Exploring the Multifaceted Potential of a Peptide Fraction Derived from Saccharomyces cerevisiae Metabolism: Antimicrobial, Antioxidant, Antidiabetic, and Anti-Inflammatory Properties
Abstract
1. Introduction
2. Results
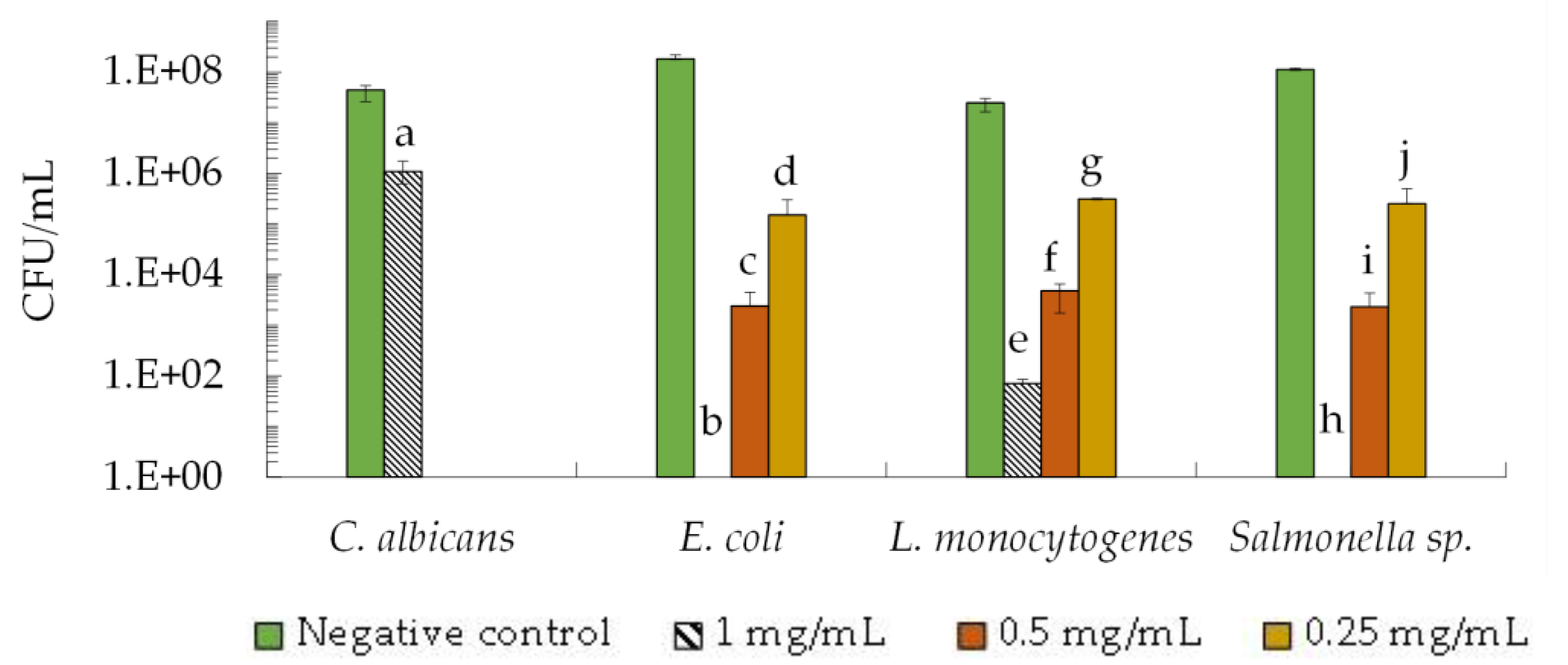
2.1. Antimicrobial Activity of the 2–10 kDa Fraction against FOODBORNE Pathogens
2.1.1. Minimal Inhibitory Concentration (MIC)
2.1.2. Fungicidal and Bactericidal Activity
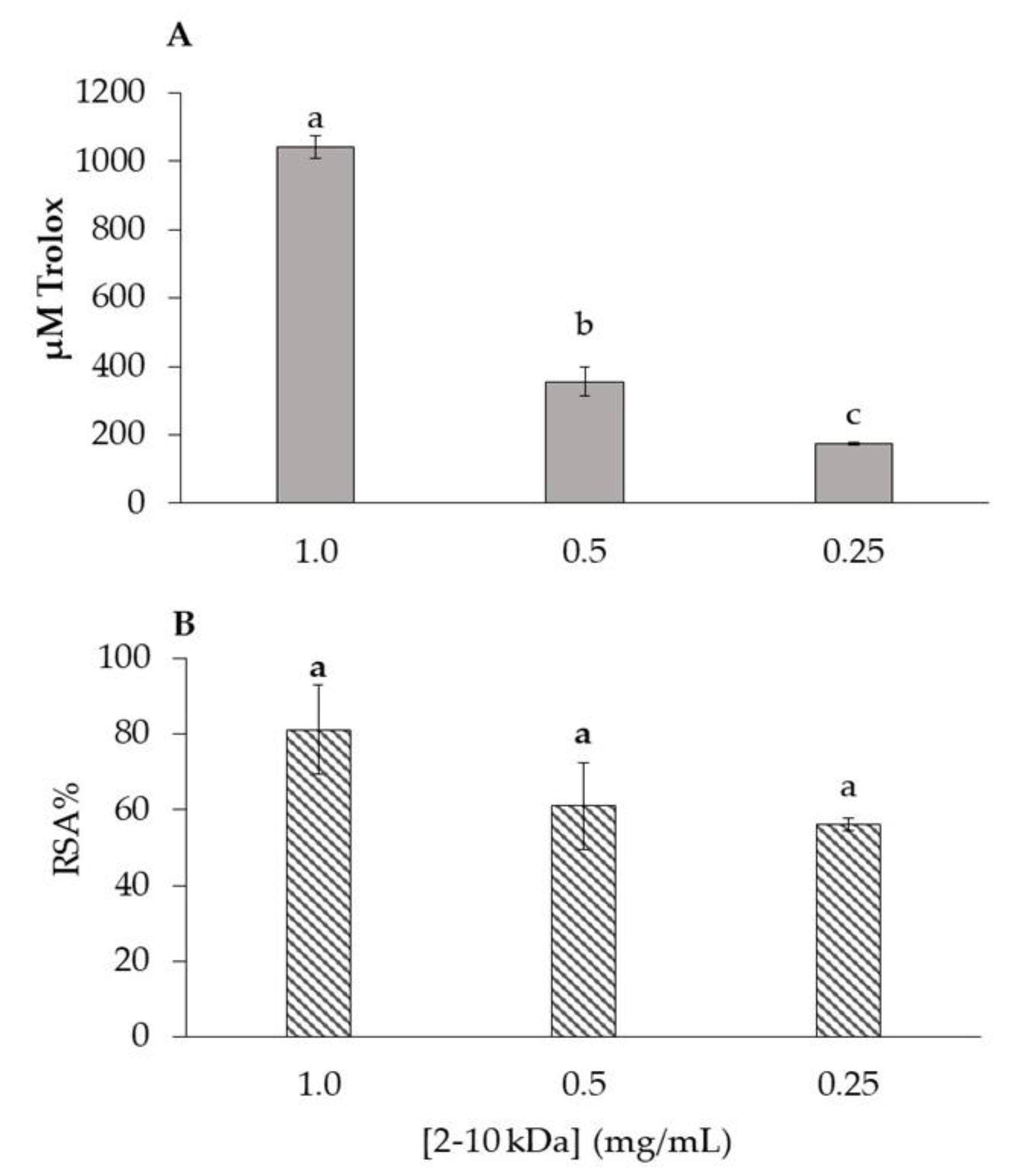
2.2. Antioxidant Activity of 2–10 kDa Fraction
2.3. Cytotoxicity of 2–10 kDa Fraction
2.4. Analysis of Anti-Inflammatory Effect of Peptide Fraction 2–10 kDa
2.5. Antidiabetic Effect of Peptide Fraction 2–10 kDa
3. Discussion
4. Materials and Methods
4.1. Production of the 2–10 kDa Fraction
4.2. Antimicrobial-Assays
4.2.1. Microorganisms and Media
4.2.2. Determination of Minimum Inhibitory Concentrations
4.2.3. Evaluation of Fungicidal and Bactericidal Activity
4.3. Antioxidant Activity
4.3.1. Ferric Reducing Antioxidant Power (FRAP)
4.3.2. DPPH Radical Scavenging Assay (2,2-Diphenyl-1-picrylhydrazyl)
4.4. Cytotoxicity Assay
4.5. Inflammatory Effect
4.6. α-Amylase Inhibitory Assay
4.7. α-Glucosidase Inhibitory Assay
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bhunia, A.K. Foodborne Microbial Pathogens: Mechanisms and Pathogenesis, 2nd ed.; Springer: New York, NY, USA, 2018; pp. 1–23. [Google Scholar]
- Brenier-Pinchart, M.P.; Faure, O.; Garban, F.; Fricker-Hidalgo, H.; Mallaret, M.R.; Trens, A.; Lebeau, B.; Pelloux, H.; Grillot, R. Ten-year surveillance of fungal contamination of food within a protected haematological unit. Mycoses 2006, 49, 421–425. [Google Scholar] [CrossRef] [PubMed]
- Pitt, I.J.; Hocking, A.D. Fungi and Food Spoilage, 3rd ed.; Springer: New York, NY, USA, 2009; p. 519. [Google Scholar]
- Hussain, M.A.; Dawson, C.O. Economic Impact of Food Safety Outbreaks on Food Businesses. Foods 2013, 2, 585–589. [Google Scholar] [CrossRef] [PubMed]
- Rasko, D.A.; Webster, D.R.; Sahl, J.W.; Bashir, A.; Boisen, N.; Scheutz, F.; Nataro, J.P. Origins of the E. coli strain causing an outbreak of hemolytic–uremic syndrome in Germany. N. Engl. J. Med. 2011, 365, 709–717. [Google Scholar] [CrossRef]
- Majowicz, S.E.; Scallan, E.; Jones-Bitton, A.; Sargeant, J.M.; Stapleton, J.; Angulo, F.J.; O’Brien, S.J. Global incidence of human Shiga toxin–producing Escherichia coli infections and deaths: A systematic review and knowledge synthesis. Foodborne Pathog. Dis. 2014, 11, 447–455. [Google Scholar] [CrossRef]
- Farber, J.M.; Peterkin, P.I. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 1991, 55, 476–511. [Google Scholar] [CrossRef]
