Antimicrobial Activity and Molecular Docking Studies of the Biotransformation of Diterpene Acanthoic Acid Using the Fungus Xylaria sp.
Abstract
:1. Introduction
2. Results
2.1. Identification of Chemical Constituents
2.2. Antimicrobial Activity
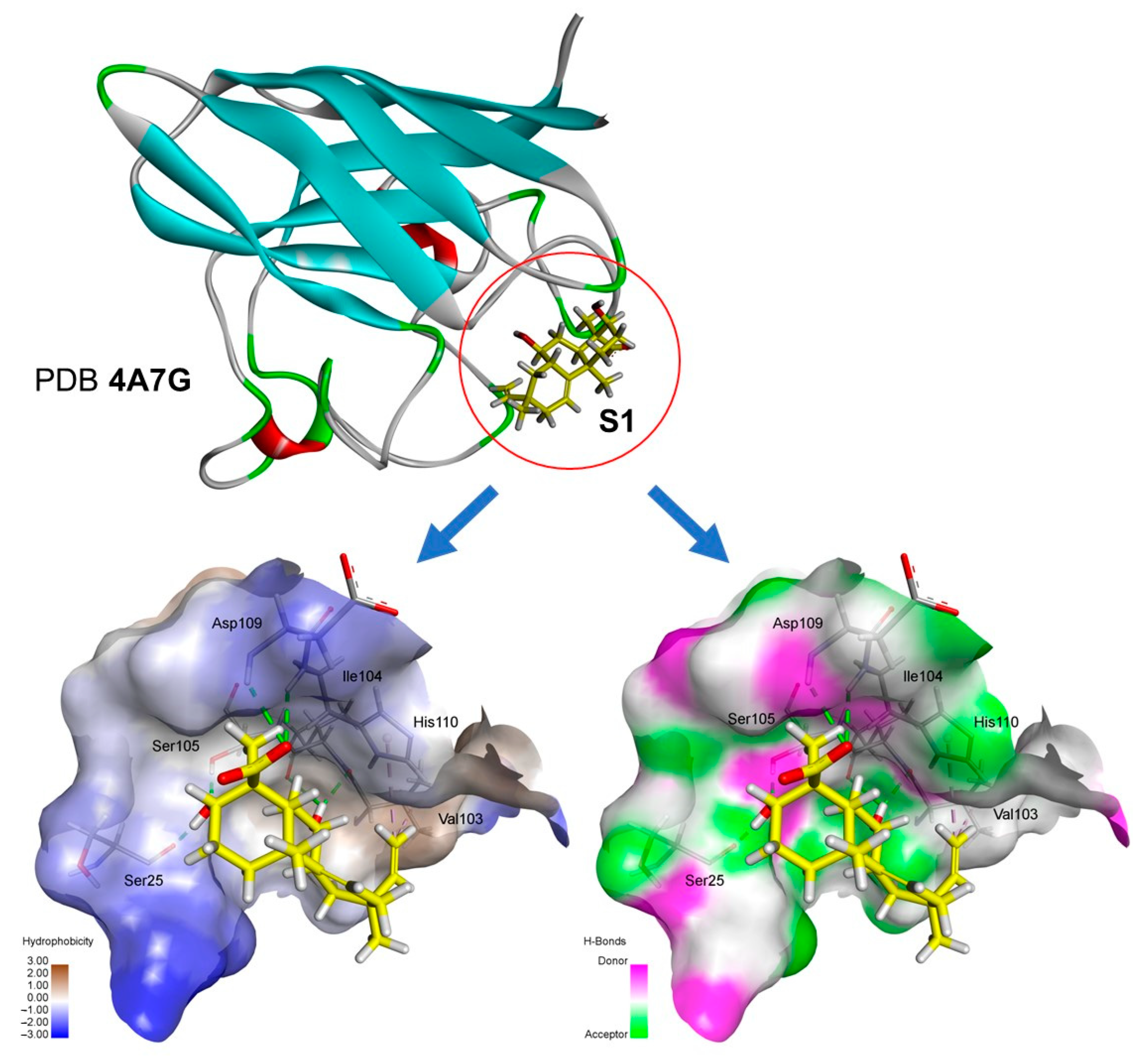
2.3. Molecular Docking
3. Discussion
4. Materials and Methods
4.1. Microorganisms
4.2. Acanthoic Acid
4.3. Biotransformation Reactions Procedure
4.4. Fractionation of PEABE Extract and Biotransformation Product Purification
4.5. Characterisation of the Compounds
4.6. Antimicrobial Assay
4.7. Molecular Docking
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Shakour, Z.T.A.; Fayek, N.M.; Farag, M.A. How Do Biocatalysis and Biotransformation Affect Citrus Dietary Flavonoids Chemistry and Bioactivity? A Review. Crit. Rev. Biotechnol. 2020, 40, 689–714. [Google Scholar] [CrossRef]
- Adams, J.P.; Brown, M.J.B.; Diaz-Rodriguez, A.; Lloyd, R.C.; Roiban, G. Biocatalysis: A Pharma Perspective. Adv. Synth. Catal. 2019, 361, 2421–2432. [Google Scholar] [CrossRef]
- Pan, T.; Wang, Z. Microbial Biocatalysis. Catalysts 2023, 13, 629. [Google Scholar] [CrossRef]
- Liu, X.; Zhou, Z.-Y.; Cui, J.-L.; Wang, M.-L.; Wang, J.-H. Biotransformation Ability of Endophytic Fungi: From Species Evolution to Industrial Applications. Appl. Microbiol. Biotechnol. 2021, 105, 7095–7113. [Google Scholar] [CrossRef] [PubMed]
- Macías-Rubalcava, M.L.; Sánchez-Fernández, R.E. Secondary Metabolites of Endophytic Xylaria Species with Potential Applications in Medicine and Agriculture. World J. Microbiol. Biotechnol. 2017, 33, 15. [Google Scholar] [CrossRef]
- Arunrattiyakorn, P.; Kuno, M.; Aree, T.; Laphookhieo, S.; Sriyatep, T.; Kanzaki, H.; Garcia Chavez, M.A.; Wang, Y.A.; Andersen, R.J. Biotransformation of β-Mangostin by an Endophytic Fungus of Garcinia Mangostana to Furnish Xanthenes with an Unprecedented Heterocyclic Skeleton. J. Nat. Prod. 2018, 81, 2244–2250. [Google Scholar] [CrossRef]
- Rusch, M.; Spielmeyer, A.; Zorn, H.; Hamscher, G. Biotransformation of Ciprofloxacin by Xylaria Longipes: Structure Elucidation and Residual Antibacterial Activity of Metabolites. Appl. Microbiol. Biotechnol. 2018, 102, 8573–8584. [Google Scholar] [CrossRef] [PubMed]
- Murgu, M.; Santos, L.F.A.; de Souza, G.D.; Daolio, C.; Schneider, B.; Ferreira, A.G.; Rodrigues-Filho, E. Hydroxylation of a Hederagenin Derived Saponin by a Xylareaceous Fungus Found in Fruits of Sapindus Saponaria. J. Braz. Chem. Soc. 2008, 19, 831–835. [Google Scholar] [CrossRef]
