Untargeted MS-Based Metabolomic Analysis of Termite Gut-Associated Streptomycetes with Antifungal Activity against Pyrrhoderma noxium
Abstract
1. Introduction
2. Results and Discussion
2.1. Phylogeny of Termite Gut-Associated Actinomycetes Based on the 16S rRNA Oligonucleotide Analysis
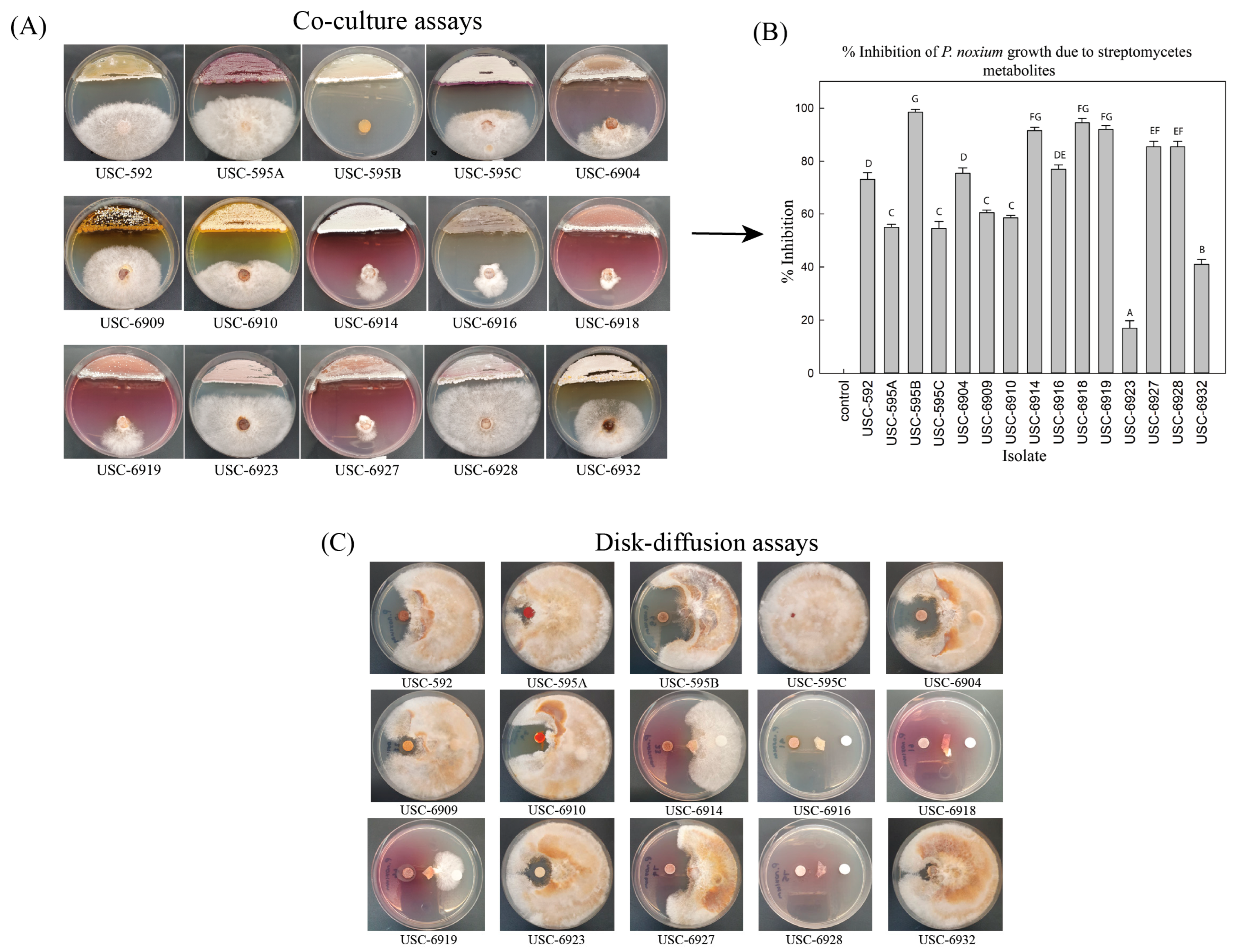
2.2. Screening for Antifungal Activity from Termite Gut-Associated Actinomycete Isolates against P. noxium
2.3. Antifungal Activity from Streptomycetes Fermentation Extracts against P. noxium
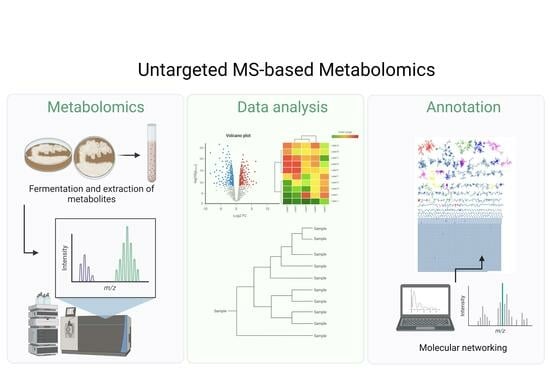
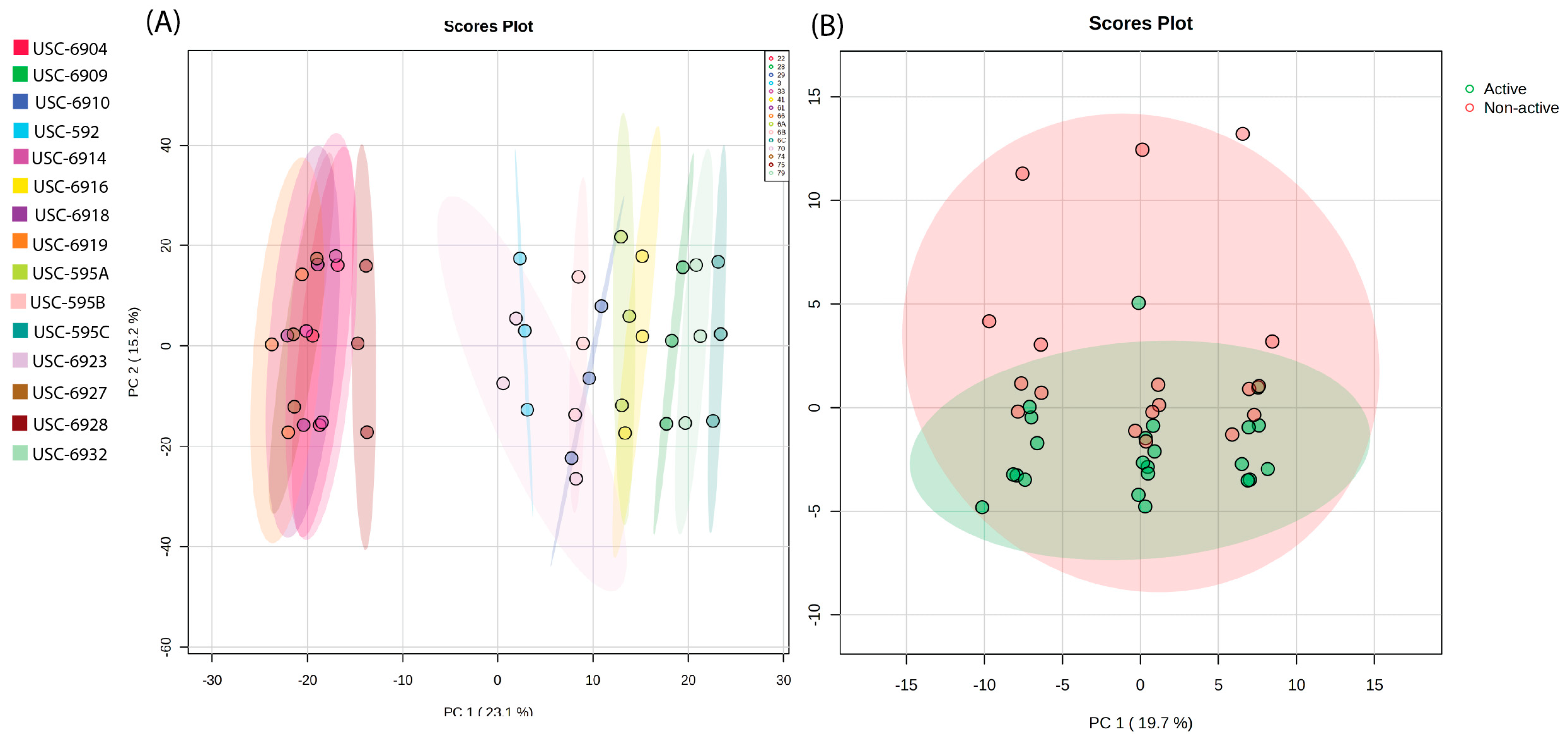
2.4. Metabolite Diversity Analysis of Streptomycetes Fermentation Extracts Using Untargeted MS-Based Metabolomic Approach
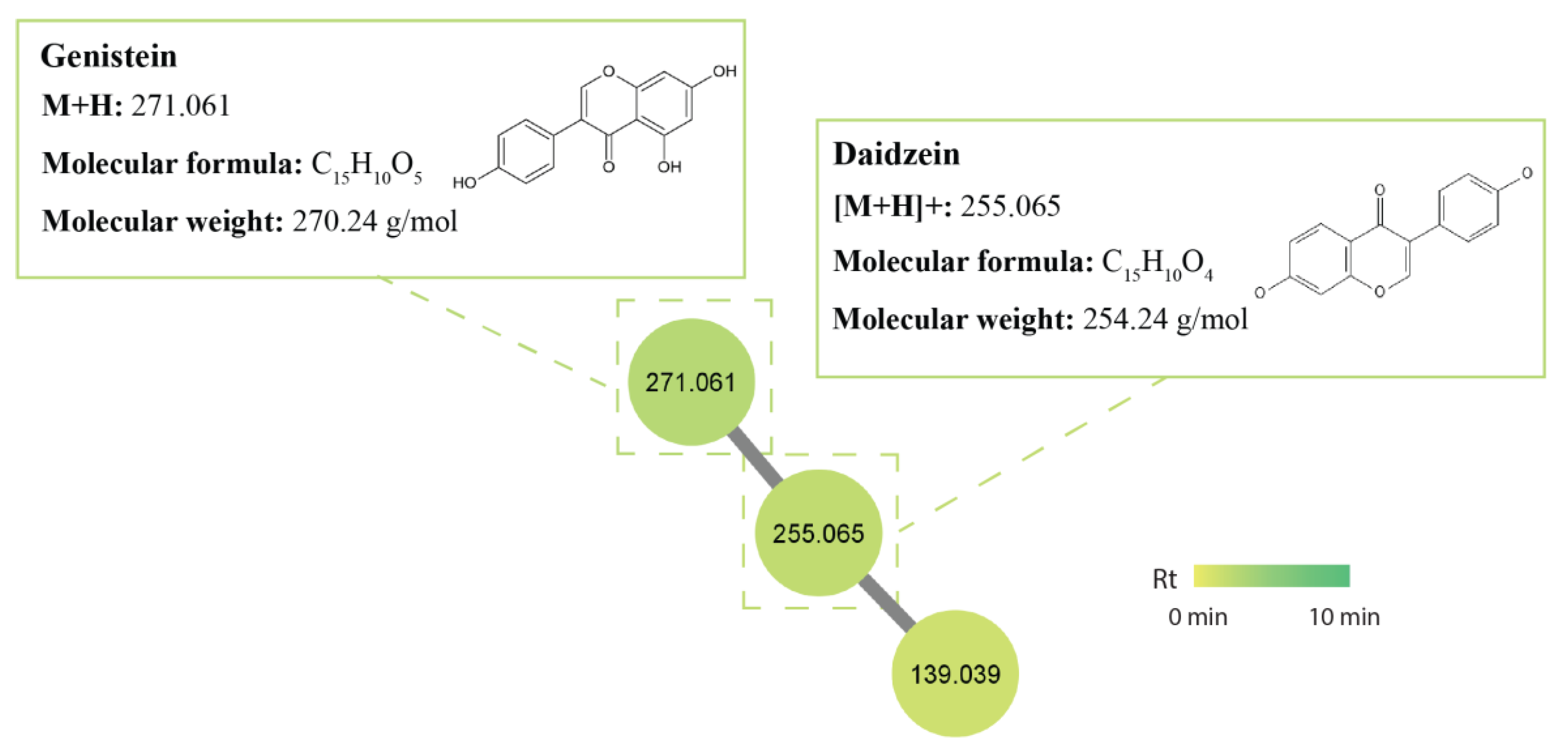
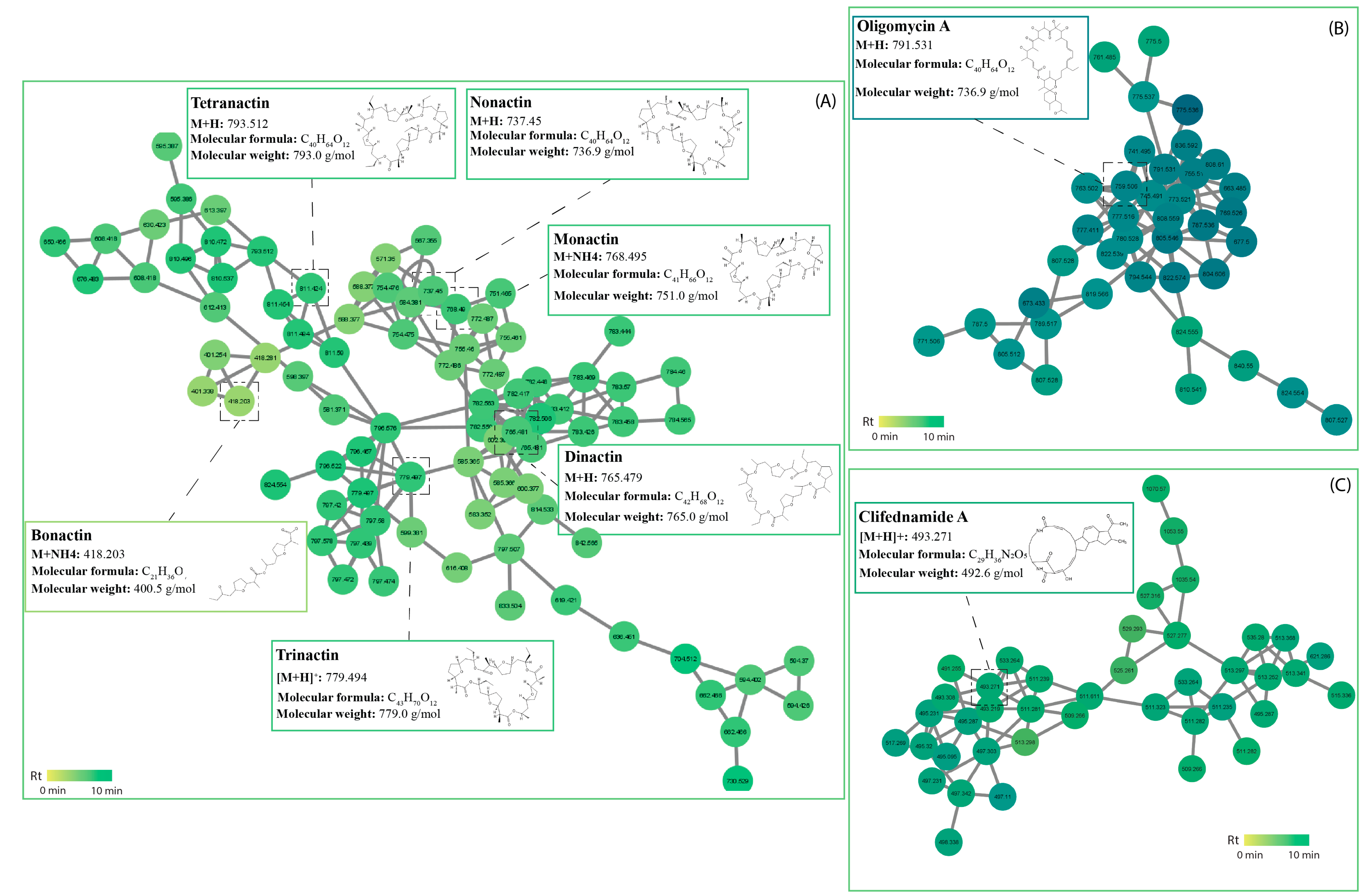
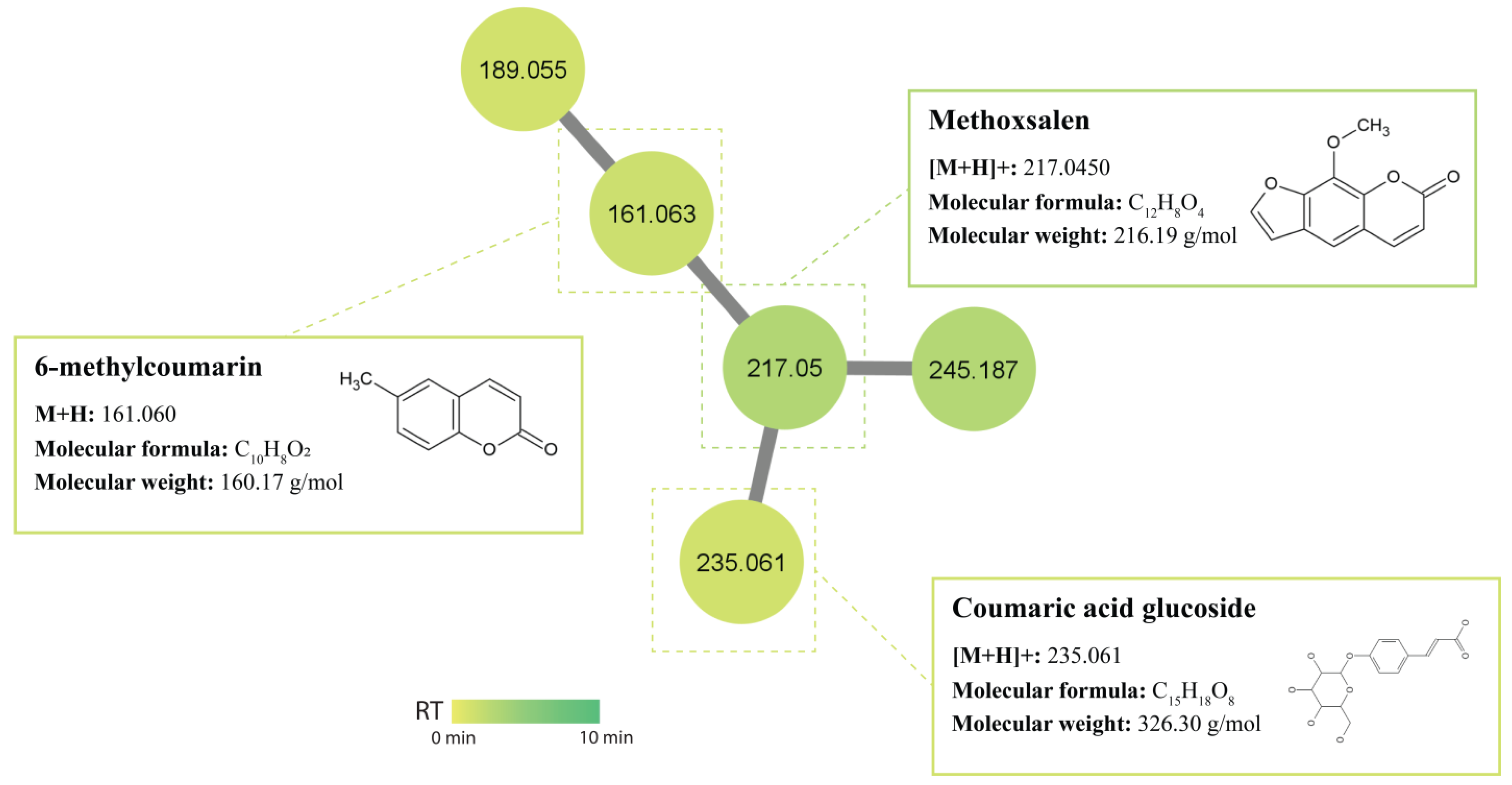
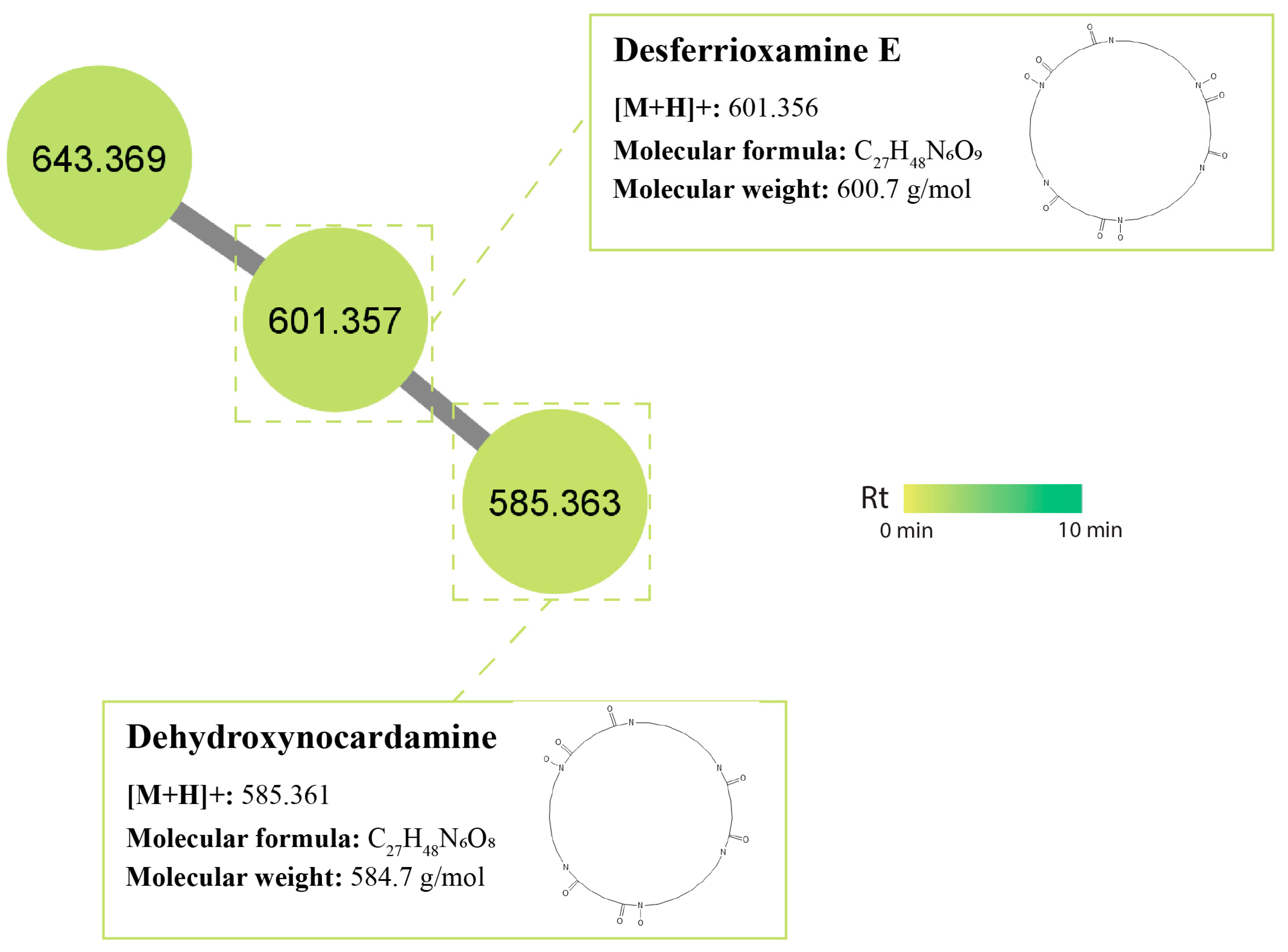
2.5. Molecular Networking and Identification of Bioactive Compounds within the Streptomycetes Fermentation Extracts
2.5.1. Isoflavonoids
2.5.2. Macrolides
2.5.3. Coumarins
2.5.4. Cyclic Peptides (Nocardamine)/Siderophores
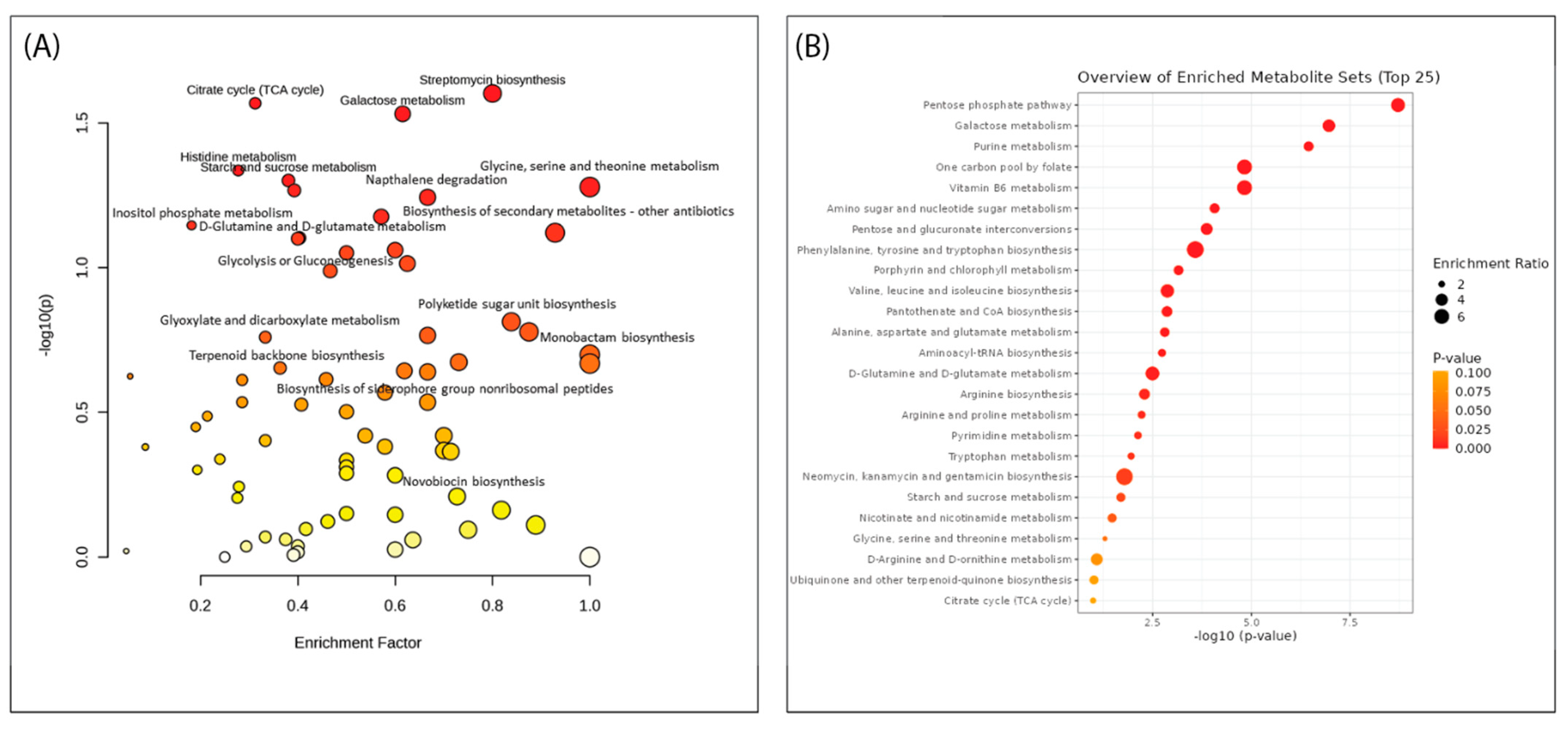
2.6. Pathway Enrichment Analysis within the Streptomycetes Fermentation Extracts
3. Materials and Methods
3.1. Cultures and Culture Media
3.2. 16S rDNA Oligonucleotide Sequencing of Actinomycete Isolates
3.3. Evaluation of Antifungal Activity of the Actinomycete Isolates
3.4. Fermentation and Extraction of Metabolites Produced by Streptomyces Isolates
3.5. Bioassays of the Streptomycetes Fermentation Extracts Using Disk-Diffusion Method
3.6. QTOF Chemical Profiling of the Metabolites within the Streptomyces Fermentation Extracts
3.7. MS Data Analysis and Annotation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fones, H.N.; Bebber, D.P.; Chaloner, T.M.; Kay, W.T.; Steinberg, G.; Gurr, S.J. Threats to global food security from emerging fungal and oomycete crop pathogens. Nat. Food 2020, 1, 332–342. [Google Scholar] [CrossRef]
- Thambugala, K.M.; Daranagama, D.A.; Phillips, A.J.; Kannangara, S.D.; Promputtha, I. Fungi vs. fungi in biocontrol: An overview of fungal antagonists applied against fungal plant pathogens. Front. Cell. Infect. Microbiol. 2020, 10, 604923. [Google Scholar] [CrossRef]
- Stewart, J.; Kim, M.-S.; Ota, Y.; Sahashi, N.; Hanna, J.; Akiba, M.; Ata, J.; Atibalentja, N.; Brooks, F.; Chung, C.-L. Phylogenetic and population genetic analyses reveal three distinct lineages of the invasive brown root-rot pathogen, Phellinus noxius, and bioclimatic modeling predicts differences in associated climate niches. Eur. J. Plant Pathol. 2020, 156, 751–766. [Google Scholar] [CrossRef]
- Ann, P.; Lee, H.; Tsai, J. Survey of brown root disease of fruit and ornamental trees caused by Phellinus noxius in Taiwan. Plant Pathol. Bull. 1999, 8, 51–60. [Google Scholar]
- Chou, H.; Xiao, Y.-T.; Tsai, J.-N.; Li, T.-T.; Wu, H.-Y.; Liu, L.-y.D.; Tzeng, D.-S.; Chung, C.-L. In Vitro and in planta evaluation of Trichoderma asperellum TA as a biocontrol agent against Phellinus noxius, the cause of brown root rot disease of trees. Plant Dis. 2019, 103, 2733–2741. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, W.-W.; Ting-Hsuan, H.; En-Jang, S. The pathogenicity of basidiospores of Phellinus noxius which causes brown root rot disease in Taiwan. Taiwania 2019, 64, 189–194. [Google Scholar]