- Todd, E.C. Epidemiology of foodborne diseases: A worldwide review. World Health Stat. Q. 1997, 50, 30–50. [Google Scholar] [PubMed]
- Munck, N.; Leekitcharoenphon, P.; Litrup, E.; Kaas, R.; Meinen, A.; Guillier, L.; Tang, Y.; Malorny, B.; Palma, F.; Borowiak, M.; et al. Four European Salmonella typhimurium datasets collected to develop WGS-based source attribution methods. Sci. Data 2020, 7, 75. [Google Scholar] [CrossRef]
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne illness acquired in the United States—Major pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Q.; Liu, X.; Liu, Y.; Wang, Q.; Zhang, Z. Occurrence and characteristics of Candida spp. in vegetables and fresh-cut fruits. J. Food Saf. 2020, 40, e12717. [Google Scholar] [CrossRef]
- Diba, K.; Ehsani, A.; Khodadadi, E. Prevalence of Candida albicans in raw meat products in Tehran. Iran. J. Food Saf. 2020, 40, e12760. [Google Scholar]
- Khodavaisy, S.; Nabili, M.; Davari, B.; Vahedi, M.; Aslani, N. The prevalence of Candida species in vegetables and characterization of isolates by ITS-RFLP analysis. Curr. Med. Mycol. 2017, 3, 17–22. [Google Scholar] [CrossRef]
- Deak, T.; Beuchat, L.R. Handbook of Food Spoilage Yeasts; CRS Press: New York, NY, USA, 1996; pp. 111–154. [Google Scholar]
- Casey, G.D.; Dobson, A.D. Molecular detection of Candida krusei contamination in fruit juice using the citrate synthase gene cs1 and a potential role for this gene in the adaptive response to acetic acid. J. Appl. Microbiol. 2003, 95, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Pelletier, R.; Alarie, I.; Lagacé, R.; Walsh, T.J. Emergence of disseminated candidiasis caused by Candida krusei during treatment with caspofungin: Case report and review of literature. Med. Mycol. J. 2005, 43, 559–564. [Google Scholar] [CrossRef][Green Version]
- Freedman, B.J. Sulphur dioxide in foods and beverages: Its use as a preservative and its effect on asthma. Br. J. Dis. Chest 1980, 74, 128–134. [Google Scholar] [CrossRef]
- Piper, J.D.; Piper, P.W. Benzoate and Sorbate Salts: A Systematic Review of the Potential Hazards of These Invaluable Preservatives and the Expanding Spectrum of Clinical Uses for Sodium. Compr. Rev. Food Sci. Food Saf. 2017, 16, 868–880. [Google Scholar] [CrossRef]
- Vally, H.; Misso, N.L.; Madan, V. Clinical effects of sulphite additives. Clinical and experimental allergy. Clin. Exp. Allergy 2009, 39, 1643–1651. [Google Scholar] [CrossRef]
- Asioli, D.; Aschemann-Witzel, J.; Caputo, V.; Vecchio, R.; Annunziata, A.; Næs, T.; Varela, P. Making sense of the “clean label” trends: A review of consumer food choice behavior and discussion of industry implications. Food Res. Int. 2017, 99 Pt 1, 58–71. [Google Scholar] [CrossRef]