- Pina, J.R.S.; Silva-Silva, J.V.; Carvalho, J.M.; Bitencourt, H.R.; Watanabe, L.A.; Fernandes, J.M.P.; de Souza, G.E.; Aguiar, A.C.C.; Guido, R.V.C.; Almeida-Souza, F.; et al. Antiprotozoal and Antibacterial Activity of Ravenelin, a Xanthone Isolated from the Endophytic Fungus Exserohilum Rostratum. Molecules 2021, 26, 3339. [Google Scholar] [CrossRef]
- Carneiro, V.A.; dos Santos, H.S.; Arruda, F.V.S.; Bandeira, P.N.; Albuquerque, M.R.J.R.; Pereira, M.O.; Henriques, M.; Cavada, B.S.; Teixeira, E.H. Casbane Diterpene as a Promising Natural Antimicrobial Agent against Biofilm-Associated Infections. Molecules 2011, 16, 190–201. [Google Scholar] [CrossRef]
- Chen, H.-Y.; Liu, T.-K.; Shi, Q.; Yang, X.-L. Sesquiterpenoids and Diterpenes with Antimicrobial Activity from Leptosphaeria sp. XL026, an Endophytic Fungus in Panax Notoginseng. Fitoterapia 2019, 137, 104243. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.-J.; Zhao, C.-L.; Ku, C.F.; Zhu, Y.; Zhu, X.-J.; Zhang, J.-J.; Deyrup, S.T.; Pan, L.-T.; Zhang, H.-J. Two New Bioactive Diterpenes Identified from Isodon Interruptus. Bioorg. Chem. 2020, 95, 103512. [Google Scholar] [CrossRef] [PubMed]
- Ullah, A.; Munir, S.; Mabkhot, Y.; Badshah, S. Bioactivity Profile of the Diterpene Isosteviol and Its Derivatives. Molecules 2019, 24, 678. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, T.; Khalid, S.; Romanha, A.; Alves, T.; Biavatti, M.; Brun, R.; Da Costa, F.; de Castro, S.; Ferreira, V.; de Lacerda, M.; et al. The Potential of Secondary Metabolites from Plants as Drugs or Leads Against Protozoan Neglected Diseases—Part I. Curr. Med. Chem. 2012, 19, 2128–2175. [Google Scholar] [CrossRef]
- Ribeiro, V.; Oliveira, L.; Santos, M.; Ambrósio, S. Biotransformation of Diterpenes from Brazilian Brown Propolis by Cunninghamella Echinulata. Planta Med. 2022, 88, 1538–1539. [Google Scholar] [CrossRef]
- Dou, J.-Y.; Jiang, Y.-C.; Cui, Z.-Y.; Lian, L.-H.; Nan, J.-X.; Wu, Y.-L. Acanthoic Acid, Unique Potential Pimaradiene Diterpene Isolated from Acanthopanax Koreanum Nakai (Araliaceae): A Review on Its Pharmacology, Molecular Mechanism, and Structural Modification. Phytochemistry 2022, 200, 113247. [Google Scholar] [CrossRef]
- Han, X.; Cui, Z.-Y.; Song, J.; Piao, H.-Q.; Lian, L.-H.; Hou, L.-S.; Wang, G.; Zheng, S.; Dong, X.-X.; Nan, J.-X.; et al. Acanthoic Acid Modulates Lipogenesis in Nonalcoholic Fatty Liver Disease via FXR/LXRs-Dependent Manner. Chem. Biol. Interact 2019, 311, 108794. [Google Scholar] [CrossRef]
