: Geneva, Switzerland, 2018. [Google Scholar]
- WHO. Ranking of medically important antimicrobials for risk management of antimicrobial resistance due to non-human use. In Critically Important Antimicrobials for Human Medicine, 6th Revision 2018; WHO: Geneva, Switzerland, 2019. [Google Scholar]
- Rawat, D.; Nair, D. Extended-spectrum beta-lactamases in Gram negative bacteria. J. Glob. Infect. Dis. 2010, 2, 263–274. [Google Scholar] [CrossRef]
- Bush, K.; Jacoby, G.A. Updated functional classification of beta-lactamases. Antimicrob. Agents Chemother. 2010, 54, 969–976. [Google Scholar] [CrossRef]
- Kiratisin, P.; Chattammanat, S.; Sa-Nguansai, S.; Dansubutra, B.; Nangpatharapornthawee, P.; Patthamalai, P.; Tirachaimongkol, N.; Nunthanasup, T. A 2-year trend of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in Thailand: An alert for infection control. Trans. R. Soc. Trop. Med. Hyg. 2008, 102, 460–464. [Google Scholar] [CrossRef]
- Irrgang, A.; Hammerl, J.A.; Falgenhauer, L.; Guiral, E.; Schmoger, S.; Imirzalioglu, C.; Fischer, J.; Guerra, B.; Chakraborty, T.; Kasbohrer, A. Diversity of CTX-M-1-producing E. coli from German food samples and genetic diversity of the bla(CTX-M-1) region on IncI1 ST3 plasmids. Vet. Microbiol. 2018, 221, 98–104. [Google Scholar] [CrossRef]
- Fielding, B.C.; Mnabisa, A.; Gouws, P.A.; Morris, T. Antimicrobial-resistant Klebsiella species isolated from free-range chicken samples in an informal settlement. Arch. Med. Sci. 2012, 8, 39–42. [Google Scholar] [CrossRef]
- Aly, M.M.; Khalil, S.; Metwaly, A. Isolation and molecular identification of Klebsiella spp. microbe isolated from chicks. Alex. J. Vet. Sci. 2014, 43, 97–103. [Google Scholar]
- Diab, M.; Hamze, M.; Bonnet, R.; Saras, E.; Madec, J.Y.; Haenni, M. OXA-48 and CTX-M-15 extended-spectrum beta-lactamases in raw milk in Lebanon: Epidemic spread of dominant Klebsiella pneumoniae clones. J. Med. Microbiol. 2017, 66, 1688–1691. [Google Scholar] [CrossRef]
- Guo, Y.; Zhou, H.; Qin, L.; Pang, Z.; Qin, T.; Ren, H.; Pan, Z.; Zhou, J. Frequency, antimicrobial resistance and genetic diversity of Klebsiella pneumoniae in food samples. PLoS ONE 2016, 11, e0153561. [Google Scholar] [CrossRef]
- Zhang, S.; Yang, G.; Ye, Q.; Wu, Q.; Zhang, J.; Huang, Y. Phenotypic and genotypic characterization of Klebsiella pneumoniae isolated from retail foods in China. Front. Microbiol. 2018, 9, 289. [Google Scholar] [CrossRef]
- Overdevest, I.; Willemsen, I.; Rijnsburger, M.; Eustace, A.; Xu, L.; Hawkey, P.; Heck, M.; Savelkoul, P.; Vandenbroucke-Grauls, C.; van der Zwaluw, K.; et al. Extended-spectrum beta-lactamase genes of Escherichia coli in chicken meat and humans, The Netherlands. Emerg. Infect. Dis. 2011, 17, 1216–1222. [Google Scholar] [CrossRef]
- Projahn, M.; von Tippelskirch, P.; Semmler, T.; Guenther, S.; Alter, T.; Roesler, U. Contamination of chicken meat with extended-spectrum beta-lactamase producing- Klebsiella pneumoniae and Escherichia coli during scalding and defeathering of broiler carcasses. Food Microbiol. 2019, 77, 185–191. [Google Scholar] [CrossRef]
- Savin, M.; Bierbaum, G.; Schmithausen, R.M.; Heinemann, C.; Kreyenschmidt, J.; Schmoger, S.; Akbaba, I.; Kasbohrer, A.; Hammerl, J.A. Slaughterhouse wastewater as a reservoir for extended-spectrum beta-lactamase (ESBL)-producing, and colistin-resistant Klebsiella spp. and their impact in a “One Health” perspective. Sci. Total Environ. 2022, 804, 150000. [Google Scholar] [CrossRef]