- Hancock, R.E.; Sahl, H.G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 2006, 24, 1551–1557. [Google Scholar] [CrossRef]
- Cotter, P.D.; Hill, C.; Ross, R.P. Bacteriocins: Developing innate immunity for food. Nat. Rev. Microbiol. 2005, 3, 777–788. [Google Scholar] [CrossRef]
- Nascimento, B.L.; Delabeneta, M.F.; Rosseto, L.R.B.; Junges, D.S.B.; Paris, A.P.; Persel, C.; Gandra, R.F. Yeast Mycocins: A great potential for application in health. FEMS Yeast Res. 2020, 20, foaa016. [Google Scholar] [CrossRef]
- Albergaria, H.; Francisco, D.; Gori, K.; Arneborg, N.; Gírio, F. Saccharomyces cerevisiae CCMI 885 secretes peptides that inhibit the growth of some non-Saccharomyces wine-related strains. Appl. Microbiol. Biotechnol. 2010, 86, 965–972. [Google Scholar] [CrossRef] [PubMed]
- Branco, P.; Francisco, D.; Chambon, C.; Hébraud, M.; Arneborg, N.; Almeida, M.G.; Caldeira, J.; Albergaria, H. Identification of novel GAPDH-derived antimicrobial peptides secreted by Saccharomyces cerevisiae and involved in wine microbial interactions. Appl. Microbiol. Biotechnol. 2014, 98, 843–853. [Google Scholar] [CrossRef] [PubMed]
- Branco, P.; Francisco, D.; Monteiro, M.; Almeida, M.G.; Caldeira, J.; Arneborg, N.; Prista, C.; Albergaria, H. Antimicrobial properties and death-inducing mechanisms of Saccharomycin, a biocide secreted by Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2017, 101, 159–171. [Google Scholar] [CrossRef]
- Branco, P.; Coutinho, R.; Malfeito-Ferreira, M.; Prista, C.; Albergaria, H. Wine spoilage control: Impact of Saccharomycin on Brettanomyces bruxellensis and its conjugated effect with sulfur dioxide. Microorganisms 2021, 9, 2528. [Google Scholar] [CrossRef]
- Al-Sahlany, S.T.G.; Altemimi, A.B.; Al-Manhel, A.J.A.; Niamah, A.K.; Lakhssassi, N.; Ibrahim, S.A. Purification of Bioactive Peptide with Antimicrobial Properties Produced by Saccharomyces cerevisiae. Foods 2020, 9, 324. [Google Scholar] [CrossRef]
- Akbarian, M.; Khani, A.; Eghbalpour, S.; Uversky, V.N. Bioactive Peptides: Synthesis, Sources, Applications, and Proposed Mechanisms of Action. Int. J. Mol. Sci. 2022, 23, 1445. [Google Scholar] [CrossRef]
- Mudgil, P.; Kamal, H.; Yuen, G.C.; Maqsood, S. Characterization and identification of novel antidiabetic and anti-obesity peptides from camel milk protein hydrolysates. Food Chem. 2018, 259, 46–54. [Google Scholar] [CrossRef]
- Sun, J.; He, H.; Xie, B.J. Novel Antioxidant Peptides from Fermented Mushroom Ganoderma lucidum. J. Agric. Food Chem. 2004, 52, 6646–6652. [Google Scholar] [CrossRef] [PubMed]