- Kasemsuk, T.; Saehlim, N.; Arsakhant, P.; Sittithumcharee, G.; Okada, S.; Saeeng, R. A Novel Synthetic Acanthoic Acid Analogues and Their Cytotoxic Activity in Cholangiocarcinoma Cells. Bioorg. Med. Chem. 2021, 29, 115886. [Google Scholar] [CrossRef]
- Kohanski, M.A.; Dwyer, D.J.; Collins, J.J. How Antibiotics Kill Bacteria: From Targets to Networks. Nat. Rev. Microbiol. 2010, 8, 423–435. [Google Scholar] [CrossRef]
- Vaishampayan, A.; Grohmann, E. Antimicrobials Functioning through ROS-Mediated Mechanisms: Current Insights. Microorganisms 2021, 10, 61. [Google Scholar] [CrossRef]
- Levy, N.; Bruneau, J.-M.; Le Rouzic, E.; Bonnard, D.; Le Strat, F.; Caravano, A.; Chevreuil, F.; Barbion, J.; Chasset, S.; Ledoussal, B.; et al. Structural Basis for E. coli Penicillin Binding Protein (PBP) 2 Inhibition, a Platform for Drug Design. J. Med. Chem. 2019, 62, 4742–4754. [Google Scholar] [CrossRef] [PubMed]
- Brvar, M.; Perdih, A.; Renko, M.; Anderluh, G.; Turk, D.; Solmajer, T. Structure-Based Discovery of Substituted 4,5′-Bithiazoles as Novel DNA Gyrase Inhibitors. J. Med. Chem. 2012, 55, 6413–6426. [Google Scholar] [CrossRef] [PubMed]
- Tari, L.W.; Trzoss, M.; Bensen, D.C.; Li, X.; Chen, Z.; Lam, T.; Zhang, J.; Creighton, C.J.; Cunningham, M.L.; Kwan, B.; et al. Pyrrolopyrimidine Inhibitors of DNA Gyrase B (GyrB) and Topoisomerase IV (ParE). Part I: Structure Guided Discovery and Optimization of Dual Targeting Agents with Potent, Broad-Spectrum Enzymatic Activity. Bioorg. Med. Chem. Lett. 2013, 23, 1529–1536. [Google Scholar] [CrossRef]
- Kershaw, N.M.; Wright, G.S.A.; Sharma, R.; Antonyuk, S.V.; Strange, R.W.; Berry, N.G.; O’Neill, P.M.; Hasnain, S.S. X-ray Crystallography and Computational Docking for the Detection and Development of Protein–Ligand Interactions. Curr. Med. Chem. 2013, 20, 569–575. [Google Scholar] [CrossRef]
- Asili, J.; Lambert, M.; Ziegler, H.L.; Stærk, D.; Sairafianpour, M.; Witt, M.; Asghari, G.; Ibrahimi, I.S.; Jaroszewski, J.W. Labdanes and Isopimaranes from Platycladus orientalis and Their Effects on Erythrocyte Membrane and on Plasmodium falciparum Growth in the Erythrocyte Host Cells. J. Nat. Prod. 2004, 67, 631–637. [Google Scholar] [CrossRef]
- Fan, S.-Y.; Pei, Y.-H.; Zeng, H.-W.; Zhang, S.-D.; Li, Y.-L.; Li, L.; Ye, J.; Pan, Y.-X.; Li, H.-L.; Zhang, W.-D. Compounds from Platycladus orientalis and Their Inhibitory Effects on Nitric Oxide and TNF-α Production. Planta Med. 2011, 77, 1623–1630. [Google Scholar] [CrossRef] [PubMed]