- Overdevest, I.T.; Heck, M.; van der Zwaluw, K.; Huijsdens, X.; van Santen, M.; Rijnsburger, M.; Eustace, A.; Xu, L.; Hawkey, P.; Savelkoul, P.; et al. Extended-spectrum beta-lactamase producing Klebsiella spp. in chicken meat and humans: A comparison of typing methods. Clin. Microbiol. Infect. 2014, 20, 251–255. [Google Scholar] [CrossRef][Green Version]
- Ewers, C.; Stamm, I.; Pfeifer, Y.; Wieler, L.H.; Kopp, P.A.; Schonning, K.; Prenger-Berninghoff, E.; Scheufen, S.; Stolle, I.; Gunther, S.; et al. Clonal spread of highly successful ST15-CTX-M-15 Klebsiella pneumoniae in companion animals and horses. J. Antimicrob. Chemother. 2014, 69, 2676–2680. [Google Scholar] [CrossRef]
- Haenni, M.; Ponsin, C.; Metayer, V.; Medaille, C.; Madec, J.Y. Veterinary hospital-acquired infections in pets with a ciprofloxacin-resistant CTX-M-15-producing Klebsiella pneumoniae ST15 clone. J. Antimicrob. Chemother. 2012, 67, 770–771. [Google Scholar] [CrossRef]
- Donati, V.; Feltrin, F.; Hendriksen, R.S.; Svendsen, C.A.; Cordaro, G.; Garcia-Fernandez, A.; Lorenzetti, S.; Lorenzetti, R.; Battisti, A.; Franco, A. Extended-spectrum-beta-lactamases, AmpC beta-lactamases and plasmid mediated quinolone resistance in Klebsiella spp. from companion animals in Italy. PLoS ONE 2014, 9, e90564. [Google Scholar] [CrossRef]
- Cardozo, M.V.; Liakopoulos, A.; Brouwer, M.; Kant, A.; Pizauro, L.J.L.; Borzi, M.M.; Mevius, D.; de Avila, F.A. Occurrence and molecular characteristics of extended-spectrum beta-lactamase-producing Enterobacterales recovered from chicken, chicken meat, and human infections in Sao Paulo State, Brazil. Front. Microbiol. 2021, 12, 628738. [Google Scholar] [CrossRef]
- Kürekci, C.; Osek, J.; Aydın, M.; Ozan Tekeli, I.; Kurpas, M.; Wieczorek, K.; Sakin, F. Evaluation of bulk tank raw milk and raw chicken meat samples as source of ESBL producing Escherichia coli in Turkey: Recent insights. J. Food Saf. 2019, 39, e12605. [Google Scholar] [CrossRef]
- Pehlivanlar Onen, S.; Aslantas, O.; Sebnem Yilmaz, E.; Kurekci, C. Prevalence of beta-lactamase producing Escherichia coli from retail meat in Turkey. J. Food Sci. 2015, 80, M2023–M2029. [Google Scholar] [CrossRef]
- Hiroi, M.; Yamazaki, F.; Harada, T.; Takahashi, N.; Iida, N.; Noda, Y.; Yagi, M.; Nishio, T.; Kanda, T.; Kawamori, F.; et al. Prevalence of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in food-producing animals. J. Vet. Med. Sci. 2012, 74, 189–195. [Google Scholar] [CrossRef]
- Chenouf, N.S.; Carvalho, I.; Messai, C.R.; Ruiz-Ripa, L.; Mama, O.M.; Titouche, Y.; Zitouni, A.; Hakem, A.; Torres, C. Extended spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae from broiler liver in the Center of Algeria, with detection of CTX-M-55 and B2/ST131-CTX-M-15 in Escherichia coli. Microb. Drug Resist. 2021, 27, 268–276. [Google Scholar] [CrossRef]
- Sivaraman, G.K.; Sudha, S.; Muneeb, K.H.; Shome, B.; Holmes, M.; Cole, J. Molecular assessment of antimicrobial resistance and virulence in multi drug resistant ESBL-producing Escherichia coli and Klebsiella pneumoniae from food fishes, Assam, India. Microb. Pathog. 2020, 149, 104581. [Google Scholar] [CrossRef]
- Diaz-Jimenez, D.; Garcia-Menino, I.; Fernandez, J.; Garcia, V.; Mora, A. Chicken and turkey meat: Consumer exposure to multidrug-resistant Enterobacteriaceae including mcr-carriers, uropathogenic E. coli and high-risk lineages such as ST131. Int. J. Food Microbiol. 2020, 331, 108750. [Google Scholar] [CrossRef]