- Akiba, M.; Ota, Y.; Tsai, I.J.; Hattori, T.; Sahashi, N.; Kikuchi, T. Genetic differentiation and spatial structure of Phellinus noxius, the causal agent of brown root rot of woody plants in Japan. PLoS ONE 2015, 10, e0141792. [Google Scholar] [CrossRef] [PubMed]
- Sahashi, N.; Akiba, M.; Ishihara, M.; Ota, Y.; Kanzaki, N. Brown root rot of trees caused by Phellinus noxius in the Ryukyu Islands, subtropical areas of Japan. For. Pathol. 2012, 42, 353–361. [Google Scholar] [CrossRef]
- Wang, Y.-F.; Meng, H.; Gu, V.W.; Gu, J.-D. Molecular diagnosis of the brown root rot disease agent Phellinus noxius on trees and in soil by rDNA ITS analysis. Appl. Environ. Biotechnol. 2016, 1, 81–91. [Google Scholar] [CrossRef]
- Zhang, H.; Ng, T.K.; Lee, K.C.; Leung, Z.W.; Yau, W.F.; Wong, W.S. Development and evaluation of loop-mediated isothermal amplification (LAMP) as a preliminary diagnostic tool for brown root rot disease caused by Phellinus noxius (Corner) GH cunningham in hong kong urban tree management. Sustainability 2022, 14, 9708. [Google Scholar] [CrossRef]
- Adra, C.; Panchalingam, H.; Kurtböke, D.İ. Termite–gut-associated actinomycetes as biological control agents against phytopathogen Pyrrhoderma noxium. Microbiol. Aust. 2022, 43, 190–193. [Google Scholar] [CrossRef]
- Dann, K.; Smith, L.; Shuey, L.; Begum, F. Phellinus noxius: A basidiomycete fungus impacting productivity of Australian avocados. In Proceedings of the 7th World Avocado Congress, Cairns, Australia, 5–9 September 2011. [Google Scholar]
- Panchalingam, H.; Ashfield-Crook, N.; Naik, V.; Frenken, R.; Foster, K.; Tomlin, R.; Shapcott, A.; Kurtböke, D.İ. Testing the biocontrol ability of a Trichoderma-Streptomycetes consortium against Pyrrhoderma noxium (Corner) LW Zhou and YC Dai in soil. J. Fungi 2022, 9, 67. [Google Scholar] [CrossRef] [PubMed]
- Schwarze, F.W.; Jauss, F.; Spencer, C.; Hallam, C.; Schubert, M. Evaluation of an antagonistic Trichoderma strain for reducing the rate of wood decomposition by the white rot fungus Phellinus noxius. Biol. Control 2012, 61, 160–168. [Google Scholar] [CrossRef]
- Caballero, J.R.I.; Ata, J.P.; Leddy, K.; Glenn, T.C.; Kieran, T.J.; Klopfenstein, N.B.; Kim, M.-S.; Stewart, J.E. Genome comparison and transcriptome analysis of the invasive brown root rot pathogen, Phellinus noxius, from different geographic regions reveals potential enzymes associated with degradation of different wood substrates. Fungal Biol. 2020, 124, 144–154. [Google Scholar] [CrossRef]
- Liu, T.Y.; Chen, C.H.; Yang, Y.L.; Tsai, I.J.; Ho, Y.N.; Chung, C.L. The brown root rot fungus Phellinus noxius affects microbial communities in different root-associated niches of Ficus trees. Environ. Microbiol. 2022, 24, 276–297. [Google Scholar] [CrossRef]
- Wu, Z.-C.; Chang, Y.-Y.; Lai, Q.-J.; Lin, H.-A.; Tzean, S.-S.; Liou, R.-F.; Tsai, I.J.; Chung, C.-L. Soil is not a reservoir for Phellinus noxius. Phytopathology 2020, 110, 362–369. [Google Scholar] [CrossRef]
