FTY720 Reduces the Biomass of Biofilms in Pseudomonas aeruginosa in a Dose-Dependent Manner
Abstract
1. Introduction
2. Results
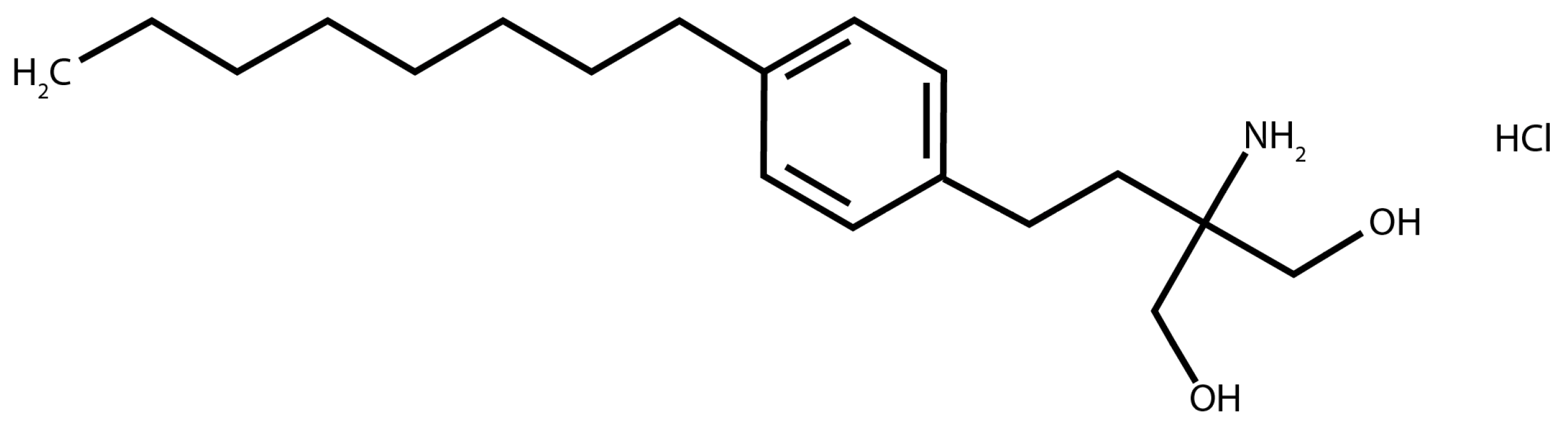
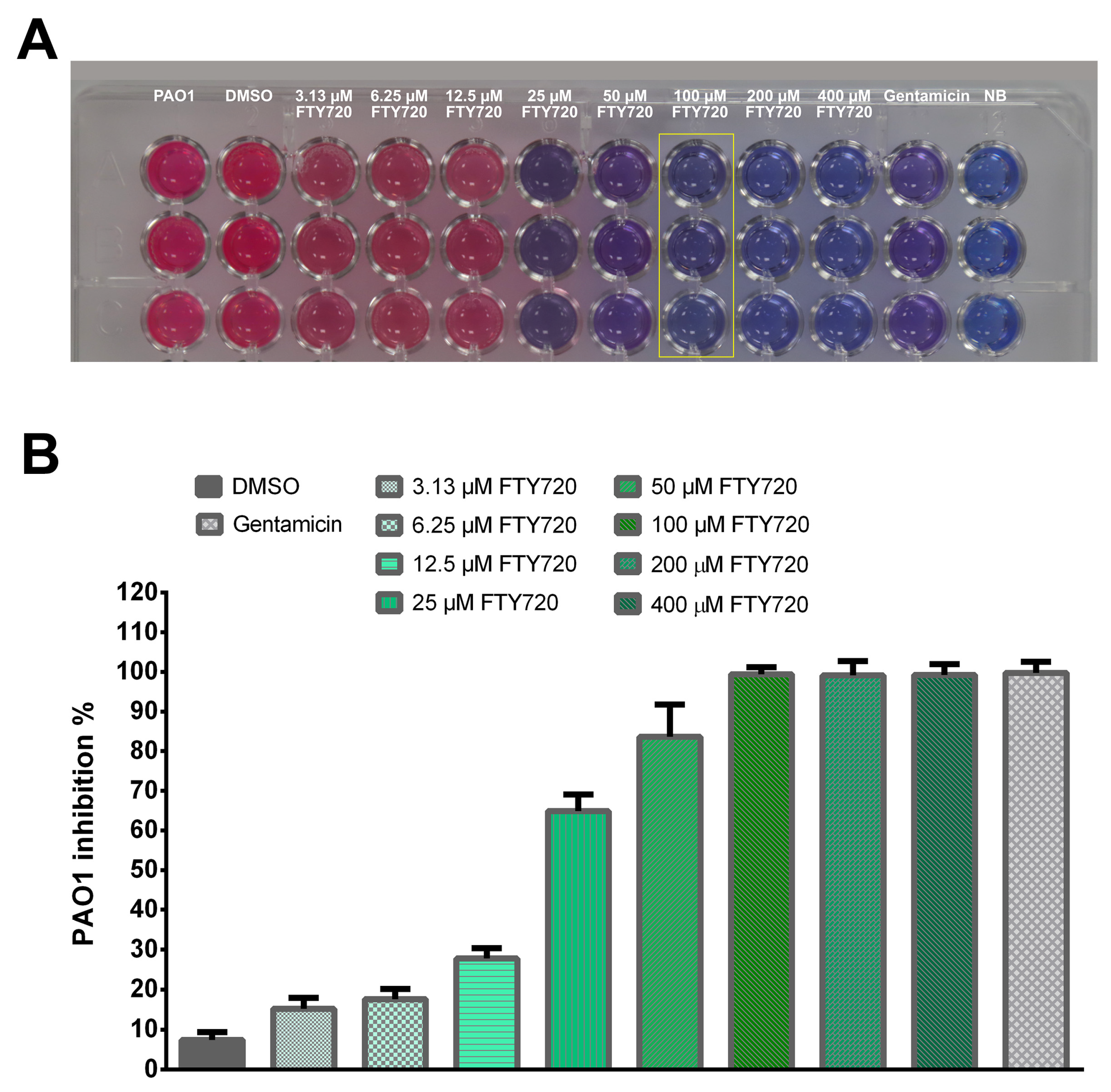
2.1. Resazurin-Based Turbidometric Assay for Minimum Inhibitory Concentration (MIC) Determination
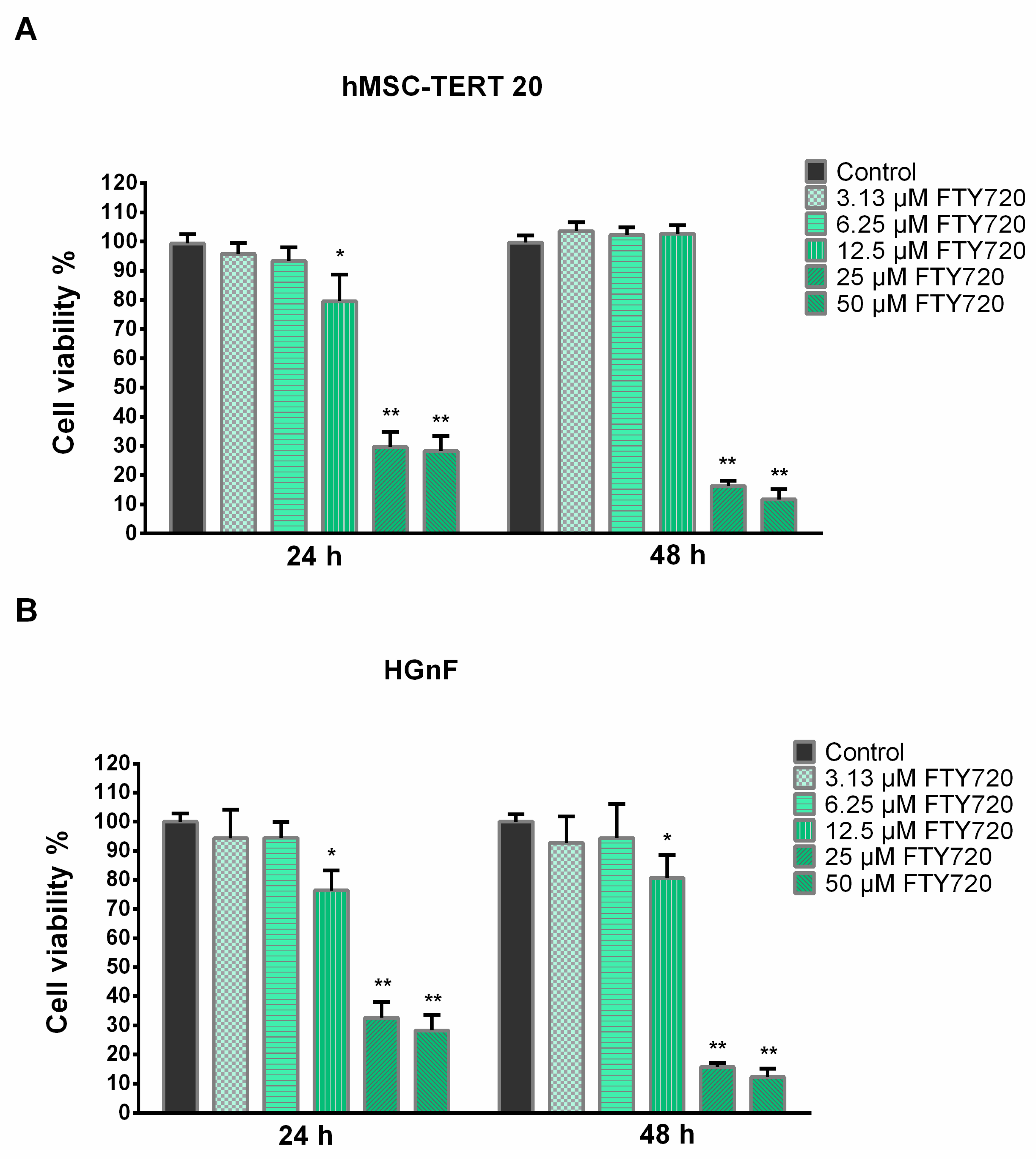
2.2. Cytotoxicity
2.3. Growth Curve and Metabolic Activities
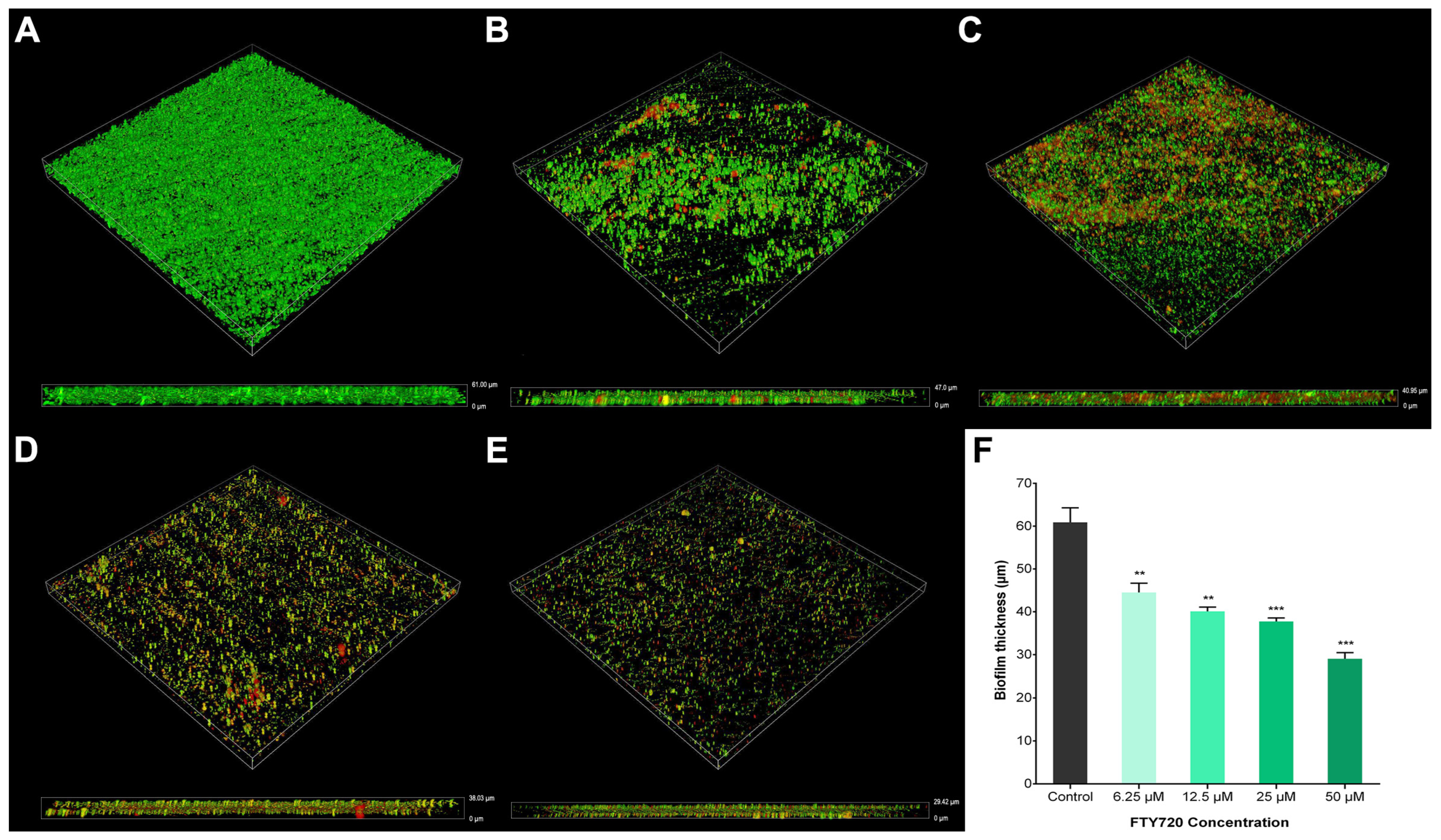
2.4. Biofilm Formation
2.5. Biomass in Biofilms
2.6. Antibacterial Activity
2.7. Pigment Production
2.8. Expression of Biofilm-Relevant Genes
3. Discussion
4. Materials and Methods
4.1. Bacterial Strain, Culture Conditions, and Chemicals
4.2. Resazurin-Based Turbidometric Assay and MIC Determination
4.3. Cytotoxicity Studies on Human Cell Lines
4.4. Growth Curve
4.5. Biofilm Inhibition Assay
4.6. CLSM Analysis of Biofilm Morphology
4.7. Pigment Production
4.7.1. Pyocyanin Assay
4.7.2. Pyoverdine Assay
4.8. RT-qPCR Analysis
4.9. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Sauer, K.; Stoodley, P.; Goeres, D.M.; Hall-Stoodley, L.; Burmolle, M.; Stewart, P.S.; Bjarnsholt, T. The biofilm life cycle: Expanding the conceptual model of biofilm formation. Nat. Rev. Microbiol. 2022, 20, 608–620. [Google Scholar] [CrossRef] [PubMed]
- Vestby, L.K.; Gronseth, T.; Simm, R.; Nesse, L.L. Bacterial Biofilm and its Role in the Pathogenesis of Disease. Antibiotics 2020, 9, 59. [Google Scholar] [CrossRef] [PubMed]
