New Preventive Strategy against Oral Biofilm Formation in Caries-Active Children: An In Vitro Study
Abstract
1. Introduction
2. Results and Discussion
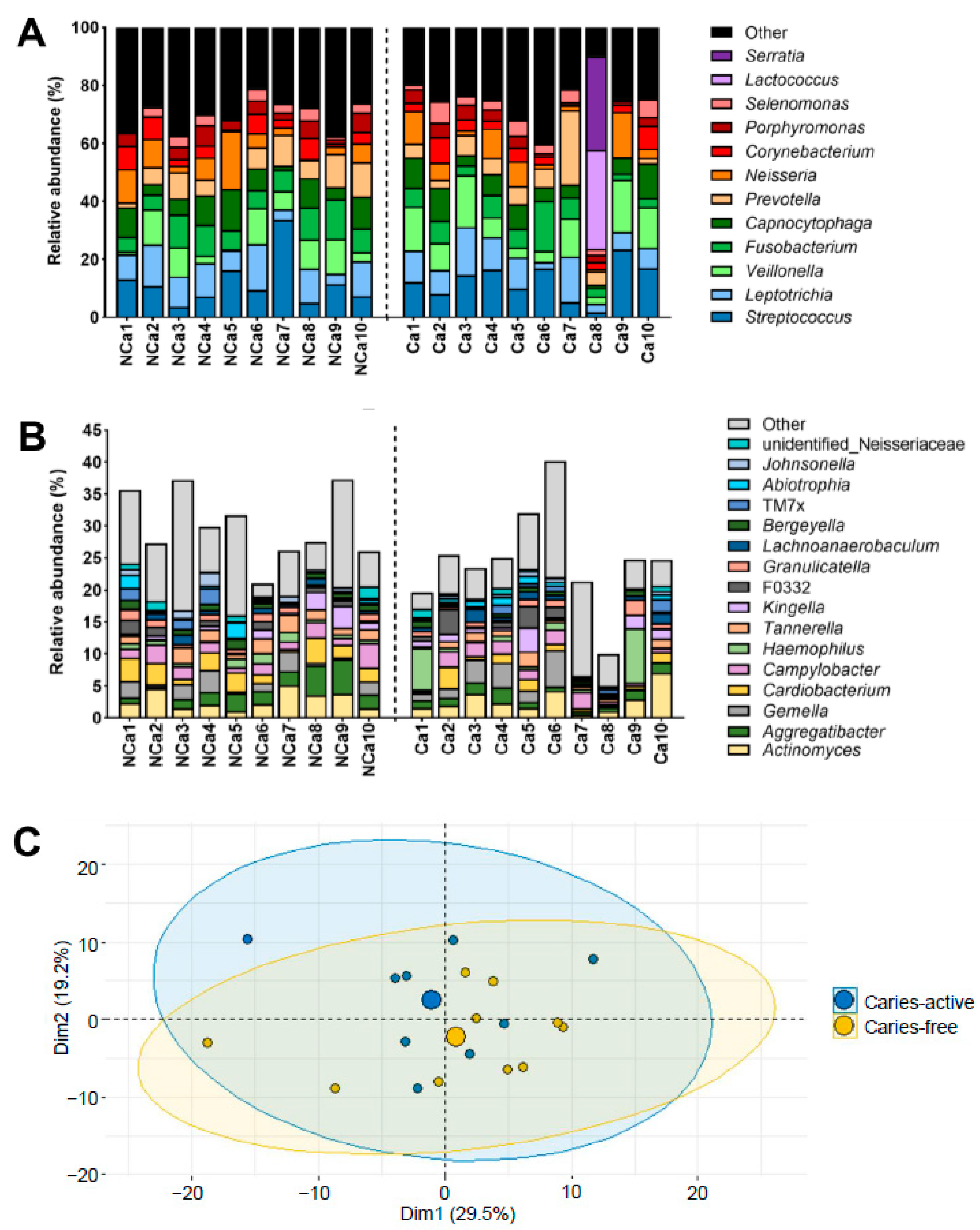
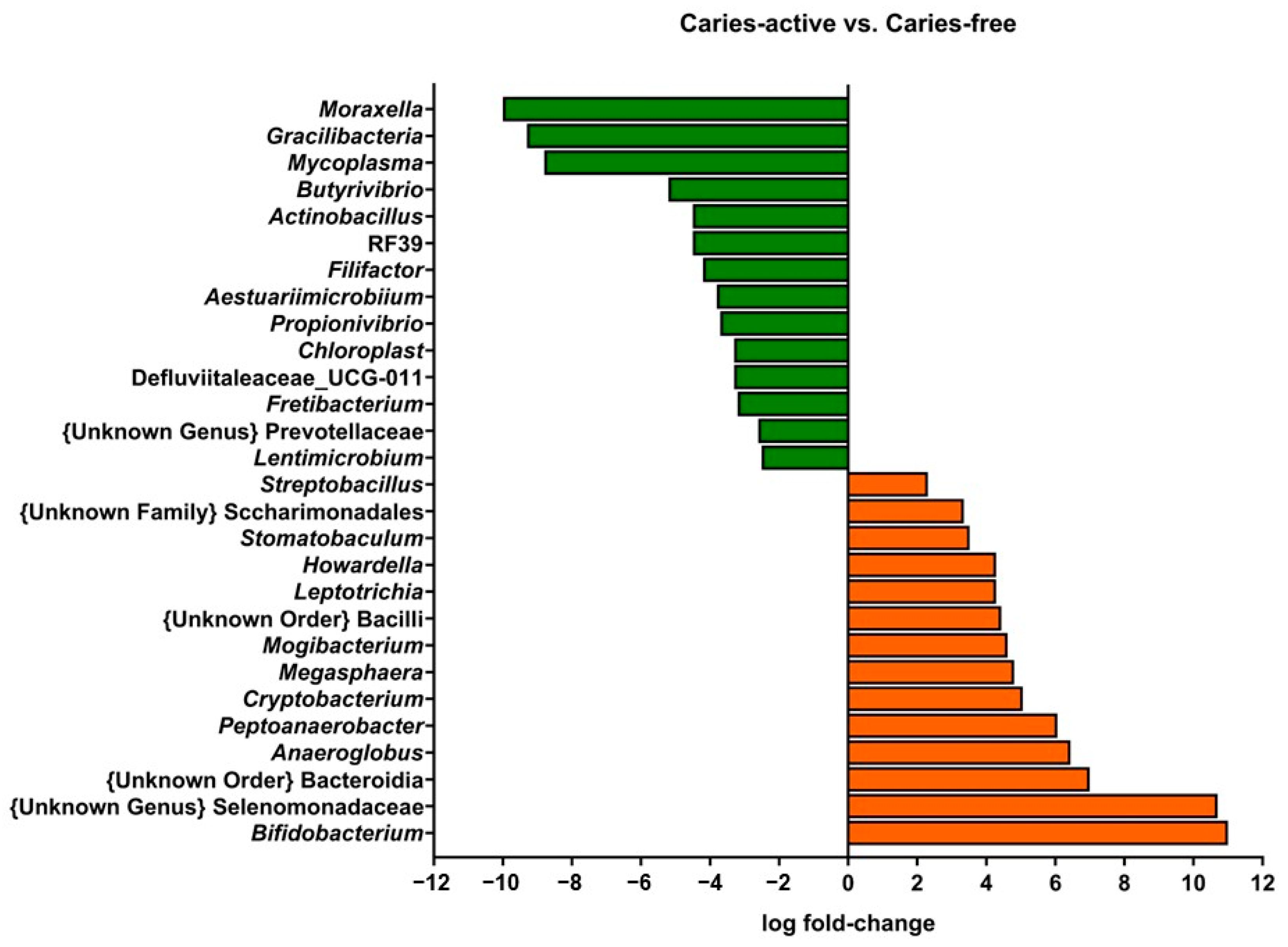
2.1. Microbial Composition of Supragingival Biofilm Samples from Caries-Free and Caries-Active Children
2.2. In Vitro Biofilm Formation from Caries-Free and Caries-Active Children
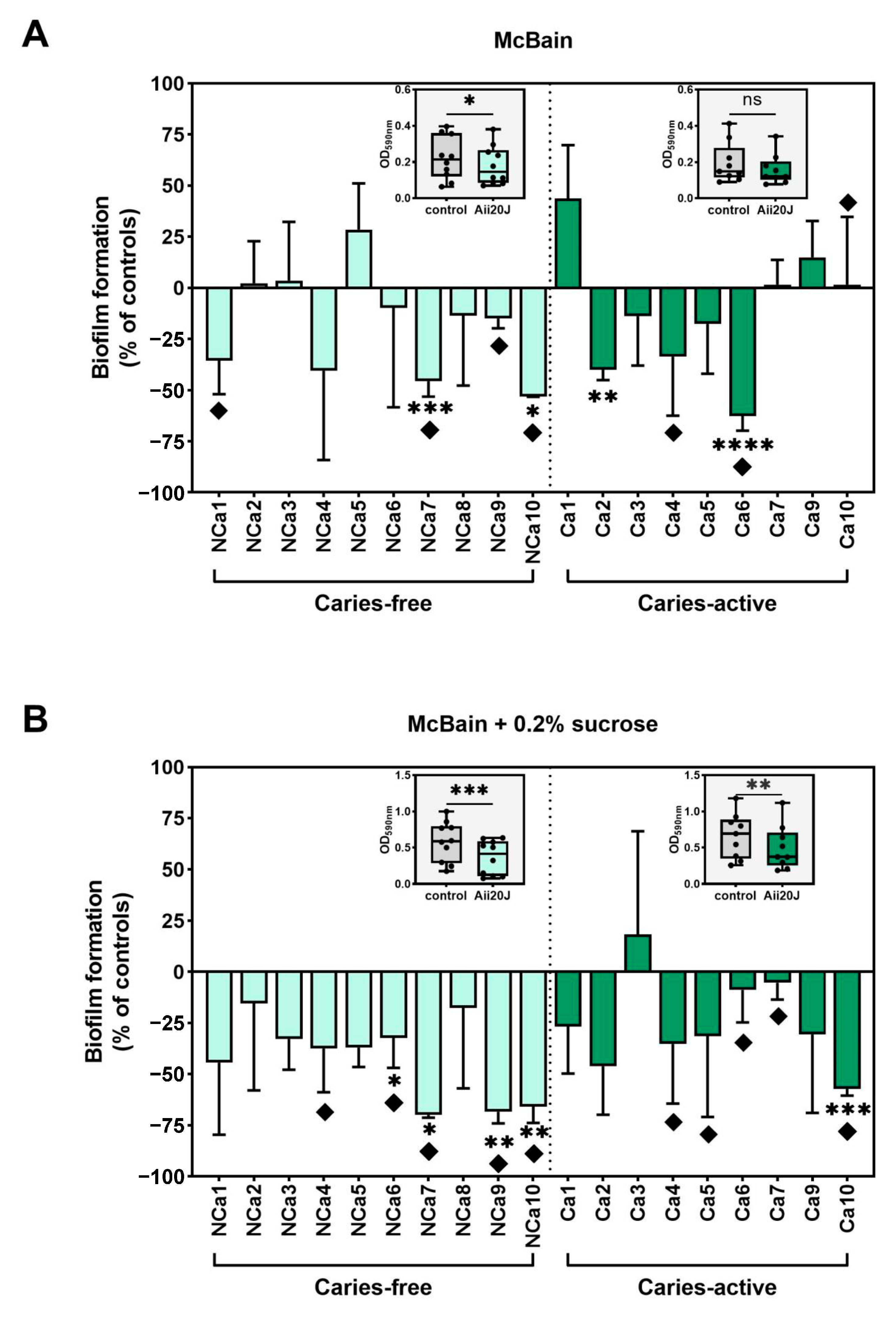
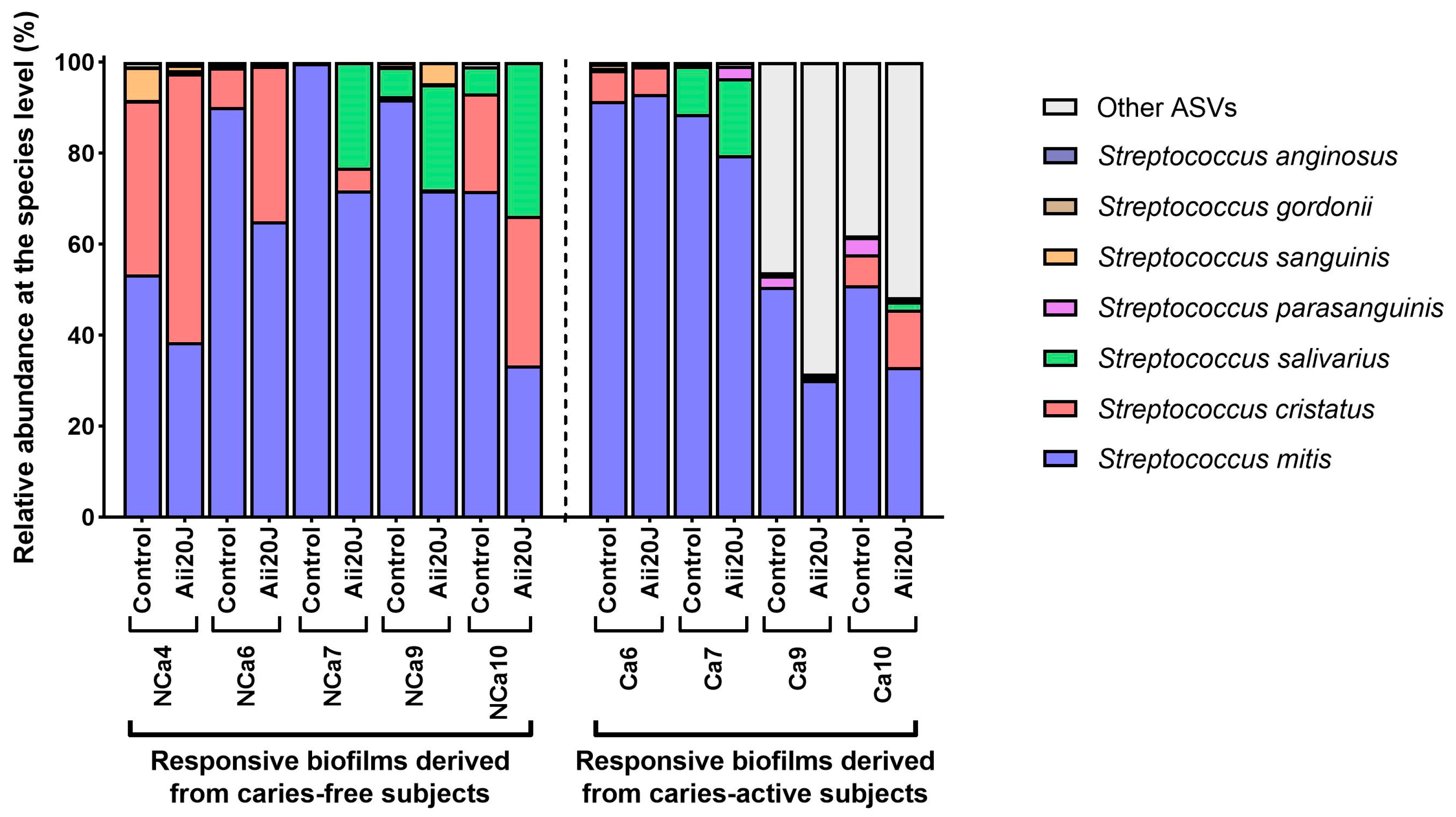
2.3. Effect of Aii20J on Supragingival-Derived Biofilms from Caries-Free and Caries-Active Children
3. Materials and Methods
3.1. Subject Recruitment and Ethics Statements
3.2. Sample Collection and Growth Conditions
3.3. Strains and Culture Conditions
3.4. Production and Purification of the QQ Enzyme Aii20J
3.5. Quorum Quenching Activity Solid Plate Assay
3.6. Biofilm Generation and Quantification
3.7. Investigation of the Microbial Composition of Biofilms
3.7.1. Genomic DNA Extraction
3.7.2. Library Preparation and Microbiome Analysis
3.8. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Global Burden of Disease Collaborative Network. Global Burden of Disease Study 2019 (GBD 2019); Institute of Health Metrics and Evaluation (IHME): Seattle, WA, USA, 2020. [Google Scholar]
- Marsh, P.D.; Lewis, M.A.O.; Rogers, H.; Williams, D.W.; Wilson, M. Marsh and Martin’s Oral Microbiology, 6th ed.; Elsevier: Edinburgh, UK; New York, NY, USA, 2016; p. 261. [Google Scholar]
- Simón-Soro, A.; Mira, A. Solving the etiology of dental caries. Trends Microbiol. 2015, 23, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, D.J.; Lynch, R.J.M. Diet and the microbial aetiology of dental caries: New paradigms. Int. Dent. J. 2013, 63, 64–72. [Google Scholar] [CrossRef]