- Tsang, K.S.W.; Cheung, M.K.; Lam, R.Y.C.; Kwan, H.S. A preliminary examination of the bacterial, archaeal, and fungal rhizosphere microbiome in healthy and Phellinus noxius-infected trees. MicrobiologyOpen 2020, 9, e1115. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.-L.; Huang, S.-Y.; Huang, Y.-C.; Tzean, S.-S.; Ann, P.-J.; Tsai, J.-N.; Yang, C.-C.; Lee, H.-H.; Huang, T.-W.; Huang, H.-Y. The genetic structure of Phellinus noxius and dissemination pattern of brown root rot disease in Taiwan. PLoS ONE 2015, 10, e0139445. [Google Scholar] [CrossRef]
- Tchotet Tchoumi, J.M.; Coetzee, M.P.A.; Vivas, M.; Rajchenberg, M.; Roux, J. Wood-rotting basidiomycetes associated with declining native trees in timber-harvesting compartments of the Garden Route National Park of South Africa. Austral Ecol. 2017, 42, 947–963. [Google Scholar] [CrossRef]
- Poveda, J.; Abril-Urias, P.; Escobar, C. Biological control of plant-parasitic nematodes by filamentous fungi inducers of resistance: Trichoderma, mycorrhizal and endophytic fungi. Front. Microbiol. 2020, 11, 992. [Google Scholar] [CrossRef]
- Ghanem, G.A.; Gebily, D.A.; Ragab, M.M.; Ali, A.M.; Soliman, N.E.-D.K.; El-Moity, T.H.A. Efficacy of antifungal substances of three Streptomyces spp. against different plant pathogenic fungi. Egypt. J. Biol. Pest Control 2022, 32, 1–13. [Google Scholar] [CrossRef]
- Wang, D.; Li, Y.; Yuan, Y.; Chu, D.; Cao, J.; Sun, G.; Ai, Y.; Cui, Z.; Zhang, Y.; Wang, F. Identification of non-volatile and volatile organic compounds produced by Bacillus siamensis LZ88 and their antifungal activity against Alternaria alternata. Biol. Control 2022, 169, 104901. [Google Scholar] [CrossRef]
- Donald, L.; Pipite, A.; Subramani, R.; Owen, J.; Keyzers, R.A.; Taufa, T. Streptomyces: Still the biggest producer of new natural secondary metabolites, a current perspective. Microbiol. Res. 2022, 13, 418–465. [Google Scholar] [CrossRef]
- Kurtböke, D.İ. Bioactive actinomycetes: Reaching rarity through sound understanding of selective culture and molecular diversity. In Microbial Resources; Kurtböke, D.İ., Ed.; Academic Press: Cambridge, MA, USA; Elsevier: Amsterdam, The Netherlands, 2017; pp. 45–76. [Google Scholar]
- Ding, T.; Yang, L.-J.; Zhang, W.-D.; Shen, Y.-H. The secondary metabolites of rare actinomycetes: Chemistry and bioactivity. RSC Adv. 2019, 9, 21964–21988. [Google Scholar] [CrossRef]
- Chen, S.; Cheng, D.; Liu, Z.; Hassan, B.; Xu, Y. Community structure and antifungal activity of actinobacteria in a fungus-growing termite. Ecol. Entomol. 2023, 48, 251–262. [Google Scholar] [CrossRef]
- Kurtböke, D.İ.; French, J.R.; Hayes, R.A.; Quinn, R.J. Eco-taxonomic insights into actinomycete symbionts of termites for discovery of novel bioactive compounds. Biotechnol. Appl. Biodivers. 2015, 111–135. [Google Scholar]
- Ayu Krishanti, N.P.R.; Zulfiana, D.; Wikantyoso, B.; Zulfitri, A.; Yusuf, S. Antimicrobial production by an actinomycetes isolated from the termite nest. J. Trop. Life Sci. 2018, 8. [Google Scholar]