- Di Domenico, E.G.; Oliva, A.; Guembe, M. The Current Knowledge on the Pathogenesis of Tissue and Medical Device-Related Biofilm Infections. Microorganisms 2022, 10, 1259. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Moser, C.; Wang, H.Z.; Hoiby, N.; Song, Z.J. Strategies for combating bacterial biofilm infections. Int. J. Oral Sci. 2015, 7, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Rice, L.B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: No ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef] [PubMed]
- Kamurai, B.; Mombeshora, M.; Mukanganyama, S. Repurposing of Drugs for Antibacterial Activities on Selected ESKAPE Bacteria Staphylococcus aureus and Pseudomonas aeruginosa. Int. J. Microbiol. 2020, 2020, 8885338. [Google Scholar] [CrossRef] [PubMed]
- Di Bonaventura, G.; Lupetti, V.; Di Giulio, A.; Muzzi, M.; Piccirilli, A.; Cariani, L.; Pompilio, A. Repurposing High-Throughput Screening Identifies Unconventional Drugs with Antibacterial and Antibiofilm Activities against Pseudomonas aeruginosa under Experimental Conditions Relevant to Cystic Fibrosis. Microbiol. Spectr. 2023, 11, e0035223. [Google Scholar] [CrossRef] [PubMed]
- Pushpakom, S.; Iorio, F.; Eyers, P.A.; Escott, K.J.; Hopper, S.; Wells, A.; Doig, A.; Guilliams, T.; Latimer, J.; McNamee, C.; et al. Drug repurposing: Progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019, 18, 41–58. [Google Scholar] [CrossRef] [PubMed]
- Barbarossa, A.; Rosato, A.; Corbo, F.; Clodoveo, M.L.; Fracchiolla, G.; Carrieri, A.; Carocci, A. Non-Antibiotic Drug Repositioning as an Alternative Antimicrobial Approach. Antibiotics 2022, 11, 816. [Google Scholar] [CrossRef]
- Hegazy, W.A.H.; Khayat, M.T.; Ibrahim, T.S.; Nassar, M.S.; Bakhrebah, M.A.; Abdulaal, W.H.; Alhakamy, N.A.; Bendary, M.M. Repurposing Anti-diabetic Drugs to Cripple Quorum Sensing in Pseudomonas aeruginosa. Microorganisms 2020, 8, 1285. [Google Scholar] [CrossRef]