- Sheiham, A.; James, W.P.T. Diet and Dental Caries: The Pivotal Role of Free Sugars Reemphasized. J. Dent. Res. 2015, 94, 1341–1347. [Google Scholar] [CrossRef] [PubMed]
- Boisen, G.; Davies, J.R.; Neilands, J. Acid tolerance in early colonizers of oral biofilms. BMC Microbiol. 2021, 21, 45. [Google Scholar] [CrossRef] [PubMed]
- Duran-Pinedo, A.E.; Frias-Lopez, J. Beyond microbial community composition: Functional activities of the oral microbiome in health and disease. Microbes Infect. 2015, 17, 505–516. [Google Scholar] [CrossRef] [PubMed]
- Bowen, W.H.; Burne, R.A.; Wu, H.; Koo, H. Oral Biofilms: Pathogens, Matrix, and Polymicrobial Interactions in Microenvironments. Trends Microbiol. 2018, 26, 229–242. [Google Scholar] [CrossRef]
- Muras, A.; Mayer, C.; Romero, M.; Camino, T.; Ferrer, M.D.; Mira, A.; Otero, A. Inhibition of Steptococcus mutans biofilm formation by extracts of Tenacibaculum sp. 20J, a bacterium with wide-spectrum quorum quenching activity. J. Oral Microbiol. 2018, 10, 1429788. [Google Scholar] [CrossRef]
- Philip, N.; Suneja, B.; Walsh, L.J. Ecological Approaches to Dental Caries Prevention: Paradigm Shift or Shibboleth? Caries Res. 2018, 52, 153–165. [Google Scholar] [CrossRef] [PubMed]
- Muras, A.; Otero, A. Breaking Bad: Understanding How Bacterial Communication Regulates Biofilm-Related Oral Diseases. In Trends in Quorum Sensing and Quorum Quenching: New Perspectives and Applications, 1st ed.; Rai, V.R., Bai, J.A., Eds.; CRC Press/Taylor & Francis Group: Boca Raton, FL, USA, 2020. [Google Scholar]
- Muras, A.; Otero-Casal, P.; Blanc, V.; Otero, A. Acyl homoserine lactone-mediated quorum sensing in the oral cavity: A paradigm revisited. Sci. Rep. 2020, 10, 9800. [Google Scholar] [CrossRef]
- Nascimento, M.M. Approaches to Modulate Biofilm Ecology. Dent. Clin. N. Am. 2019, 63, 581–594. [Google Scholar] [CrossRef]