- Malavika, S.; Kranthi, S.; Shishira Rao, H.; Moily, S.S.; Martin Paul, A. Isolation of gut Actinobacteria from termites. In Methods in Actinobacteriology; Dharumadurai, D., Ed.; Springer Protocols Handbooks; Humana: New York, NY, USA, 2022; pp. 51–60. [Google Scholar] [CrossRef]
- Zeki, Ö.C.; Eylem, C.C.; Reçber, T.; Kır, S.; Nemutlu, E. Integration of GC–MS and LC–MS for untargeted metabolomics profiling. J. Pharm. Biomed. Anal. 2020, 190, 113509. [Google Scholar] [CrossRef]
- Abdelrahman, S.M.; Dosoky, N.S.; Hanora, A.M.; Lopanik, N.B. Metabolomic profiling and molecular networking of nudibranch-associated Streptomyces sp. SCSIO 001680. Molecules 2022, 27, 4542. [Google Scholar] [CrossRef]
- Nothias, L.-F.; Petras, D.; Schmid, R.; Dührkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H.; Fleischauer, M. Feature-based molecular networking in the GNPS analysis environment. Nat. Methods 2020, 17, 905–908. [Google Scholar] [CrossRef]
- Tinte, M.M.; Masike, K.; Steenkamp, P.A.; Huyser, J.; van der Hooft, J.J.; Tugizimana, F. Computational metabolomics tools reveal metabolic reconfigurations underlying the effects of biostimulant seaweed extracts on maize plants under drought stress conditions. Metabolites 2022, 12, 487. [Google Scholar] [CrossRef]
- Kurtböke, D.İ. Actino-Rush on the Sunshine Coast: Prospects for Bioprospecting. In Proceedings of the 10th International Congress on Culture Collections, Tsukuba, Japan, 10–15 October 2004; pp. 319–322. [Google Scholar]
- Kurtböke, D.İ.; French, J.R. Use of phage battery to investigate the actinofloral layers of termite gut microflora. J. Appl. Microbiol. 2007, 103, 722–734. [Google Scholar] [CrossRef]
- Kurtböke, D.İ.; French, J.R. Actinobacterial resources from termite guts for regional bioindustries. Microbiol. Aust. 2008, 29, 42–44. [Google Scholar] [CrossRef][Green Version]
- Berasategui, A.; Shukla, S.; Salem, H.; Kaltenpoth, M. Potential applications of insect symbionts in biotechnology. Appl. Microbiol. Biotechnol. 2016, 100, 1567–1577. [Google Scholar] [CrossRef] [PubMed]
- Kaltenpoth, M. Actinobacteria as mutualists: General healthcare for insects? Trends Microbiol. 2009, 17, 529–535. [Google Scholar] [CrossRef] [PubMed]
- Sujada, N.; Sungthong, R.; Lumyong, S. Termite nests as an abundant source of cultivable actinobacteria for biotechnological purposes. Microbes Environ. 2014, 29, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Shinzato, N.; Fukatsu, T. Isolation of actinomycetes from termites’ guts. Biosci. Biotechnol. Biochem. 2003, 67, 1797–1801. [Google Scholar] [CrossRef]
- Ayed, A.; Kalai-Grami, L.; Ben Slimene, I.; Chaouachi, M.; Mankai, H.; Karkouch, I.; Djebali, N.; Elkahoui, S.; Tabbene, O.; Limam, F. Antifungal activity of volatile organic compounds from Streptomyces sp. strain S97 against Botrytis cinerea. Biocontrol Sci. Technol. 2021, 31, 1330–1348. [Google Scholar] [CrossRef]
- Lyu, A.; Liu, H.; Che, H.; Yang, L.; Zhang, J.; Wu, M.; Chen, W.; Li, G. Reveromycins A and B from Streptomyces sp. 3–10: Antifungal activity against plant pathogenic fungi in vitro and in a strawberry food model system. Front. Microbiol. 2017, 8, 550. [Google Scholar] [CrossRef]