- Mishra, A.S.; Vasanthan, M.; Malliappan, S.P. Drug Repurposing: A Leading Strategy for New Threats and Targets. ACS Pharmacol. Transl. Sci. 2024, 7, 915–932. [Google Scholar] [CrossRef]
- Rivera, A.; Heitman, J. Natural product ligands of FKBP12: Immunosuppressive antifungal agents FK506, rapamycin, and beyond. PLoS Pathog. 2023, 19, e1011056. [Google Scholar] [CrossRef]
- Gracia, J.; Perumal, D.; Dhandapani, P.; Ragunathan, P. Systematic identification and repurposing of FDA-approved drugs as antibacterial agents against Streptococcus pyogenes: In silico and in vitro studies. Int. J. Biol. Macromol. 2024, 257, 128667. [Google Scholar] [CrossRef]
- Sharma, S.; Mathur, A.G.; Pradhan, S.; Singh, D.B.; Gupta, S. Fingolimod (FTY720): First approved oral therapy for multiple sclerosis. J. Pharmacol. Pharmacother. 2011, 2, 49–51. [Google Scholar] [CrossRef]
- Brinkmann, V.; Billich, A.; Baumruker, T.; Heining, P.; Schmouder, R.; Francis, G.; Aradhye, S.; Burtin, P. Fingolimod (FTY720): Discovery and development of an oral drug to treat multiple sclerosis. Nat. Rev. Drug Discov. 2010, 9, 883–897. [Google Scholar] [CrossRef]
- Zecri, F.J. From Natural Product to the First Oral Treatment for Multiple Sclerosis: The Discovery of FTY720 (Gilenya)? Curr. Opin. Chem. Biol. 2016, 32, 60–66. [Google Scholar] [CrossRef]
- Baer, A.; Colon-Moran, W.; Bhattarai, N. Characterization of the effects of immunomodulatory drug fingolimod (FTY720) on human T cell receptor signaling pathways. Sci. Rep. 2018, 8, 10910. [Google Scholar] [CrossRef]