- Savin, M.; Alexander, J.; Bierbaum, G.; Hammerl, J.A.; Hembach, N.; Schwartz, T.; Schmithausen, R.M.; Sib, E.; Voigt, A.; Kreyenschmidt, J. Antibiotic-resistant bacteria, antibiotic resistance genes, and antibiotic residues in wastewater from a poultry slaughterhouse after conventional and advanced treatments. Sci. Rep. 2021, 11, 16622. [Google Scholar] [CrossRef]
- Savin, M.; Bierbaum, G.; Kreyenschmidt, J.; Schmithausen, R.M.; Sib, E.; Schmoger, S.; Kasbohrer, A.; Hammerl, J.A. Clinically relevant Escherichia coli isolates from process waters and wastewater of poultry and pig slaughterhouses in Germany. Microorganisms 2021, 9, 698. [Google Scholar] [CrossRef]
- Davis, G.S.; Waits, K.; Nordstrom, L.; Weaver, B.; Aziz, M.; Gauld, L.; Grande, H.; Bigler, R.; Horwinski, J.; Porter, S.; et al. Intermingled Klebsiella pneumoniae populations between retail meats and human urinary tract infections. Clin. Infect. Dis. 2015, 61, 892–899. [Google Scholar] [CrossRef]
- Ludden, C.; Lotsch, F.; Alm, E.; Kumar, N.; Johansson, K.; Albiger, B.; Huang, T.D.; Denis, O.; Hammerum, A.M.; Hasman, H.; et al. Cross-border spread of bla(NDM-1)- and bla(OXA-48)-positive Klebsiella pneumoniae: A European collaborative analysis of whole genome sequencing and epidemiological data, 2014 to 2019. Euro Surveill. 2020, 25, 2000627. [Google Scholar] [CrossRef]
- Kurittu, P.; Khakipoor, B.; Aarnio, M.; Nykasenoja, S.; Brouwer, M.; Myllyniemi, A.L.; Vatunen, E.; Heikinheimo, A. Plasmid-borne and chromosomal ESBL/AmpC genes in Escherichia coli and Klebsiella pneumoniae in global food products. Front. Microbiol. 2021, 12, 592291. [Google Scholar] [CrossRef]
- Damjanova, I.; Toth, A.; Paszti, J.; Hajbel-Vekony, G.; Jakab, M.; Berta, J.; Milch, H.; Fuzi, M. Expansion and countrywide dissemination of ST11, ST15 and ST147 ciprofloxacin-resistant CTX-M-15-type beta-lactamase-producing Klebsiella pneumoniae epidemic clones in Hungary in 2005—The new ‘MRSAs’? J. Antimicrob. Chemother. 2008, 62, 978–985. [Google Scholar] [CrossRef]
- Gondal, A.J.; Saleem, S.; Jahan, S.; Choudhry, N.; Yasmin, N. Novel carbapenem-resistant Klebsiella pneumoniae ST147 coharboring bla(NDM-1), bla(OXA-48) and extended-spectrum beta-lactamases from Pakistan. Infect. Drug Resist. 2020, 13, 2105–2115. [Google Scholar] [CrossRef]
- Suzuki, Y.; Nazareno, P.J.; Nakano, R.; Mondoy, M.; Nakano, A.; Bugayong, M.P.; Bilar, J.; Perez, M.T.; Medina, E.J.; Saito-Obata, M.; et al. Environmental presence and genetic characteristics of carbapenemase-producing Enterobacteriaceae from hospital sewage and river water in the Philippines. Appl. Environ. Microbiol. 2020, 86, e01906-19. [Google Scholar] [CrossRef]
- Peirano, G.; Chen, L.; Kreiswirth, B.N.; Pitout, J.D. Genomic diversity and global epidemiology of carbapenem-resistant K. pneumoniae (CRKp) clonal group 147 (CG147). Abstract 25. In Proceedings of the ASM Microbe, New Orleans, LA, USA, 8 June 2017. [Google Scholar]
- World Health Organization. Global Priority List of Antibiotic Resistant Bacteria to Guide Research, Discovery and Development of New Antibiotics; WHO: Geneva, Switzerland, 2017; Available online: https://remed.org/wp-content/uploads/2017/03/lobal-priority-list-of-antibiotic-resistant-bacteria-2017.pdf (accessed on 28 November 2023).