- Marquina, S.; Parra, J.L.; González, M.; Zamilpa, A.; Escalante, J.; Trejo-Hernández, M.R.; Álvarez, L. Hydroxylation of the Diterpenes Ent-Kaur-16-En-19-Oic and Ent-Beyer-15-En-19-Oic Acids by the Fungus Aspergillus Niger. Phytochemistry 2009, 70, 2017–2022. [Google Scholar] [CrossRef]
- Nogueira, M.; Da Costa, F.; Brun, R.; Kaiser, M.; Schmidt, T. Ent-Pimarane and Ent-Kaurane Diterpenes from Aldama Discolor (Asteraceae) and Their Antiprotozoal Activity. Molecules 2016, 21, 1237. [Google Scholar] [CrossRef]
- Porto, T.S.; Simão, M.R.; Carlos, L.Z.; Martins, C.H.G.; Furtado, N.A.J.C.; Said, S.; Arakawa, N.S.; dos Santos, R.A.; Veneziani, R.C.S.; Ambrósio, S.R. Pimarane-Type Diterpenes Obtained by Biotransformation: Antimicrobial Properties Against Clinically Isolated Gram-Positive Multidrug-Resistant Bacteria. Phytother. Res. 2013, 27, 1502–1507. [Google Scholar] [CrossRef]
- Matsumori, N.; Nonomura, T.; Sasaki, M.; Murata, M.; Tachibana, K.; Satake, M.; Yasumoto, T. Long-Range Carbon-Proton Coupling Constants for Stereochemical Assignment of Acyclic Structures in Natural Products: Configuration of the C5-C9 Portion of Maitotoxin. Tetrahedron. Lett. 1996, 37, 1269–1272. [Google Scholar] [CrossRef]
- López-Vallejo, F.; Fragoso-Serrano, M.; Suárez-Ortiz, G.A.; Hernández-Rojas, A.C.; Cerda-García-Rojas, C.M.; Pereda-Miranda, R. Vicinal 1 H–1 H NMR Coupling Constants from Density Functional Theory as Reliable Tools for Stereochemical Analysis of Highly Flexible Multichiral Center Molecules. J. Org. Chem. 2011, 76, 6057–6066. [Google Scholar] [CrossRef] [PubMed]
- Silva, D.M.; Costa, E.V.; de Nogueira, P.C.L.; de Moraes, V.R.S.; de Cavalcanti, S.C.H.; Salvador, M.J.; Ribeiro, L.H.G.; Gadelha, F.R.; Barison, A.; Ferreira, A.G. Ent-Kaurane Diterpenoids and Other Constituents from the Stem of Xylopia Laevigata (Annonaceae). Quim. Nova 2012, 35, 1570–1576. [Google Scholar] [CrossRef]
- Li, J.-C.; Dai, W.-F.; Liu, D.; Jiang, M.-Y.; Zhang, Z.-J.; Chen, X.-Q.; Chen, C.-H.; Li, R.-T.; Li, H.-M. Bioactive Ent-Isopimarane Diterpenoids from Euphorbia Neriifolia. Phytochemistry 2020, 175, 112373. [Google Scholar] [CrossRef]
- Thongnest, S.; Mahidol, C.; Sutthivaiyakit, S.; Ruchirawat, S. Oxygenated Pimarane Diterpenes from Kaempferia marginata. J. Nat. Prod. 2005, 68, 1632–1636. [Google Scholar] [CrossRef]
- Saha, P.; Rahman, F.I.; Hussain, F.; Rahman, S.M.A.; Rahman, M.M. Antimicrobial Diterpenes: Recent Development from Natural Sources. Front. Pharmacol. 2022, 12, 820312. [Google Scholar] [CrossRef]