- Cvitkovitch, D.G.; Li, Y.-H.; Ellen, R.P. Quorum sensing and biofilm formation in Streptococcal infections. J. Clin. Investig. 2003, 112, 1626–1632. [Google Scholar] [CrossRef]
- Qi, F.; Kreth, J.; Lévesque, C.M.; Kay, O.; Mair, R.W.; Shi, W.; Cvitkovitch, D.G.; Goodman, S.D. Peptide pheromone induced cell death of Streptococcus mutans. FEMS Microbiol. Lett. 2005, 251, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Marsh, P.D.; Zaura, E. Dental biofilm: Ecological interactions in health and disease. J. Clin. Periodontol. 2017, 44 (Suppl. S18), S12–S22. [Google Scholar] [CrossRef] [PubMed]
- Frias, J.; Olle, E.; Alsina, M. Periodontal Pathogens Produce Quorum Sensing Signal Molecules. Infect. Immun. 2001, 69, 3431–3434. [Google Scholar] [CrossRef] [PubMed]
- Rickard, A.H.; Palmer, R.J.; Blehert, D.S.; Campagna, S.R.; Semmelhack, M.F.; Egland, P.G.; Bassler, B.L.; Kolenbrander, P.E. Autoinducer 2: A concentration-dependent signal for mutualistic bacterial biofilm growth. Mol. Microbiol. 2006, 60, 1446–1456. [Google Scholar] [CrossRef]
- Jakubovics, N.; Kolenbrander, P. The road to ruin: The formation of disease-associated oral biofilms: Formation of oral biofilms. Oral Dis. 2010, 16, 729–739. [Google Scholar] [CrossRef]
- Guo, L.; He, X.; Shi, W. Intercellular communications in multispecies oral microbial communities. Front. Microbiol. 2014, 5, 328. [Google Scholar] [CrossRef] [PubMed]
- Kumari, A.; Pasini, P.; Daunert, S. Detection of bacterial quorum sensing N-acyl homoserine lactones in clinical samples. Anal. Bioanal. Chem. 2008, 391, 1619–1627. [Google Scholar] [CrossRef] [PubMed]
- Goh, S.Y.; Tan, W.-S.; Khan, S.A.; Chew, H.P.; Kasim, N.H.A.; Yin, W.-F.; Chan, K.-G. Unusual multiple production of N-acylhomoserine lactones a by Burkholderia sp. strain C10B isolated from dentine caries. Sensors 2014, 14, 8940–8949. [Google Scholar] [CrossRef]