- Heleno, S.A.; Barros, L.; Martins, A.; Queiroz, M.J.R.P.; Santos-Buelga, C.; Ferreira, I.C.F.R. Fruiting body, spores and in vitro produced mycelium of Ganoderma lucidum from Northeast Portugal: A comparative study of the antioxidant potential of phenolic and polysaccharidic extracts. Food Res. Int. 2012, 46, 135–140. [Google Scholar] [CrossRef]
- Mishra, J.; Rajput, R.; Singh, K. Antibacterial Natural Peptide Fractions from Indian Ganoderma lucidum. Int. J. Pept. Res. Ther. 2018, 24, 543–554. [Google Scholar] [CrossRef]
- Kelebek, H.; Selli, S.; Canbas, A.; Cabaroglu, T. HPLC determination of organic acids, sugars, phenolic compositions and antioxidant capacity of orange juice and orange wine made from a Turkish cv. Kozan. Microchem. J. 2009, 91, 187–192. [Google Scholar] [CrossRef]
- Stinco, C.M.; Fernández-Vázquez, R.; Hernanz, D.; Heredia, F.J.; Meléndez-Martínez, A.J.; Vicario, I.M. Industrial orange juice debittering: Impact on bioactive compounds and nutritional value. J. Food Eng. 2013, 116, 155–161. [Google Scholar] [CrossRef]
- Muchagato Maurício, E.; Rosado, C.; Duarte, M.P.; Fernando, A.L.; Díaz-Lanza, A.M. Evaluation of industrial sour cherry liquor wastes as an ecofriendly source of added value chemical compounds and energy. Waste Biomass Valorization 2020, 11, 201–210. [Google Scholar] [CrossRef]
- Correia, R.; Duarte, M.P.; Maurício, E.M.; Brinco, J.; Quintela, J.C.; da Silva, M.G.; Gonçalves, M. Chemical and Functional Characterization of Extracts from Leaves and Twigs of Acacia dealbata. Processes 2022, 10, 2429. [Google Scholar] [CrossRef]
- Guo, Q.; Chen, P.; Chen, X. Bioactive Peptides Derived from Fermented Foods: Preparation and Biological Activities. J. Funct. Foods 2023, 101, 105422. [Google Scholar] [CrossRef]
- Oliveira, A.S.; Pereira, J.O.; Ferreira, C.; Faustino, M.; Durão, J.; Pintado, M.E.; Carvalho, A.P. Peptide-rich extracts from spent yeast waste streams as a source of bioactive compounds for the nutraceutical market. Innov. Food Sci. Emerg. Technol. 2022, 81, 103148. [Google Scholar] [CrossRef]
- Lopes, A.; Azevedo-Silva, J.; Carsanba, E.; Pintado, M.; Oliveira, A.S.; Ferreira, C.; Oliveira, C. Peptide extract from spent yeast improves resistance of Saccharomyces cerevisiae to oxidative stress. Appl. Microbiol. Biotechnol. 2023, 107, 3405–3417. [Google Scholar] [CrossRef]
- Diplock, A.T. Antioxidants and disease prevention. Mol. Aspects Med. 1994, 15, 293–376. [Google Scholar] [CrossRef]
- Halliwell, B. Free radicals and antioxidants: Updating a personal view. Nutr. Rev. 2012, 70, 257–265. [Google Scholar] [CrossRef]