- Peirano, G.; Chen, L.; Kreiswirth, B.N.; Pitout, J.D.D. Emerging antimicrobial-resistant high-risk Klebsiella pneumoniae clones ST307 and ST147. Antimicrob. Agents Chemother. 2020, 64, e01148-20. [Google Scholar] [CrossRef]
- Ozad Duzgun, A. From Turkey: First report of KPC-3- and CTX-M-27-producing multidrug-resistant Klebsiella pneumoniae ST147 clone carrying OmpK36 and Ompk37 porin mutations. Microb. Drug Resist. 2021, 27, 1265–1270. [Google Scholar] [CrossRef]
- Ovejero, C.M.; Escudero, J.A.; Thomas-Lopez, D.; Hoefer, A.; Moyano, G.; Montero, N.; Martin-Espada, C.; Gonzalez-Zorn, B. Highly tigecycline-resistant Klebsiella pneumoniae sequence type 11 (ST11) and ST147 isolates from companion animals. Antimicrob. Agents Chemother. 2017, 61, e02640-16. [Google Scholar] [CrossRef]
- Surleac, M.; Czobor Barbu, I.; Paraschiv, S.; Popa, L.I.; Gheorghe, I.; Marutescu, L.; Popa, M.; Sarbu, I.; Talapan, D.; Nita, M.; et al. Whole genome sequencing snapshot of multi-drug resistant Klebsiella pneumoniae strains from hospitals and receiving wastewater treatment plants in Southern Romania. PLoS ONE 2020, 15, e0228079. [Google Scholar] [CrossRef]
- Fajardo-Lubian, A.; Ben Zakour, N.L.; Agyekum, A.; Qi, Q.; Iredell, J.R. Host adaptation and convergent evolution increases antibiotic resistance without loss of virulence in a major human pathogen. PLoS Pathog. 2019, 15, e1007218. [Google Scholar] [CrossRef]
- Rocker, A.; Lacey, J.A.; Belousoff, M.J.; Wilksch, J.J.; Strugnell, R.A.; Davies, M.R.; Lithgow, T. Global trends in proteome remodeling of the outer membrane modulate antimicrobial permeability in Klebsiella pneumoniae. mBio 2020, 11, e00603-20. [Google Scholar] [CrossRef]