- Goh, S.-Y.; Khan, S.A.; Tee, K.K.; Abu Kasim, N.H.; Yin, W.-F.; Chan, K.-G. Quorum sensing activity of Citrobacter amalonaticus L8A, a bacterium isolated from dental plaque. Sci. Rep. 2016, 6, 20702. [Google Scholar] [CrossRef] [PubMed]
- Aleti, G.; Baker, J.L.; Tang, X.; Alvarez, R.; Dinis, M.; Tran, N.C.; Melnik, A.V.; Zhong, C.; Ernst, M.; Dorrestein, P.C.; et al. Identification of the Bacterial Biosynthetic Gene Clusters of the Oral Microbiome Illuminates the Unexplored Social Language of Bacteria during Health and Disease. mBio 2019, 10, e00321-19. [Google Scholar] [CrossRef] [PubMed]
- Janus, M.M.; Crielaard, W.; Zaura, E.; Keijser, B.J.; Brandt, B.W.; Krom, B.P. A novel compound to maintain a healthy oral plaque ecology in vitro. J. Oral Microbiol. 2016, 8, 32513. [Google Scholar] [CrossRef]
- Muras, A.; Mayer, C.; Otero-Casal, P.; Exterkate, R.A.M.; Brandt, B.W.; Crielaard, W.; Otero, A.; Krom, B.P. Short-Chain N -Acylhomoserine Lactone Quorum-Sensing Molecules Promote Periodontal Pathogens in In Vitro Oral Biofilms. Appl. Environ. Microbiol. 2020, 86, e01941-19. [Google Scholar] [CrossRef]
- Parga, A.; Muras, A.; Otero-Casal, P.; Arredondo, A.; Soler-Ollé, A.; Àlvarez, G.; Alcaraz, L.D.; Mira, A.; Blanc, V.; Otero, A. The quorum quenching enzyme Aii20J modifies in vitro periodontal biofilm formation. Front. Cell. Infect. Microbiol. 2023, 13, 1118630. [Google Scholar] [CrossRef]
- Eren, A.M.; Borisy, G.G.; Huse, S.M.; Mark Welch, J.L. Oligotyping analysis of the human oral microbiome. Proc. Natl. Acad. Sci. USA 2014, 111, E2875–E2884. [Google Scholar] [CrossRef]
- Segata, N.; Haake, S.K.; Mannon, P.; Lemon, K.P.; Waldron, L.; Gevers, D.; Huttenhower, C.; Izard, J. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 2012, 13, R42. [Google Scholar] [CrossRef] [PubMed]