- Morén, C.; deSouza, R.M.; Giraldo, D.M.; Uff, C. Antioxidant Therapeutic Strategies in Neurodegenerative Diseases. Int. J. Mol. Sci. 2022, 23, 9328. [Google Scholar] [CrossRef] [PubMed]
- Duarte, M.C.T.; Leme, E.E.; Delarmelina, C.; Soares, A.A.; Figueira, G.M.; Sartoratto, A. Activity of essential oils from Brazilian medicinal plants on Escherichia coli. J. Ethnopharmacol. 2007, 111, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Vieitez, I.; Maceiras, L.; Jachmanián, I.; Alborés, S. Antioxidant and antibacterial activity of different extracts from herbs obtained by maceration or supercritical technology. J. Supercrit. Fluids 2018, 133, 58–64. [Google Scholar] [CrossRef]
- Pereira, P.; Mauricio, E.M.; Duarte, M.P.; Lima, K.; Fernandes, A.S.; Bernardo-Gil, G.; Cebola, M.J. Potential of supercritical fluid myrtle extracts as an active ingredient and co-preservative for cosmetic and topical pharmaceutical applications. Sustain. Chem. Pharm. 2022, 28, 100739. [Google Scholar] [CrossRef]
- Sharma, S. Food Preservatives and their harmful effects. Int. J. Sci. Res. Publ. 2015, 5, 1–2. [Google Scholar]
- EFSA Panel on Additives and Products or Substances used in Animal Feed. Scientific opinion on the safety and efficacy of nisin (E 234) as a feed additive for all animal species. EFSA J. 2017, 15, 4749. [Google Scholar] [CrossRef]
- Bodley, M.D. Application of Bacteriocins in the Preservation of Fruit Juice. Ph.D. Thesis, Faculty of Science at the Nelson Mandela Metropolitan University, Gqeberha, South Africa, January 2015. [Google Scholar]
- Gharsallaoui, A.; Oulahal, N.; Joly, C.; Degraeve, P. Nisin as a Food Preservative: Part 1: Physicochemical Properties, Antimicrobial Activity, and Main Uses. Crit. Rev. Food Sci. Nutr. 2016, 56, 1262–1274. [Google Scholar] [CrossRef] [PubMed]
- Campion, A.; Casey, P.G.; Field, D.; Cotter, P.D.; Hill, C.; Ross, R.P. In vivo activity of nisin A and nisin V against Listeria monocytogenes in mice. BMC Microbiol. 2013, 13, 23. [Google Scholar] [CrossRef]
- Madrazo, A.L.; Segura Campos, M.R. Review of antimicrobial peptides as promoters of food safety: Limitations and possibilities within the food industry. J. Food Saf. 2020, 40, e12854. [Google Scholar] [CrossRef]
- Maurício, E.; Rosado, C.; Duarte, M.P.; Verissimo, J.; Bom, S.; Vasconcelos, L. Efficiency of Nisin as Preservative in Cosmetics and Topical Products. Cosmetics 2017, 4, 41. [Google Scholar] [CrossRef]
- Delves-Broughton, J.P.; Blackburn, P.; Evans, R.J.; Hugenholtz, J. Application of the bacteriocin, nisin. Anton Leeuw 1996, 69, 193–202. [Google Scholar] [CrossRef]
- Golubev, W.I. Mycocins (killer toxins). In The Yeasts, 4th ed.; Kurtzman, C.P., Fell, J.W., Eds.; Elsevier: Amsterdam, The Netherlands, 1998; pp. 55–62. [Google Scholar]