- Shankar, C.; Kumar, S.; Venkatesan, M.; Veeraraghavan, B. Emergence of ST147 Klebsiella pneumoniae carrying bla(NDM-7) on IncA/C2 with ompK35 and ompK36 mutations in India. J. Infect. Public Health 2019, 12, 741–743. [Google Scholar] [CrossRef]
- Chapman, J.S. Disinfectant resistance mechanisms, cross-resistance, and co-resistance. Int. Biodeterior. Biodegrad. 2003, 51, 271–279. [Google Scholar] [CrossRef]
- Lam, M.M.C.; Wick, R.R.; Wyres, K.L.; Gorrie, C.L.; Judd, L.M.; Jenney, A.W.J.; Brisse, S.; Holt, K.E. Genetic diversity, mobilisation and spread of the yersiniabactin-encoding mobile element ICEKp in Klebsiella pneumoniae populations. Microb. Genom. 2018, 4, e000196. [Google Scholar] [CrossRef]
- Lam, M.M.C.; Wyres, K.L.; Judd, L.M.; Wick, R.R.; Jenney, A.; Brisse, S.; Holt, K.E. Tracking key virulence loci encoding aerobactin and salmochelin siderophore synthesis in Klebsiella pneumoniae. Genome Med. 2018, 10, 77. [Google Scholar] [CrossRef]
- Lan, P.; Jiang, Y.; Zhou, J.; Yu, Y. A global perspective on the convergence of hypervirulence and carbapenem resistance in Klebsiella pneumoniae. J. Glob. Antimicrob. Resist. 2021, 25, 26–34. [Google Scholar] [CrossRef]
- Lan, P.; Yan, R.; Lu, Y.; Zhao, D.; Shi, Q.; Jiang, Y.; Yu, Y.; Zhou, J. Genetic diversity of siderophores and hypermucoviscosity phenotype in Klebsiella pneumoniae. Microb. Pathog. 2021, 158, 105014. [Google Scholar] [CrossRef]
- El Fertas-Aissani, R.; Messai, Y.; Alouache, S.; Bakour, R. Virulence profiles and antibiotic susceptibility patterns of Klebsiella pneumoniae strains isolated from different clinical specimens. Pathol. Biol. 2013, 61, 209–216. [Google Scholar] [CrossRef]