- Belda-Ferre, P.; Williamson, J.; Simón-Soro, Á.; Artacho, A.; Jensen, O.N.; Mira, A. The human oral metaproteome reveals potential biomarkers for caries disease. Proteomics 2015, 15, 3497–3507. [Google Scholar] [CrossRef]
- Meurman, J.H. Probiotics: Do they have a role in oral medicine and dentistry? Eur. J. Oral Sci. 2005, 113, 188–196. [Google Scholar] [CrossRef]
- Belda-Ferre, P.; Alcaraz, L.D.; Cabrera-Rubio, R.; Romero, H.; Simón-Soro, A.; Pignatelli, M.; Mira, A. The oral metagenome in health and disease. ISME J. 2012, 6, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Exterkate, R.A.M.; Crielaard, W.; Ten Cate, J.M. Different Response to Amine Fluoride by Streptococcus mutans and Polymicrobial Biofilms in a Novel High-Throughput Active Attachment Model. Caries Res. 2010, 44, 372–379. [Google Scholar] [CrossRef]
- Janus, M.M.; Keijser, B.J.F.; Bikker, F.J.; Exterkate, R.A.M.; Crielaard, W.; Krom, B.P. In vitro phenotypic differentiation towards commensal and pathogenic oral biofilms. Biofouling 2015, 31, 503–510. [Google Scholar] [CrossRef] [PubMed]
- Solbiati, J.; Frias-Lopez, J. Metatranscriptome of the oral microbiome in health and disease. J. Dent. Res. 2018, 97, 492–500. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Lin, X.; Wang, B.-Y.; Wu, J.; Lamont, R.J.Y. Identification of a signalling molecule involved in bacterial intergeneric communication. Microbiology 2007, 153, 3228–3234. [Google Scholar] [CrossRef] [PubMed]
- López-Santacruz, H.D.; López-López, A.; Revilla-Guarinos, A.; Camelo-Castillo, A.; Esparza-Villalpando, V.; Mira, A.; Aranda-Romo, S. Streptococcus dentisani is a common inhabitant of the oral microbiota worldwide and is found at higher levels in caries-free individuals. Int. Microbiol. Off. J. Span. Soc. Microbiol. 2021, 24, 619–629. [Google Scholar] [CrossRef]