- Izgü, F.; Altinbay, D.; Türeli, A.E. In vitro susceptibilities of Candida spp. to Panomycocin, a novel exo-beta-1,3-glucanase isolated from Pichia anomala NCYC 434. Microbiol. Immunol. 2017, 51, 797–803. [Google Scholar] [CrossRef] [PubMed]
- Al-Qaysi, S.; Al-Haideri, H.; Thabit, Z.A.; Al-Kubaisy, W.; Ibrahim, J. Production, Characterization, and Antimicrobial Activity of Mycocin Produced by Debaryomyces hansenii DSMZ70238. Int. J. Microbiol. 2017, 2017, 2605382. [Google Scholar] [CrossRef] [PubMed]
- Comitini, F.; Pietro, N.D.; Zacchi, L.; Mannazzu, I.; Ciani, M. Kluyveromyces phaffii killer toxin active against wine spoilage yeasts: Purification and characterization. Microbiology 2004, 150, 2535–2541. [Google Scholar] [CrossRef]
- Ciani, M.; Fatichenti, F. Killer toxin of Kluyveromyces phaffii DBVPG 6076 as a biopreservative agent to control apiculate wine Yeasts. Appl. Environ. Microbiol. 2001, 67, 3058–3063. [Google Scholar] [CrossRef]
- De Ingeniis, J.; Raffaelli, N.; Ciani, M.; Mannazzu, I. Pichia anomala DBVPG 3003 secretes a ubiquitin-like protein that has antimicrobial activity. Appl. Environ. Microbiol. 2009, 75, 1129–1134. [Google Scholar] [CrossRef]
- Mirzaei, M.; Mirdamadi, S.; Ehsani, M.R.; Aminlari, M.; Hosseini, E. Purification and Identification of Antioxidant and ACE-Inhibitory Peptide from Saccharomyces cerevisiae Protein Hydrolysate. J. Funct. Foods 2015, 19, 259–268. [Google Scholar] [CrossRef]
- Pereira, P.R.; Freitas, C.S.; Paschoalin, V.M.F. Saccharomyces cerevisiae Biomass as a Source of Next-generation Food Preservatives: Evaluating Potential Proteins as a Source of Antimicrobial Peptides. Compr. Rev. Food Sci. Food Saf. 2021, 20, 4450–4479. [Google Scholar] [CrossRef]
- Pereira, P.; Bernardo-Gil, M.G.; Cebola, M.J.; Mauricio, E.; Romano, A. Supercritical fluid extracts with antioxidant and antimicrobial activities from myrtle (Myrtus communis L.) leaves. Response surface optimization. J. Supercrit. Fluids 2013, 83, 57–64. [Google Scholar] [CrossRef]
- Loebler, M.; Sánchez, C.; Muchagato Maurício, E.; Diogo, E.; Santos, M.; Vasilenko, P.; Duarte, M.P. Potential application of propolis extracts to control the growth of Stemphylium vesicarium in “Rocha” pear. Appl. Sci. 2020, 10, 1990. [Google Scholar] [CrossRef]
- Zou, T.B.; He, T.P.; Li, H.B.; Tang, H.W.; Xia, E.Q. The Structure-Activity Relationship of the Antioxidant Peptides from Natural Proteins. Molecules 2016, 21, 72. [Google Scholar] [CrossRef]