- Kus, H.; Arslan, U.; Turk Dagi, H.; Findik, D. Investigation of various virulence factors of Klebsiella pneumoniae strains isolated from nosocomial infections. Mikrobiyol. Bul. 2017, 51, 329–339. [Google Scholar] [CrossRef]
- Zhu, C.; Liyanapathirana, V.; Li, C.; Pinto, V.; Hui, M.; Lo, N.; Wong, K.T.; Dissanayake, N.; Ip, M. Characterizing mobilized virulence factors and multidrug resistance genes in carbapenemase-producing Klebsiella pneumoniae in a Sri Lankan Hospital. Front. Microbiol. 2018, 9, 2044. [Google Scholar] [CrossRef]
- Cortazzo, V.; Giordano, L.; D’Inzeo, T.; Fiori, B.; Brigante, G.; Luzzaro, F.; Liotti, F.M.; Menchinelli, G.; Sanguinetti, M.; Spanu, T.; et al. EUCAST rapid antimicrobial susceptibility testing of blood cultures positive for Escherichia coli or Klebsiella pneumoniae: Experience of three laboratories in Italy. J. Antimicrob. Chemother. 2021, 76, 1110–1112. [Google Scholar] [CrossRef]
- Hammerl, J.A.; Klein, I.; Lanka, E.; Appel, B.; Hertwig, S. Genetic and functional properties of the self-transmissible Yersinia enterocolitica plasmid pYE854, which mobilizes the virulence plasmid pYV. J. Bacteriol. 2008, 190, 991–1010. [Google Scholar] [CrossRef]
- Irrgang, A.; Tenhagen, B.A.; Pauly, N.; Schmoger, S.; Kaesbohrer, A.; Hammerl, J.A. Characterization of VIM-1-producing E. coli isolated from a German fattening pig farm by an improved isolation procedure. Front. Microbiol. 2019, 10, 2256. [Google Scholar] [CrossRef]
- Hammerl, J.A.; Vom Ort, N.; Barac, A.; Jackel, C.; Grund, L.; Dreyer, S.; Heydel, C.; Kuczka, A.; Peters, H.; Hertwig, S. Analysis of Yersinia pseudotuberculosis isolates recovered from deceased mammals of a German zoo animal collection. J. Clin. Microbiol. 2021, 59, e03125-20. [Google Scholar] [CrossRef]