- Wan, M.; Li, G.; Zhang, J.; Jiang, D.; Huang, H.-C. Effect of volatile substances of Streptomyces platensis F-1 on control of plant fungal diseases. Biol. Control 2008, 46, 552–559. [Google Scholar] [CrossRef]
- Igarashi, M.; Kinoshita, N.; Ikeda, T.; Kameda, M.; Hamada, M.; Takeuchi, T. Resormycin, a novel herbicidal and antifungal antibiotic produced by a strain of Streptomyces platensis I. Taxonomy, production, isolation and biological properties. J. Antibiot. 1997, 50, 1020–1025. [Google Scholar] [CrossRef][Green Version]
- Chen, Y.-L.; Zhao, J.; Liu, W.; Gao, J.-F.; Tao, L.-M.; Pan, H.-X.; Tang, G.-L. Identification of phoslactomycin biosynthetic gene clusters from Streptomyces platensis SAM-0654 and characterization of PnR1 and PnR2 as positive transcriptional regulators. Gene 2012, 509, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Lai, B.; Dai, R.; Zeng, T.; Yao, J. Optimization of Fermentation Conditions and Properties of Secondary Metabolite of Streptomyces catenulae XG40. Chin. J. Biol. Control 2022, 38, 1242. [Google Scholar]
- Budi, M.B.S.; Giyanto, G.; Tondok, E.T. Isolation of actinomycetes from peatland to suppress the growth of Ganoderma boninense the causal agent of basal stem rot disease in oil palm. Biodiversitas J. Biol. Divers. 2022, 23. [Google Scholar] [CrossRef]
- Zhang, R.; Han, X.; Xia, Z.; Luo, X.; Wan, C.; Zhang, L. Streptomyces luozhongensis sp. nov., a novel actinomycete with antifungal activity and antibacterial activity. Antonie Van Leeuwenhoek 2017, 110, 195–203. [Google Scholar] [CrossRef]
- Marian, M.; Ohno, T.; Suzuki, H.; Kitamura, H.; Kuroda, K.; Shimizu, M. A novel strain of endophytic Streptomyces for the biocontrol of strawberry anthracnose caused by Glomerella cingulata. Microbiol. Res. 2020, 234, 126428. [Google Scholar] [CrossRef]
- Yang, P.; Li, M.; Zhao, J.; Zhu, M.; Shang, H.; Li, J.; Cui, X.; Huang, R.; Wen, M. Oligomycins A and C, major secondary metabolites isolated from the newly isolated strain Streptomyces diastaticus. Folia Microbiol. 2010, 55, 10–16. [Google Scholar] [CrossRef]
- De Simeis, D.; Serra, S. Actinomycetes: A never-ending source of bioactive compounds—An overview on antibiotics production. Antibiotics 2021, 10, 483. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, D.; Bi, Y.; Yu, Z. Macrolides from Streptomyces sp. SN5452 and their antifungal activity against Pyricularia oryzae. Microorganisms 2022, 10, 1612. [Google Scholar] [CrossRef] [PubMed]
- Risdian, C.; Mozef, T.; Wink, J. Biosynthesis of polyketides in Streptomyces. Microorganisms 2019, 7, 124. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Yang, B.; Tan, G.-Y.; Ouyang, L.-M.; Qiu, S.; Wang, W.; Xiang, W.; Zhang, L. Polyketide pesticides from actinomycetes. Curr. Opin. Biotechnol. 2021, 69, 299–307. [Google Scholar] [CrossRef]
- Wang, J.-F.; Liu, S.-S.; Song, Z.-Q.; Xu, T.-C.; Liu, C.-S.; Hou, Y.-G.; Huang, R.; Wu, S.-H. Naturally occurring flavonoids and isoflavonoids and their microbial transformation: A review. Molecules 2020, 25, 5112. [Google Scholar] [CrossRef] [PubMed]