- Hur, S.J.; Lee, S.Y.; Kim, Y.C.; Choi, I.; Kim, G.B. Effect of Fermentation on the Antioxidant Activity in Plant-Based Foods. Food Chem. 2014, 160, 346–356. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, M.; Shavandi, A.; Mirdamadi, S.; Soleymanzadeh, N.; Motahari, P.; Mirdamadi, N.; Moser, M.; Subra, G.; Alimoradi, H.; Goriely, S. Bioactive Peptides from Yeast: A Comparative Review on Production Methods, Bioactivity, Structure-Function Relationship, and Stability. Trends Food Sci. Technol. 2021, 118, 297–315. [Google Scholar] [CrossRef]
- Brand-Williams, W.; Cuvelier, M.E.; Berset, C. Use of a Free Radical Method to Evaluate Antioxidant Activity. LWT Food Sci. Technol. 1995, 28, 25–30. [Google Scholar] [CrossRef]
- Benzie, I.F.F.; Strain, J.J. The Ferric Reducing Ability of Plasma (FRAP) as a Measure of “Antioxidant Power”: The FRAP Assay. Anal. Biochem. 1996, 239, 70–76. [Google Scholar] [CrossRef]
- Maher, S.; McClean, S. Investigation of the cytotoxicity of eukaryotic and prokaryotic antimicrobial peptides in intestinal epithelial cells in vitro. Biochem. Pharmacol. 2006, 71, 1289–1298. [Google Scholar] [CrossRef]
- Antony, P.; Vijayan, R. Bioactive peptides as potential nutraceuticals for diabetes therapy: A comprehensive review. Int. J. Mol. Sci. 2021, 22, 9059. [Google Scholar] [CrossRef] [PubMed]
- Rivero-Pino, F.; Espejo-Carpio, F.J.; Guadix, E.M. Antidiabetic food-derived peptides for functional feeding: Production, functionality and in vivo evidences. Foods 2020, 9, 983. [Google Scholar] [CrossRef] [PubMed]
- Romeiras, M.M.; Essoh, A.P.; Catarino, S.; Silva, J.; Lima, K.; Varela, E.; Moura, M.; Gomes, I.; Duarte, M.C.; Duarte, M.P. Diversity and biological activities of medicinal plants of Santiago island (Cabo Verde). Heliyon 2023, 9, e14651. [Google Scholar] [CrossRef]
- Zhou, H.; Safdar, B.; Li, H.; Yang, L.; Ying, Z.; Liu, X. Identification of a novel α-amylase inhibitory activity peptide from quinoa protein hydrolysate. Food Chem. 2023, 403, 134434. [Google Scholar] [CrossRef]
- González-Montoya, M.; Hernández-Ledesma, B.; Mora-Escobedo, R.; Martínez-Villaluenga, C. Bioactive peptides from germinated soybean with anti-diabetic potential by inhibition of dipeptidyl peptidase-IV, α-amylase, and α-glucosidase enzymes. Int. J. Mol. Sci. 2018, 19, 2883. [Google Scholar] [CrossRef]
- Wang, R.; Zhao, H.; Pan, X.; Orfila, C.; Lu, W.; Ma, Y. Preparation of bioactive peptides with antidiabetic, antihypertensive, and antioxidant activities and identification of α-glucosidase inhibitory peptides from soy protein. Food Sci. Nutr. 2019, 7, 1848–1856. [Google Scholar] [CrossRef] [PubMed]
- CLSI Document M07-A9; Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically. Approved Standard—Ninth Edition. Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012.