- Deneke, C.; Brendebach, H.; Uelze, L.; Borowiak, M.; Malorny, B.; Tausch, S.H. Species-specific quality control, assembly and contamination detection in microbial isolate sequences with AQUAMIS. Genes 2021, 12, 644. [Google Scholar] [CrossRef]
- Feldgarden, M.; Brover, V.; Haft, D.H.; Prasad, A.B.; Slotta, D.J.; Tolstoy, I.; Tyson, G.H.; Zhao, S.; Hsu, C.H.; McDermott, P.F.; et al. Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob. Agents Chemother. 2019, 63, e00483-19. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; Garcia-Fernandez, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Moller Aarestrup, F.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Chen, L.; Yang, J.; Yu, J.; Yao, Z.; Sun, L.; Shen, Y.; Jin, Q. VFDB: A reference database for bacterial virulence factors. Nucleic Acids Res. 2005, 33, D325–D328. [Google Scholar] [CrossRef] [PubMed]
- Jolley, K.A.; Maiden, M.C. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinform. 2010, 11, 595. [Google Scholar] [CrossRef]
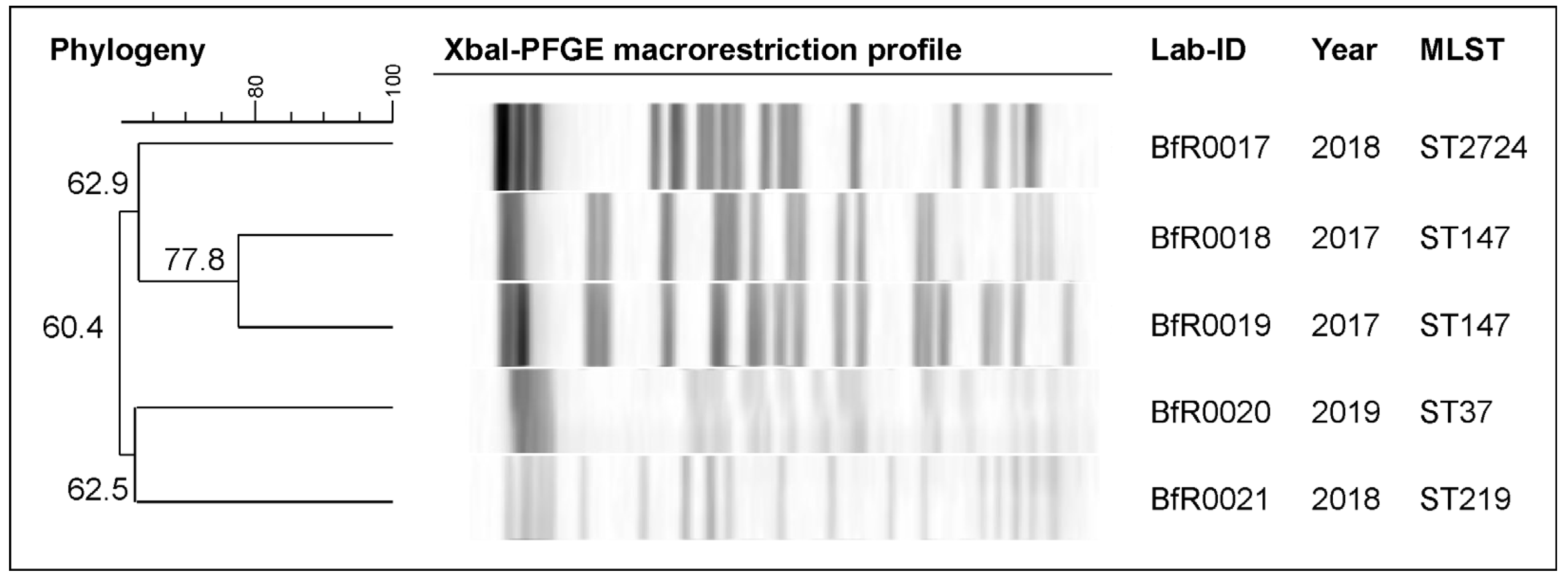
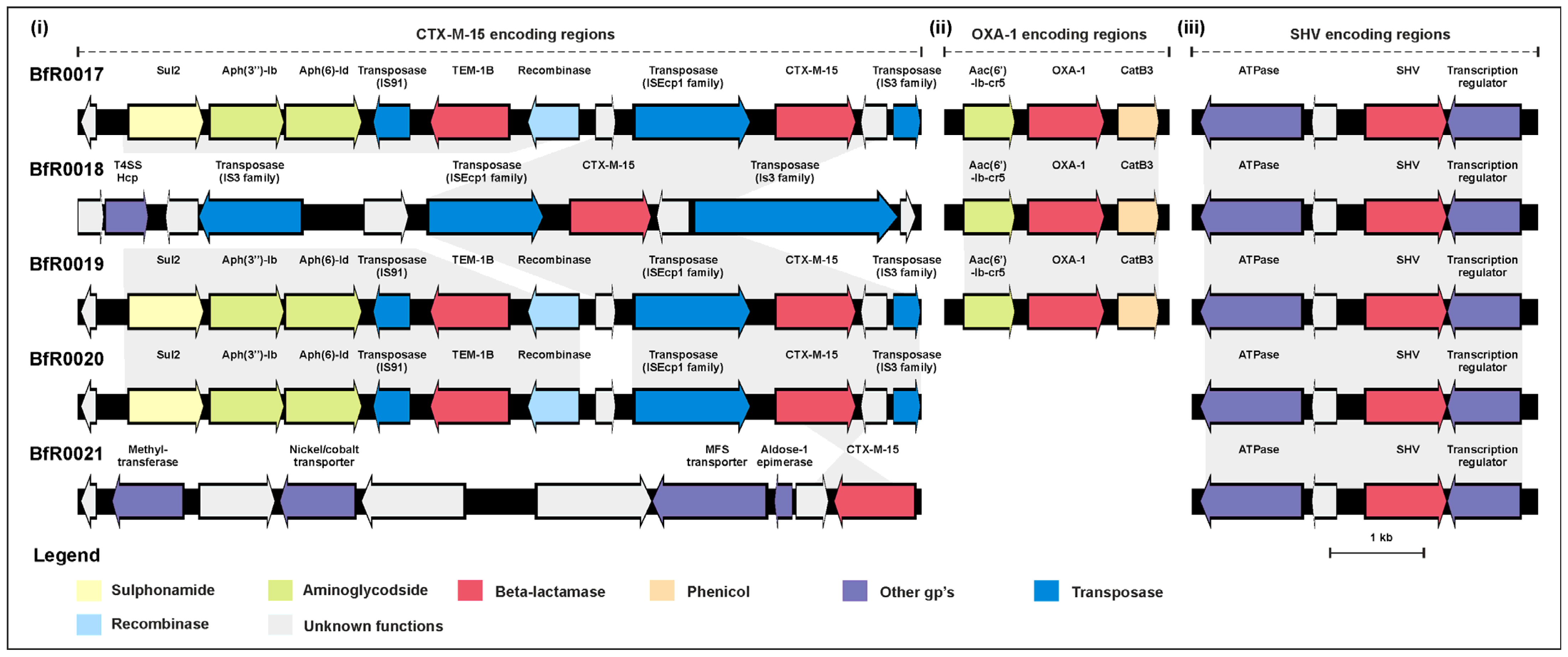
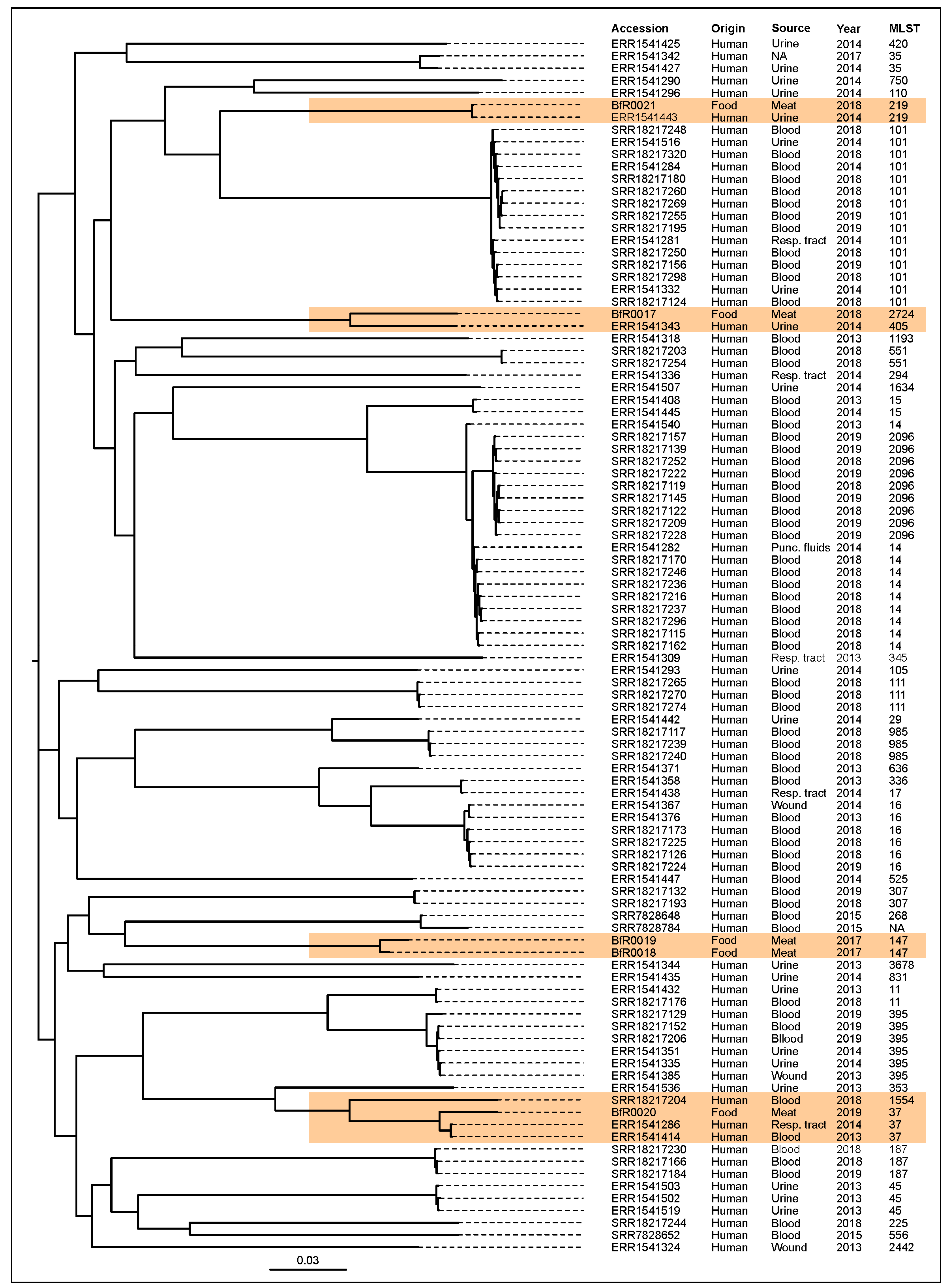
| Lab ID | AMP | TRM | CIP | NAL | GEN | AZI | TET | TGC | COL | CHL | TMP | SMX |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BfR0017 | >64 | 8 | 2 | 16 | 32 | 32 | >64 | 0.5 | ≤1 | ≤8 | >32 | >1024 |
| BfR0018 | >64 | 4 | >8 | >128 | 1 | 16 | >64 | 0.5 | ≤1 | ≤8 | ≤0.25 | ≤8 |
| BfR0019 | >64 | 4 | >8 | >128 | 1 | 16 | >64 | 1 | ≤1 | 16 | >32 | >1024 |
| BfR0020 | >64 | 4 | 0.06 | ≤4 | ≤0.5 | 16 | >64 | 0.5 | ≤1 | ≤8 | >32 | >1024 |