- Barbour, A.; Elebyary, O.; Fine, N.; Oveisi, M.; Glogauer, M. Metabolites of the oral microbiome: Important mediators of multikingdom interactions. FEMS Microbiol. Rev. 2022, 46, fuab039. [Google Scholar] [CrossRef]
- Ho, M.H.; Lamont, R.J.; Xie, H. Identification of Streptococcus cristatus peptides that repress expression of virulence genes in Porphyromonas gingivalis. Sci. Rep. 2017, 7, 1413. [Google Scholar] [CrossRef]
- Qazi, S.; Middleton, B.; Muharram, S.H.; Cockayne, A.; Hill, P.; O’Shea, P.; Chhabra, S.R.; Cámara, M.; Williams, P. N-acylhomoserine lactones antagonize virulence gene expression and quorum sensing in Staphylococcus aureus. Infect. Immun. 2006, 74, 910–919. [Google Scholar] [CrossRef] [PubMed]
- Parga, A.; Manoil, D.; Brundin, M.; Otero, A.; Belibasakis, G.N. Gram-negative quorum sensing signalling enhances biofilm formation and virulence traits in gram-positive pathogen Enterococcus Faecalis. J. Oral Microbiol. 2023, 15, 2208901. [Google Scholar] [CrossRef]
- Gill, E.E.; Franco, O.L.; Hancock, R.E. Antibiotic adjuvants: Diverse strategies for controlling drug-resistant pathogens. Chem. Biol. Drug. Des. 2015, 85, 56–78. [Google Scholar] [CrossRef] [PubMed]
- Mira, A.; Buetas, E.; Rosier, B.; Mazurel, D.; Villanueva-Castellote, Á.; Llena, C.; Ferrer, M.D. Development of an in vitro system to study oral biofilms in real time through impedance technology: Validation and potential applications. J. Oral Microbiol. 2019, 11, 1609838. [Google Scholar] [CrossRef] [PubMed]
- McClean, K.H.; Winson, M.K.; Fish, L.; Taylor, A.; Chhabra, S.R.; Camara, M.; Daykin, M.; Lamb, J.H.; Swift, S.; Bycroft, B.W.; et al. Quorum sensing and Chromobacterium violaceum: Exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 1997, 143, 3703–3711. [Google Scholar] [CrossRef] [PubMed]