- Ebada, S.S.; Eze, P.; Okoye, F.B.; Esimone, C.O.; Proksch, P. The fungal endophyte Nigrospora oryzae produces quercetin monoglycosides previously known only from plants. ChemistrySelect 2016, 1, 2767–2771. [Google Scholar] [CrossRef]
- Moore, B.S.; Hertweck, C.; Hopke, J.N.; Izumikawa, M.; Kalaitzis, J.A.; Nilsen, G.; O’Hare, T.; Piel, J.; Shipley, P.R.; Xiang, L. Plant-like biosynthetic pathways in bacteria: From benzoic acid to chalcone. J. Nat. Prod. 2002, 65, 1956–1962. [Google Scholar] [CrossRef]
- Matseliukh, B.; Polishchuk, L.; Lutchenko, V.; Bambura, O.; Kopiyko, O. Synthesis of daidzein and genistein by streptomycetes and their effect on production of antibiotics. Mikrobiolohichnyi Zhurnal 2005, 67, 12–21. [Google Scholar]
- Kang, H.R.; Lee, D.; Benndorf, R.; Jung, W.H.; Beemelmanns, C.; Kang, K.S.; Kim, K.H. Termisoflavones A–C, isoflavonoid glycosides from termite-associated Streptomyces sp. RB1. J. Nat. Prod. 2016, 79, 3072–3078. [Google Scholar] [CrossRef]
- Savi, D.C.; Shaaban, K.A.; Gos, F.M.; Thorson, J.S.; Glienke, C.; Rohr, J. Secondary metabolites produced by Microbacterium sp. LGMB471 with antifungal activity against the phytopathogen Phyllosticta citricarpa. Folia Microbiol. 2019, 64, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Pancu, D.F.; Scurtu, A.; Macasoi, I.G.; Marti, D.; Mioc, M.; Soica, C.; Coricovac, D.; Horhat, D.; Poenaru, M.; Dehelean, C. Antibiotics: Conventional therapy and natural compounds with antibacterial activity—A pharmaco-toxicological screening. Antibiotics 2021, 10, 401. [Google Scholar] [CrossRef]
- Bao, Y.; Li, H.; Dong, Y.; Duan, H.; Li, H.; Li, W. Genome-guided discovery of antifungal filipins from a deep-sea-derived Streptomyces antibioticus. J. Nat. Prod. 2022, 85, 365–374. [Google Scholar] [CrossRef]
- Jizba, J.; Sedmera, P.; Zima, J.; Beran, M.; Blumauerová, M.; Kandybin, N.; Samoukina, G. Macrotetrolide antibiotics produced by Streptomyces globisporus. Folia Microbiol. 1991, 36, 437–443. [Google Scholar] [CrossRef]
- Wang, J.; Tan, H.; Lu, Y.; Cao, L. Determination of ionophore antibiotics nactins produced by fecal Streptomyces from sheep. BioMetals 2014, 27, 403–407. [Google Scholar] [CrossRef]
- Shishlyannikova, T.A.; Kuzmin, A.V.; Fedorova, G.A.; Shishlyannikov, S.M.; Lipko, I.A.; Sukhanova, E.V.; Belkova, N.L. Ionofore antibiotic polynactin produced by Streptomyces sp. 156A isolated from Lake Baikal. Nat. Prod. Res. 2017, 31, 639–644. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Gu, L.; Zhang, Y.; Liu, Z.; Li, X. Dinactin from a new producer, Streptomyces badius gz-8, and its antifungal activity against the rubber anthracnose fungus Colletotrichum gloeosporioides. Microbiol. Res. 2020, 240, 126548. [Google Scholar] [CrossRef] [PubMed]
- Khebizi, N.; Boudjella, H.; Bijani, C.; Bouras, N.; Klenk, H.-P.; Pont, F.; Mathieu, F.; Sabaou, N. Oligomycins A and E, major bioactive secondary metabolites produced by Streptomyces sp. strain HG29 isolated from a Saharan soil. J. De Mycol. Medicale 2018, 28, 150–160. [Google Scholar] [CrossRef]