- Dyckman, A.J. Modulators of Sphingosine-1-phosphate Pathway Biology: Recent Advances of Sphingosine-1-phosphate Receptor 1 (S1P1) Agonists and Future Perspectives. J. Med. Chem. 2017, 60, 5267–5289. [Google Scholar] [CrossRef]
- Blanc, C.A.; Rosen, H.; Lane, T.E. FTY720 (fingolimod) modulates the severity of viral-induced encephalomyelitis and demyelination. J. Neuroinflamm. 2014, 11, 138. [Google Scholar] [CrossRef]
- Jin, H.W.; Kim, H.R.; Eom, Y.B. Fingolimod Promotes Antibacterial Effect of Doripenem against Carbapenem-Resistant Escherichia coli. Antibiotics 2022, 11, 1043. [Google Scholar] [CrossRef]
- Rumah, K.R.; Vartanian, T.K.; Fischetti, V.A. Oral Multiple Sclerosis Drugs Inhibit the In vitro Growth of Epsilon Toxin Producing Gut Bacterium, Clostridium perfringens. Front. Cell. Infect. Microbiol. 2017, 7, 11. [Google Scholar] [CrossRef] [PubMed]
- Zore, M.; Gilbert-Girard, S.; Reigada, I.; Patel, J.Z.; Savijoki, K.; Fallarero, A.; Yli-Kauhaluoma, J. Synthesis and Biological Evaluation of Fingolimod Derivatives as Antibacterial Agents. ACS Omega 2021, 6, 18465–18486. [Google Scholar] [CrossRef]
- Wei, L.Q.; Tan, J.C.; Wang, Y.; Mei, Y.K.; Xue, J.Y.; Tian, L.; Song, K.Y.; Han, L.; Cui, Y.C.; Peng, Y.B.; et al. Fingolimod Potentiates the Antifungal Activity of Amphotericin B. Front. Cell. Infect. Microbiol. 2021, 11, 627917. [Google Scholar] [CrossRef] [PubMed]