- Mayer, C.; Muras, A.; Romero, M.; López, M.; Tomás, M.; Otero, A. Multiple quorum quenching enzymes are active in the nosocomial pathogen Acinetobacter baumannii ATCC17978. Front. Cell. Infect. Microbiol. 2018, 8, 310. [Google Scholar] [CrossRef] [PubMed]
- Mayer, C.; Romero, M.; Muras, A.; Otero, A. Aii20J, a wide-spectrum thermostable N-acylhomoserine lactonase from the marine bacterium Tenacibaculum sp. 20J, can quench AHL-mediated acid resistance in Escherichia coli. Appl. Microbiol. Biotechnol. 2015, 99, 9523–9539. [Google Scholar] [CrossRef]
- Muras, A.; López-Pérez, M.; Mayer, C.; Parga, A.; Amaro-Blanco, J.; Otero, A. High Prevalence of Quorum-Sensing and Quorum-Quenching Activity among Cultivable Bacteria and Metagenomic Sequences in the Mediterranean Sea. Genes 2018, 9, 100. [Google Scholar] [CrossRef] [PubMed]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Team, R.C. R: A Language and Environment for Statistical Computing; 4.0.2; R Foundation for Statistical Computing: Vienna, Austria, 2022. [Google Scholar]
- Allaire, J.J.; Horner, J.; Marti, V.; Porte, N. Markdown: Markdown Rendering for R, 0.7.4. 2014. Available online: http://CRAN.R-project.org/package=markdown (accessed on 21 April 2020).
- Boettiger, C. Knitcitations: Citations for Knitr Markdown Files. 1.0.5. 2014. Available online: http://CRAN.R-project.org/package=knitcitations (accessed on 26 May 2020).
- Xie, Y. Dynamic Documents with R and Knitr; CRC Press, Taylor & Francis: Boca Raton, FL, USA, 2014; p. 190. [Google Scholar]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. 2.4-4. 2017. Available online: https://CRAN.R-project.org/package=vegan (accessed on 26 May 2020).
- Pagès, H.P.A. Biostrings; Bioconductor, 2017; Available online: https://bioconductor.org/packages/Biostrings (accessed on 1 June 2023).