- Chakraborty, M.; Mahmud, N.U.; Muzahid, A.N.M.; Rabby, S.F.; Islam, T. Oligomycins inhibit Magnaporthe oryzae Triticum and suppress wheat blast disease. PLoS ONE 2020, 15, e0233665. [Google Scholar] [CrossRef]
- Yu, L.; Li, H.; Zhou, Z.; Liu, F.; Du, L. An antifungal polycyclic tetramate macrolactam, heat-stable antifungal factor (HSAF), is a novel oxidative stress modulator in Lysobacter enzymogenes. Appl. Environ. Microbiol. 2021, 87, e03105–e03120. [Google Scholar] [CrossRef]
- Gray, K.C.; Palacios, D.S.; Dailey, I.; Endo, M.M.; Uno, B.E.; Wilcock, B.C.; Burke, M.D. Amphotericin primarily kills yeast by simply binding ergosterol. Proc. Natl. Acad. Sci. USA 2012, 109, 2234–2239. [Google Scholar] [CrossRef]
- Al-Amiery, A.A.; Kadhum, A.A.H.; Mohamad, A.B. Antifungal activities of new coumarins. Molecules 2012, 17, 5713–5723. [Google Scholar] [CrossRef]
- Hussain, M.I.; Syed, Q.A.; Khattak, M.N.K.; Hafez, B.; Reigosa, M.J.; El-Keblawy, A. Natural product coumarins: Biological and pharmacological perspectives. Biologia 2019, 74, 863–888. [Google Scholar] [CrossRef]
- Chen, Y.-Z.; Wang, S.-R.; Li, T.; Zhang, G.-C.; Yang, J. Antifungal activity of 6-Methylcoumarin against Valsa mali and its possible mechanism of action. J. Fungi 2022, 9, 5. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, J.L.; Gomes, D.; Rodrigues, L.R. Challenges in the heterologous production of furanocoumarins in Escherichia coli. Molecules 2022, 27, 7230. [Google Scholar] [CrossRef]
- Yamanaka, K.; Oikawa, H.; Ogawa, H.-o.; Hosono, K.; Shinmachi, F.; Takano, H.; Sakuda, S.; Beppu, T.; Ueda, K. Desferrioxamine E produced by Streptomyces griseus stimulates growth and development of Streptomyces tanashiensis. Microbiology 2005, 151, 2899–2905. [Google Scholar] [CrossRef]
- Jarmusch, S.A.; Lagos-Susaeta, D.; Diab, E.; Salazar, O.; Asenjo, J.A.; Ebel, R.; Jaspars, M. Iron-meditated fungal starvation by lupine rhizosphere-associated and extremotolerant Streptomyces sp. S29 desferrioxamine production. Mol. Omics 2021, 17, 95–107. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, E.-M.; Xu, L.-Y. Guide to metabolomics analysis: A bioinformatics workflow. Metabolites 2022, 12, 357. [Google Scholar] [CrossRef]
- Wieder, C.; Frainay, C.; Poupin, N.; Rodríguez-Mier, P.; Vinson, F.; Cooke, J.; Lai, R.P.; Bundy, J.G.; Jourdan, F.; Ebbels, T. Pathway analysis in metabolomics: Recommendations for the use of over-representation analysis. PLoS Comput. Biol. 2021, 17, e1009105. [Google Scholar] [CrossRef]
- Sivapragasam, S.; Grove, A. The link between purine metabolism and production of antibiotics in Streptomyces. Antibiotics 2019, 8, 76. [Google Scholar] [CrossRef]
- Kumavath, R.N.; Barh, D.; Azevedo, V.; Kumar, A.P. Potential pharmacological applications of enzymes associated with bacterial metabolism of aromatic compounds. J. Microbiol. Antimicrob. 2017, 9, 1–13. [Google Scholar]
- Alam, K.; Mazumder, A.; Sikdar, S.; Zhao, Y.-M.; Hao, J.; Song, C.; Wang, Y.; Sarkar, R.; Islam, S.; Zhang, Y. Streptomyces: The biofactory of secondary metabolites. Front. Microbiol. 2022, 13, 968053. [Google Scholar] [CrossRef] [PubMed]