- Hao, Y.; Wei, Z.; Wang, Z.; Li, G.; Yao, Y.; Dun, B. Biotransformation of Flavonoids Improves Antimicrobial and Anti-Breast Cancer Activities In Vitro. Foods 2021, 10, 2367. [Google Scholar] [CrossRef]
- Aminudin, N.I.; Amran, N.A.; Zainal Abidin, Z.A.; Susanti, D. Biotransformation of Curcumin and Structure–Activity Relationship (SAR) of Its Analogues: A Systematic Review. Biocatal. Biotransformation 2023, 41, 1–14. [Google Scholar] [CrossRef]
- Fàbrega, A.; Madurga, S.; Giralt, E.; Vila, J. Mechanism of Action of and Resistance to Quinolones. Microb. Biotechnol. 2009, 2, 40–61. [Google Scholar] [CrossRef] [PubMed]
- Cochrane, S.A.; Lohans, C.T. Breaking down the Cell Wall: Strategies for Antibiotic Discovery Targeting Bacterial Transpeptidases. Eur. J. Med. Chem. 2020, 194, 112262. [Google Scholar] [CrossRef]
- Li, Y.; Zhan, F.; Li, F.; Lu, Z.; Shi, F.; Xu, Z.; Yang, Y.; Zhao, L.; Qin, Z.; Lin, L. Immune Function of Cytosolic Manganese Superoxide Dismutase from Macrobrachium Rosenbergii in Response to Bacterial Infection. Aquaculture 2021, 541, 736771. [Google Scholar] [CrossRef]
- Ahmmed, F.; Islam, A.U.; Mukhrish, Y.E.; El Bakri, Y.; Ahmad, S.; Ozeki, Y.; Kawsar, S.M.A. Efficient Antibacterial/Antifungal Activities: Synthesis, Molecular Docking, Molecular Dynamics, Pharmacokinetic, and Binding Free Energy of Galactopyranoside Derivatives. Molecules 2023, 28, 219. [Google Scholar] [CrossRef]
- Kostal, J. Chapter Four—Computational Chemistry in Predictive Toxicology: Status Quo et Quo Vadis? In Advances in Molecular Toxicology; Elsevier: Amsterdam, The Netherlands, 2016; Volume 10, pp. 139–186. [Google Scholar]
- Schiebel, J.; Gaspari, R.; Wulsdorf, T.; Ngo, K.; Sohn, C.; Schrader, T.E.; Cavalli, A.; Ostermann, A.; Heine, A.; Klebe, G. Intriguing Role of Water in Protein-Ligand Binding Studied by Neutron Crystallography on Trypsin Complexes. Nat. Commun. 2018, 9, 3559. [Google Scholar] [CrossRef]
- Porto, T.S.; Furtado, N.A.J.C.; Heleno, V.C.G.; Martins, C.H.G.; Da Costa, F.B.; Severiano, M.E.; Silva, A.N.; Veneziani, R.C.S.; Ambrósio, S.R. Antimicrobial Ent-Pimarane Diterpenes from Viguiera Arenaria against Gram-Positive Bacteria. Fitoterapia 2009, 80, 432–436. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, M.L.B.; Xavier, C.M.; deSouza, A.D.L.; de Rabelo, D.M.; Batista, C.L.; Batista, R.L.; Costa, E.V.; Campos, F.R.; Barison, A.; Valdez, R.H.; et al. Acanthoic Acid and Other Constituents from the Stem of Annona Amazonica (Annonaceae). J. Braz. Chem. Soc. 2009, 20, 1095–1102. [Google Scholar] [CrossRef]