- Najarzadegan, N.; Madani, M.; Etemadifar, M.; Sedaghat, N. Immunomodulatory drug fingolimod (FTY720) restricts the growth of opportunistic yeast Candida albicans in vitro and in a mouse candidiasis model. PLoS ONE 2022, 17, e0278488. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Letsiou, E.; Wang, H.; Belvitch, P.; Meliton, L.N.; Brown, M.E.; Bandela, M.; Chen, J.; Garcia, J.G.N.; Dudek, S.M. MRSA-induced endothelial permeability and acute lung injury are attenuated by FTY720 S-phosphonate. Am. J. Physiol. Lung Cell. Mol. Physiol. 2022, 322, L149–L161. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Xie, L.; Wang, L.; Xue, M.; Xu, D.; Zhong, G. Effects of Immunomodulatory Drug Fingolimod (FTY720) on Chlamydia Dissemination and Pathogenesis. Infect. Immun. 2020, 88, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Murphy, C.T.; Hall, L.J.; Hurley, G.; Quinlan, A.; MacSharry, J.; Shanahan, F.; Nally, K.; Melgar, S. The sphingosine-1-phosphate analogue FTY720 impairs mucosal immunity and clearance of the enteric pathogen Citrobacter rodentium. Infect. Immun. 2012, 80, 2712–2723. [Google Scholar] [CrossRef] [PubMed]
- Geng, X.; Yang, Y.J.; Li, Z.; Ge, W.B.; Xu, X.; Liu, X.W.; Li, J.Y. Fingolimod Inhibits Exopolysaccharide Production and Regulates Relevant Genes to Eliminate the Biofilm of K. pneumoniae. Int. J. Mol. Sci. 2024, 25, 1397. [Google Scholar] [CrossRef] [PubMed]
- Thi, M.T.T.; Wibowo, D.; Rehm, B.H.A. Pseudomonas aeruginosa Biofilms. Int. J. Mol. Sci. 2020, 21, 8671. [Google Scholar] [CrossRef]
- Jimenez Otero, F.; Newman, D.K.; Tender, L.M. Pyocyanin-dependent electrochemical inhibition of Pseudomonas aeruginosa biofilms is synergistic with antibiotic treatment. mBio 2023, 14, e0070223. [Google Scholar] [CrossRef]