- Fernandes, A.R.; Mendonça-Martins, I.; Santos, M.F.A.; Raposo, L.R.; Mendes, R.; Marques, J.; Romão, C.C.; Romão, M.J.; Santos-Silva, T.; Baptista, P.V. Improving the Anti-inflammatory Response via Gold Nanoparticle Vectorization of CO-Releasing Molecules. ACS Biomater. Sci. Eng. 2020, 6, 1090–1101. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 4. [Google Scholar] [CrossRef] [PubMed]


| MIC (mg/mL) | ||||
|---|---|---|---|---|
| Tested Microorganisms | 2–10 kDa Fraction | Chloramphenicol a | Ketoconazole a | Ethanol b |
| C. albicans | 1.0 | - | 0.012 | n.d |
| C. krusei | >1.0 | - | 0.012 | n.d |
| E. coli | 0.25 | 0.012 | - | n.d |
| L. monocytogenes | 0.25 | 0.050 | - | n.d |
| Salmonella sp. | 0.25 | 0.025 | - | n.d |
| Cell Line | HCT116 | coN |
|---|---|---|
| IC50 (mg/mL) | 0.44 ± 0.10 | 0.42 ± 0.08 |
| Peptide Fraction | Acarbose | |
|---|---|---|
| α-Amylase (IC50 (µg/mL)) | 199.3 ± 0.9 | 11.6 ± 1.0 |
| α-Glucosidase (IC20 (µg/mL)) | 270.6 ± 6.0 | 134.1 ± 4.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Branco, P.; Maurício, E.M.; Costa, A.; Ventura, D.; Roma-Rodrigues, C.; Duarte, M.P.; Fernandes, A.R.; Prista, C. Exploring the Multifaceted Potential of a Peptide Fraction Derived from Saccharomyces cerevisiae Metabolism: Antimicrobial, Antioxidant, Antidiabetic, and Anti-Inflammatory Properties. Antibiotics 2023, 12, 1332. https://doi.org/10.3390/antibiotics12081332
Branco P, Maurício EM, Costa A, Ventura D, Roma-Rodrigues C, Duarte MP, Fernandes AR, Prista C. Exploring the Multifaceted Potential of a Peptide Fraction Derived from Saccharomyces cerevisiae Metabolism: Antimicrobial, Antioxidant, Antidiabetic, and Anti-Inflammatory Properties. Antibiotics. 2023; 12(8):1332. https://doi.org/10.3390/antibiotics12081332
Chicago/Turabian StyleBranco, Patrícia, Elisabete Muchagato Maurício, Ana Costa, Diogo Ventura, Catarina Roma-Rodrigues, Maria Paula Duarte, Alexandra R. Fernandes, and Catarina Prista. 2023. "Exploring the Multifaceted Potential of a Peptide Fraction Derived from Saccharomyces cerevisiae Metabolism: Antimicrobial, Antioxidant, Antidiabetic, and Anti-Inflammatory Properties" Antibiotics 12, no. 8: 1332. https://doi.org/10.3390/antibiotics12081332
APA StyleBranco, P., Maurício, E. M., Costa, A., Ventura, D., Roma-Rodrigues, C., Duarte, M. P., Fernandes, A. R., & Prista, C. (2023). Exploring the Multifaceted Potential of a Peptide Fraction Derived from Saccharomyces cerevisiae Metabolism: Antimicrobial, Antioxidant, Antidiabetic, and Anti-Inflammatory Properties. Antibiotics, 12(8), 1332. https://doi.org/10.3390/antibiotics12081332