- CLSI. M07: Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, 11th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018; ISBN 1562388363. [Google Scholar]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An Advanced Semantic Chemical Editor, Visualization, and Analysis Platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Silva-Silva, J.V.; Moreira, R.F.; Watanabe, L.A.; de Souza, C.d.S.F.; de Hardoim, D.J.; Taniwaki, N.N.; Bertho, A.L.; Teixeira, K.F.; Cenci, A.R.; Doring, T.H.; et al. Monomethylsulochrin Isolated from Biomass Extract of Aspergillus sp. against Leishmania amazonensis: In Vitro Biological Evaluation and Molecular Docking. Front. Cell Infect. Microbiol. 2022, 12, 974910. [Google Scholar] [CrossRef] [PubMed]
- Matin, M.M.; Hasan, M.d.S.; Uzzaman, M.; Bhuiyan, M.d.M.H.; Kibria, S.M.; Hossain, M.d.E.; Roshid, M.H.O. Synthesis, Spectroscopic Characterization, Molecular Docking, and ADMET Studies of Mannopyranoside Esters as Antimicrobial Agents. J. Mol. Struct. 2020, 1222, 128821. [Google Scholar] [CrossRef]
- Alam, A.; Hosen, M.A.; Hosen, A.; Fujii, Y.; Ozeki, Y.; Kawsar, S.M.A. Synthesis, Characterization, and Molecular Docking Against a Receptor Protein FimH of Escherichia coli (4XO8) of Thymidine Derivatives. J. Mex. Chem. Soc. 2021, 65, 256–276. [Google Scholar] [CrossRef]


| AA | S1 | |||
|---|---|---|---|---|
| n | 1H δ (Mult, J in Hz) | 13C | 1H δ (Mult, J in Hz) | 13C |
| 1 | ax 1.28 (m) eq 1.79 (m) | 41.9 | 1.55 1.70 | 34.8 |
| 2 | ax 2.19 (m) eq 1.92 (m) | 18.9 | 1.68 (m) 2.22 (m) | 27.5 |
| 3 | ax 1.05 (m) eq 2.15 (m) | 38.1 | 4.12 (t, 2.8) | 70.5 |
| 4 | - | 44.2 | - | 47.6 |
| 5 | 1.66 (dd, 12.9 and 6.0) | 48.0 | 2.30 (dd, 12.8 and 5.0) | 39.5 |
| 6 | ax 1.48 (m) eq 1.90 (m) | 20.3 | 1.70 (m) 2.75 (m) | 30.8 |
| 7 | ax 1.21 (m) eq 1.73 (m) | 27.8 | 3.65 (ddd, 10.1, 9.0 and 4.9) | 72.6 |
| 8 | 2.32 (m) | 28.7 | 2.40 (m) | 37.2 |
| 9 | - | 149.9 | - | 145.7 |
| 10 | - | 38.4 | - | 38.2 |
| 11 | 5.39 (dt, 5.0 and 2.1) | 116.6 | 5.53 (dt, 5.3 and 2.2) | 119.9 |
| 12 | ax 1.75 (m) eq 2.03 (ddd, 17.4; 4.0 and 2.3) | 37.5 | 1.85 (m) 2.08 (ddd, 17.4; 4.0 and 2.3) | 37.3 |
| 13 | - | 34.8 | - | 34.4 |
| 14 | ax 1.02 (m) eq 1.45 (m) | 41.8 | 1.15 (m) 1.26 (m) | 38.7 |
| 15 | 5.81 (dd, 17.5 and 10.7) | 150.2 | 5.86 (dd, 17.4 and 10.7) | 149.7 |
| 16 | 4.86 cis (dd, 10.7 and 1.4) 4.93 (dd, 17.5 and 1.4) | 109.1 | 4.92 (dd, 10.7 and 1.2) 4.95 (dd 17.5 and 1.2) | 109.6 |
| 17 | 0.96 (s) | 22.2 | 0.95 (s) | 21.6 |
| 18 | 1.24 (s) | 28.5 | 1.37 (s) | 23.6 |
| 19 | - | 184.3 | - | 181.0 |
| 20 | 0.99 (s) | 22.4 | 0.98 (s) | 22.5 |