- Fekete-Kertesz, I.; Berkl, Z.; Buda, K.; Fenyvesi, E.; Szente, L.; Molnar, M. Quorum quenching effect of cyclodextrins on the pyocyanin and pyoverdine production of Pseudomonas aeruginosa. Appl. Environ. 2024, 108, 271. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.; Pham, D.T.N.; Oloketuyi, S.F.; Kim, Y.M. Regulation and controlling the motility properties of Pseudomonas aeruginosa. Appl. Environ. 2020, 104, 33–49. [Google Scholar] [CrossRef] [PubMed]
- Kazmierczak, B.I.; Schniederberend, M.; Jain, R. Cross-regulation of Pseudomonas motility systems: The intimate relationship between flagella, pili and virulence. Curr. Opin. Chem. Biol. 2015, 28, 78–82. [Google Scholar] [CrossRef] [PubMed]
- Okkotsu, Y.; Tieku, P.; Fitzsimmons, L.F.; Churchill, M.E.; Schurr, M.J. Pseudomonas aeruginosa AlgR phosphorylation modulates rhamnolipid production and motility. J. Bacteriol. 2013, 195, 5499–5515. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Mawgoud, A.M.; Hausmann, R.; Lépine, F.; Müller, M.M.; Déziel, E. Rhamnolipids: Detection, analysis, biosynthesis, genetic regulation, and bioengineering of production. In Biosurfactants; Microbiology Monographs; Springer: Berlin/Heidelberg, Germany, 2011; pp. 13–55. [Google Scholar] [CrossRef]
- Stover, C.K.; Pham, X.Q.; Erwin, A.L.; Mizoguchi, S.D.; Warrener, P.; Hickey, M.J.; Brinkman, F.S.; Hufnagle, W.O.; Kowalik, D.J.; Lagrou, M.; et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 2000, 406, 959–964. [Google Scholar] [CrossRef] [PubMed]
- Grace, A.; Sahu, R.; Owen, D.R.; Dennis, V.A. Pseudomonas aeruginosa reference strains PAO1 and PA14: A genomic, phenotypic, and therapeutic review. Front. Microbiol. 2022, 13, 1023523. [Google Scholar] [CrossRef] [PubMed]