| BfR0021 | >64 | 4 | 1 | ≤4 | ≤0.5 | 64 | >64 | 0.5 | ≤1 | ≤8 | >32 | >1024 |
| Lab ID | FEB | FOT | TAZ | FOX | ETP | IMI | MER | TAX/CLA | TAZ/CLA | |||
| BfR0017 | 8 | >64 | 8 | 4 | 0.03 | 0.25 | 0.06 | ≤0.06 | 0.25 | |||
| BfR0018 | 16 | >64 | 16 | 8 | 0.12 | 0.5 | 0.12 | ≤0.06 | ≤0.12 | |||
| BfR0019 | 16 | >64 | 8 | 4 | 0.03 | 1 | 0.06 | ≤0.06 | ≤0.12 | |||
| BfR0020 | 8 | 64 | 8 | 4 | 0.03 | 0.25 | ≤0.03 | ≤0.06 | ≤0.25 | |||
| BfR0021 | 32 | >64 | 16 | 2 | 0.03 | 0.25 | ≤0.03 | ≤0.06 | 0.5 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kürekci, C.; Ünaldı, Ö.; Şahin, S.; García-Meniño, I.; Hammerl, J.A. Impact and Diversity of ESBL-Producing Klebsiella pneumoniae Recovered from Raw Chicken Meat Samples in Türkiye. Antibiotics 2024, 13, 14. https://doi.org/10.3390/antibiotics13010014
Kürekci C, Ünaldı Ö, Şahin S, García-Meniño I, Hammerl JA. Impact and Diversity of ESBL-Producing Klebsiella pneumoniae Recovered from Raw Chicken Meat Samples in Türkiye. Antibiotics. 2024; 13(1):14. https://doi.org/10.3390/antibiotics13010014
Chicago/Turabian StyleKürekci, Cemil, Özlem Ünaldı, Seyda Şahin, Isidro García-Meniño, and Jens Andre Hammerl. 2024. "Impact and Diversity of ESBL-Producing Klebsiella pneumoniae Recovered from Raw Chicken Meat Samples in Türkiye" Antibiotics 13, no. 1: 14. https://doi.org/10.3390/antibiotics13010014
APA StyleKürekci, C., Ünaldı, Ö., Şahin, S., García-Meniño, I., & Hammerl, J. A. (2024). Impact and Diversity of ESBL-Producing Klebsiella pneumoniae Recovered from Raw Chicken Meat Samples in Türkiye. Antibiotics, 13(1), 14. https://doi.org/10.3390/antibiotics13010014