| Compound | MIC (µg.mL−1) | ||
|---|---|---|---|
| Gram (+) Bacteria | Gram (−) Bacteria | ||
| Bacillus subtilis (ATCC 6633) | Escherichia coli (ATCC 25922) | Salmonella typhimurium (ATCC 14028) | |
| AA | 31.25 | 250 | 62.5 |
| S1 | 500 | 31.25 | 62.5 |
| Amoxicillin | 7.81 | 7.81 | 7.81 |
| Tetracycline | 7.81 | 7.81 | 7.81 |
| Compounds | Docking Score | Bond Category | Residues in Contact | Interaction Types | Distance (Å) |
|---|---|---|---|---|---|
| Docking inside the penicillin-binding protein 2 (PDB: 6G9S) | |||||
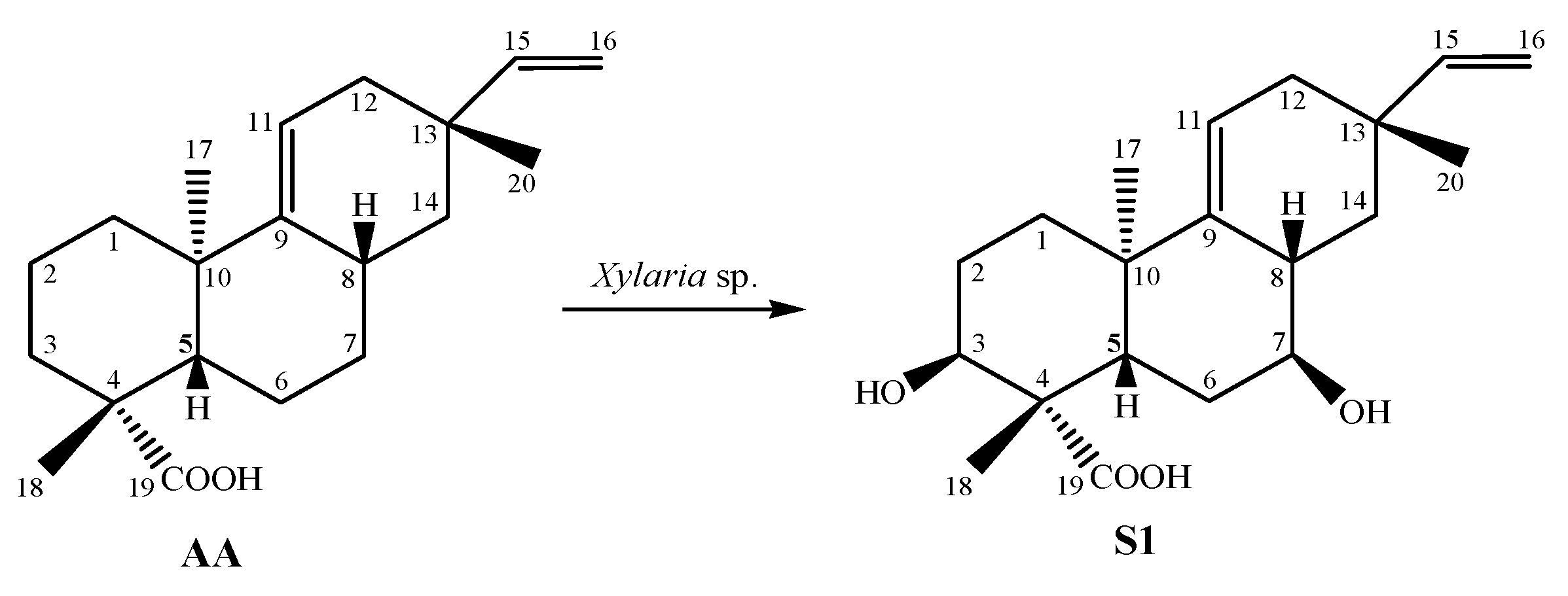
| AA | Goldscore fitness: 50.47 | Hydrophobic | TRP370 | PA | 4.02, 4.05, 5.11, 5.48 |
| Hydrophobic | TRP370 | PS | 2.55 | ||
| Hydrogen | SER387 | H | 2.43 | ||
| Hydrogen | SER545 | H | 2.54 | ||
| S1 | Goldscore fitness: 44.29 | Hydrophobic | TRP370 | PA | 3.20, 3,70, 5.15 |
| Hydrogen | SER387 | H | 2.87 | ||
| ET5 | Goldscore fitness: 49.58 | Hydrogen | SER330 | H | 2.23, 2.88 |
| Hydrogen | LYS333 | H | 3.02 | ||
| Hydrophobic | TRP370 | PA | 4.98 | ||
| Hydrophobic | TRP370 | PPS | 5.01 | ||
| Unfavorable | TYR393 | UDD | 2.38 | ||
| Hydrogen | ILE453 | H | 2.35 | ||
| Hydrogen | GLN455 | H | 2.50 | ||
| Hydrogen | LYS544 | H | 5.18 | ||
| Hydrogen | THR547 | H | 1.86 | ||
| Docking inside the DNA gyrase subunit B (PDB: 4DUH) | |||||
| AA | ChemPLP Score: 50.62 | Hydrophobic | GLY77 | A | 2.88 |
| Hydrogen | LYS103 | H | 3.81, 4.08, 5.43 | ||
| S1 | ChemPLP Score: 40.70 | Hydrogen | ASN46 | H | 1.71 |
| Hydrogen | GLY77 | H | 1.76, 2.70 | ||
| Hydrophobic | LYS103 | UB | 1.26, 1.33 | ||
| Hydrophobic | LYS103 | A | 4.36, 5.13, 5.15 | ||
| RLI | ChemPLP Score: 78.99 | Hydrophobic | VAL43 | A | 4.70 |
| Hydrogen | ASN46 | H | 3.03 | ||
| Hydrophobic | VAL71 | A | 4.17 | ||
| Hydrogen | ASP73 | H | 1.70 | ||
| Hydrogen | ARG76 | H | 2.56 | ||
| Hydrophobic | ILE78 | PA | 4.65, 4.90, 4.99 | ||
| Hydrophobic | PRO79 | PA | 4.14 | ||
| Hydrophobic | ILE94 | PA | 4.84 | ||
| Hydrophobic | LYS103 | PA | 4.64 | ||
| Hydrogen | ARG136 | H | 1.80, 2.10 | ||
| Hydrophobic | VAL167 | A | 4.21 | ||
| Docking inside the topoisomerase IV (PDB: 4HZ0) | |||||
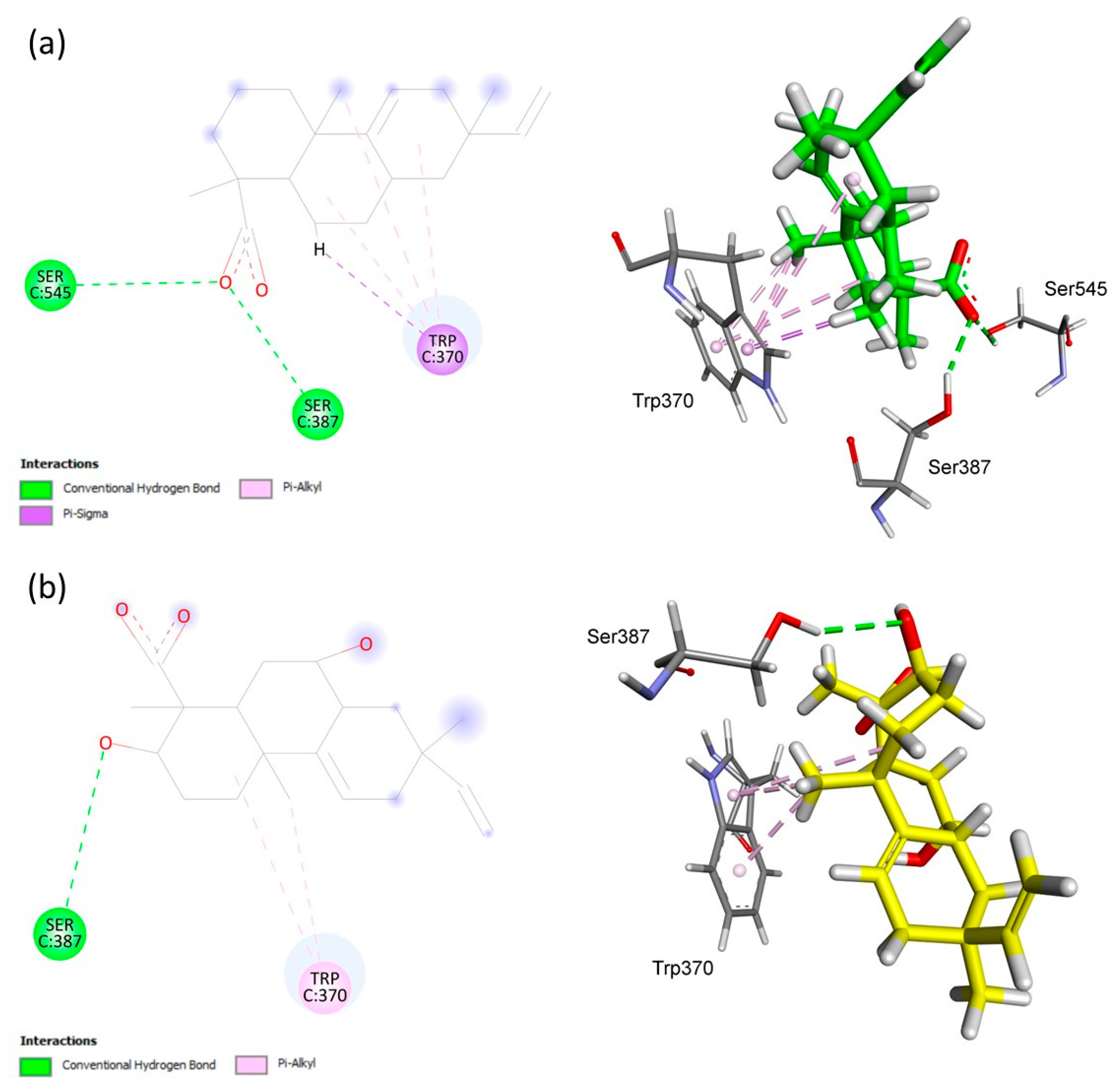
| AA | ChemPLP Score: 41.28 | Hydrophobic | HIS95 | PA | 3.82, 4.18 |
| S1 | ChemPLP Score: 43.29 | Hydrogen | ASN42 | H | 1.98 |
| 1AV | ChemPLP Score: 57.00 | Hydrophobic | ASN42 | APS | 4.67 |
| Hydrogen | ASP69 | H | 1.85 | ||
| Hydrophobic | ARG72 | PA | 5.48 | ||
| Hydrophobic | MET74 | PS | 2.76 | ||
| Hydrophobic | MET74 | PA | 4.15, 4.74 | ||