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]

| Code | Age | Sex | Classification |
|---|---|---|---|
| NCa1 | 3 | M | Caries-free |
| NCa2 | 11 | F | Caries-free |
| NCa3 | 13 | M | Caries-free |
| NCa4 | 11 | M | Caries-free |
| NCa5 | N/A | M | Caries-free |
| NCa6 | 9 | F | Caries-free |
| NCa7 | 8 | F | Caries-free |
| NCa8 | 7 | M | Caries-free |
| NCa9 | 5 | F | Caries-free |
| NCa10 | 11 | M | Caries-free |
| Ca1 | 6 | M | Caries-active |
| Ca2 | 9 | M | Caries-active |
| Ca3 | 7 | F | Caries-active |
| Ca4 | 6 | M | Caries-active |
| Ca5 | 7 | M | Caries-active |
| Ca6 | 9 | M | Caries-active |
| Ca7 | 6 | F | Caries-active |
| Ca8 | 10 | F | Caries-active |
| Ca9 | 9 | F | Caries-active |
| Ca10 | 9 | F | Caries-active |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parga, A.; Balboa, S.; Otero-Casal, P.; Otero, A. New Preventive Strategy against Oral Biofilm Formation in Caries-Active Children: An In Vitro Study. Antibiotics 2023, 12, 1263. https://doi.org/10.3390/antibiotics12081263
Parga A, Balboa S, Otero-Casal P, Otero A. New Preventive Strategy against Oral Biofilm Formation in Caries-Active Children: An In Vitro Study. Antibiotics. 2023; 12(8):1263. https://doi.org/10.3390/antibiotics12081263
Chicago/Turabian StyleParga, Ana, Sabela Balboa, Paz Otero-Casal, and Ana Otero. 2023. "New Preventive Strategy against Oral Biofilm Formation in Caries-Active Children: An In Vitro Study" Antibiotics 12, no. 8: 1263. https://doi.org/10.3390/antibiotics12081263
APA StyleParga, A., Balboa, S., Otero-Casal, P., & Otero, A. (2023). New Preventive Strategy against Oral Biofilm Formation in Caries-Active Children: An In Vitro Study. Antibiotics, 12(8), 1263. https://doi.org/10.3390/antibiotics12081263