- Baba, M.S.; Mohamad Zin, N.; Ahmad, S.J.; Mazlan, N.W.; Baharum, S.N.; Ahmad, N.; Azmi, F. Antibiotic biosynthesis pathways from endophytic Streptomyces SUK 48 through metabolomics and genomics approaches. Antibiotics 2021, 10, 969. [Google Scholar] [CrossRef]
- Zhang, B.; Zhou, Y.-T.; Jiang, S.-X.; Zhang, Y.-H.; Huang, K.; Liu, Z.-Q.; Zheng, Y.-G. Amphotericin B biosynthesis in Streptomyces nodosus: Quantitative analysis of metabolism via LC–MS/MS based metabolomics for rational design. Microb. Cell Factories 2020, 19, 1–12. [Google Scholar] [CrossRef]
- Mulks, M.; Nair, M.; Putnam, A. In vitro antibacterial activity of faeriefungin, a new broad-spectrum polyene macrolide antibiotic. Antimicrob. Agents Chemother. 1990, 34, 1762–1765. [Google Scholar] [CrossRef] [PubMed]
- Yao, T.; Liu, Z.; Li, T.; Zhang, H.; Liu, J.; Li, H.; Che, Q.; Zhu, T.; Li, D.; Li, W. Characterization of the biosynthetic gene cluster of the polyene macrolide antibiotic reedsmycins from a marine-derived Streptomyces strain. Microb. Cell Factories 2018, 17, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.; Wellington, E. Actinomycetes. In Methods of Soil Analysis: Part 2, Chemical and Microbiological Properties, Agronomy Monograph; Page, A.L., Ed.; American Society of Agronomy Inc.; Soil Science of America Inc.: Madison, WI, USA, 1983; pp. 969–987. [Google Scholar]
- Bauer, A.W.; Perry, D.M.; Kirby, W.M. Single-disk antibiotic-sensitivity testing of staphylococci: An analysis of technique and results. AMA Arch. Intern. Med. 1959, 104, 208–216. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef]
- Fiehn, O.; Robertson, D.; Griffin, J.; van der Werf, M.; Nikolau, B.; Morrison, N.; Sumner, L.W.; Goodacre, R.; Hardy, N.W.; Taylor, C. The metabolomics standards initiative (MSI). Metabolomics 2007, 3, 175–178. [Google Scholar] [CrossRef]
- Goodacre, R. Water, water, every where, but rarely any drop to drink. Metabolomics 2014, 10, 5–7. [Google Scholar] [CrossRef][Green Version]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adra, C.; Tran, T.D.; Foster, K.; Tomlin, R.; Kurtböke, D.İ. Untargeted MS-Based Metabolomic Analysis of Termite Gut-Associated Streptomycetes with Antifungal Activity against Pyrrhoderma noxium. Antibiotics 2023, 12, 1373. https://doi.org/10.3390/antibiotics12091373
Adra C, Tran TD, Foster K, Tomlin R, Kurtböke Dİ. Untargeted MS-Based Metabolomic Analysis of Termite Gut-Associated Streptomycetes with Antifungal Activity against Pyrrhoderma noxium. Antibiotics. 2023; 12(9):1373. https://doi.org/10.3390/antibiotics12091373
Chicago/Turabian StyleAdra, Cherrihan, Trong D. Tran, Keith Foster, Russell Tomlin, and D. İpek Kurtböke. 2023. "Untargeted MS-Based Metabolomic Analysis of Termite Gut-Associated Streptomycetes with Antifungal Activity against Pyrrhoderma noxium" Antibiotics 12, no. 9: 1373. https://doi.org/10.3390/antibiotics12091373
APA StyleAdra, C., Tran, T. D., Foster, K., Tomlin, R., & Kurtböke, D. İ. (2023). Untargeted MS-Based Metabolomic Analysis of Termite Gut-Associated Streptomycetes with Antifungal Activity against Pyrrhoderma noxium. Antibiotics, 12(9), 1373. https://doi.org/10.3390/antibiotics12091373