| Hydrophobic | PRO75 | PA | 4.06 | ||
| Hydrophobic | ILE90 | PA | 4.51 | ||
| Docking inside the superoxide dismutase (PDB: 4A7G) | |||||
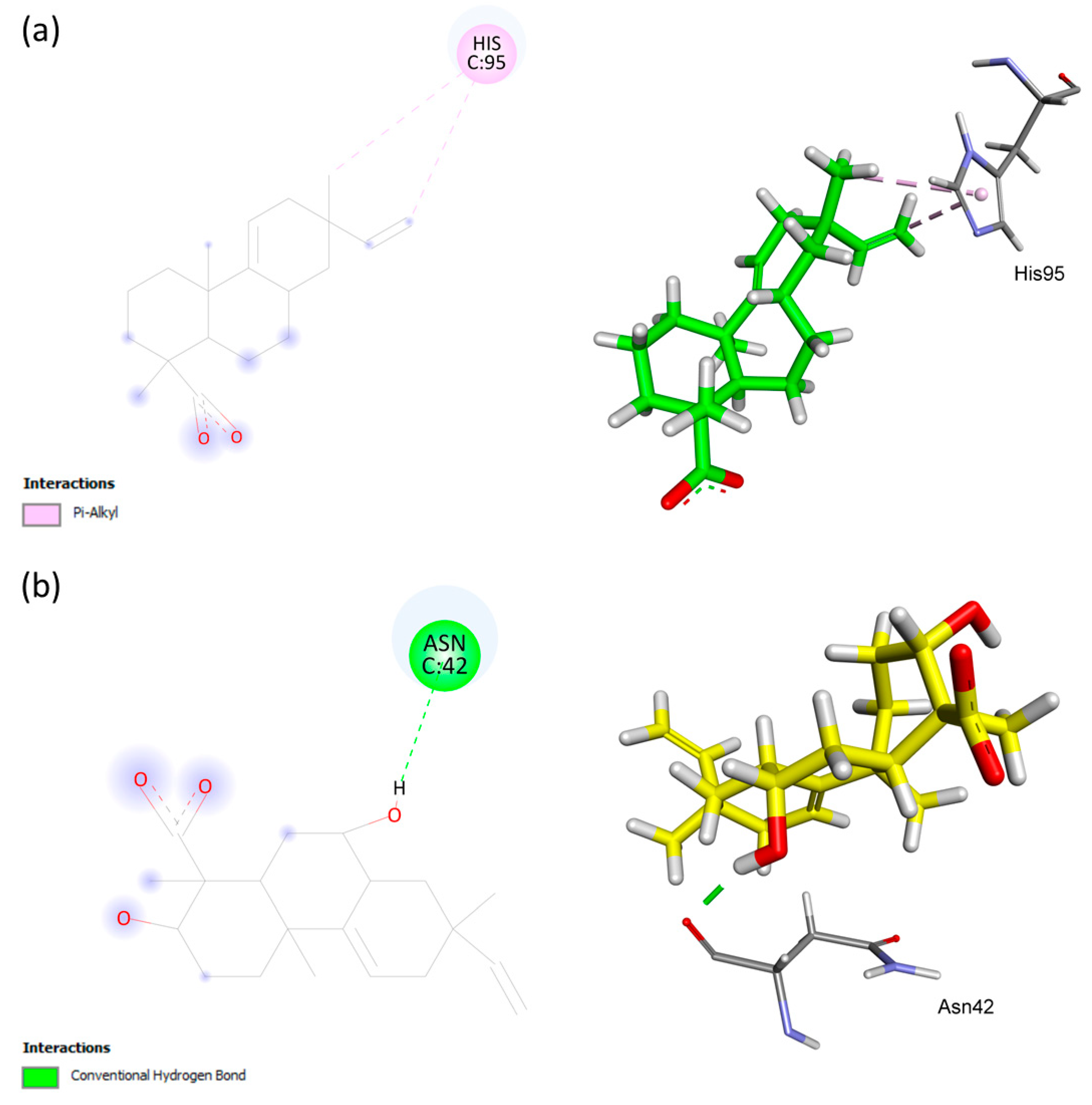
| AA | ChemPLP Score: 31.34 | Hydrogen | SER25 | H | 2.61 |
| Hydrophobic | HIS110 | PA | 4.26, 5.14 | ||
| S1 | ChemPLP Score: 42.45 | Hydrogen | SER25 | H | 1.68 |
| Hydrophobic | VAL103 | A | 3.76 | ||
| Hydrogen | VAL103 | H | 2.02 | ||
| Hydrogen | ILE104 | H | 2.99 | ||
| Hydrogen | SER105 | H | 1.89 | ||
| Hydrogen | ASP109 | H | 2.39 | ||
| Hydrophobic | HIS110 | PA | 4.58 | ||
| Hydrogen | HIS110 | H | 2.75 | ||
| 12I | ChemPLP Score: 41.74 | Hydrogen | ASP109 | H | 2.55 |
| Hydrogen | HIS110 | H | 2.13 | ||
| Hydrophobic | HIS110 | PPS | 3.68, 4.31 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marinho, A.M.d.R.; de Oliveira, C.M.S.C.; Silva-Silva, J.V.; de Jesus, S.C.A.; Siqueira, J.E.S.; de Oliveira, L.C.; Auzier, J.F.; Soares, L.N.; Pinheiro, M.L.B.; Silva, S.C.; et al. Antimicrobial Activity and Molecular Docking Studies of the Biotransformation of Diterpene Acanthoic Acid Using the Fungus Xylaria sp. Antibiotics 2023, 12, 1331. https://doi.org/10.3390/antibiotics12081331
Marinho AMdR, de Oliveira CMSC, Silva-Silva JV, de Jesus SCA, Siqueira JES, de Oliveira LC, Auzier JF, Soares LN, Pinheiro MLB, Silva SC, et al. Antimicrobial Activity and Molecular Docking Studies of the Biotransformation of Diterpene Acanthoic Acid Using the Fungus Xylaria sp. Antibiotics. 2023; 12(8):1331. https://doi.org/10.3390/antibiotics12081331
Chicago/Turabian StyleMarinho, Andrey Moacir do Rosario, Claudia Maria S. C. de Oliveira, João Victor Silva-Silva, Samara C. Anchieta de Jesus, José Edson S. Siqueira, Luana C. de Oliveira, Jéssica Fernandes Auzier, Liviane N. Soares, Maria Lúcia Belém Pinheiro, Sebastião C. Silva, and et al. 2023. "Antimicrobial Activity and Molecular Docking Studies of the Biotransformation of Diterpene Acanthoic Acid Using the Fungus Xylaria sp." Antibiotics 12, no. 8: 1331. https://doi.org/10.3390/antibiotics12081331
APA StyleMarinho, A. M. d. R., de Oliveira, C. M. S. C., Silva-Silva, J. V., de Jesus, S. C. A., Siqueira, J. E. S., de Oliveira, L. C., Auzier, J. F., Soares, L. N., Pinheiro, M. L. B., Silva, S. C., Medeiros, L. S., Costa, E. V., & Marinho, P. S. B. (2023). Antimicrobial Activity and Molecular Docking Studies of the Biotransformation of Diterpene Acanthoic Acid Using the Fungus Xylaria sp. Antibiotics, 12(8), 1331. https://doi.org/10.3390/antibiotics12081331