- Nikaido, H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 2003, 67, 593–656. [Google Scholar] [CrossRef] [PubMed]
- Tseng, B.S.; Reichhardt, C.; Merrihew, G.E.; Araujo-Hernandez, S.A.; Harrison, J.J.; MacCoss, M.J.; Parsek, M.R. A Biofilm Matrix-Associated Protease Inhibitor Protects Pseudomonas aeruginosa from Proteolytic Attack. mBio 2018, 9, 10-1128. [Google Scholar] [CrossRef]
- Gilbert-Girard, S.; Savijoki, K.; Yli-Kauhaluoma, J.; Fallarero, A. Screening of FDA-Approved Drugs Using a 384-Well Plate-Based Biofilm Platform: The Case of Fingolimod. Microorganisms 2020, 8, 1834. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Thomsen, T.R.; Winkler, H.; Xu, Y. Influence of biofilm growth age, media, antibiotic concentration and exposure time on Staphylococcus aureus and Pseudomonas aeruginosa biofilm removal in vitro. BMC Microbiol. 2020, 20, 264. [Google Scholar] [CrossRef]
- da Cruz Nizer, W.S.; Inkovskiy, V.; Overhage, J. Surviving Reactive Chlorine Stress: Responses of Gram-Negative Bacteria to Hypochlorous Acid. Microorganisms 2020, 8, 1220. [Google Scholar] [CrossRef]
- Zore, M.; Gilbert-Girard, S.; San-Martin-Galindo, P.; Reigada, I.; Hanski, L.; Savijoki, K.; Fallarero, A.; Yli-Kauhaluoma, J.; Patel, J.Z. Repurposing the Sphingosine-1-Phosphate Receptor Modulator Etrasimod as an Antibacterial Agent Against Gram-Positive Bacteria. Front. Microbiol. 2022, 13, 926170. [Google Scholar] [CrossRef]
- Duplantier, M.; Lohou, E.; Sonnet, P. Quorum Sensing Inhibitors to Quench P. aeruginosa Pathogenicity. Pharmaceuticals 2021, 14, 1262. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Perez, S.P.; Solis, C.S.; Lopez-Bucio, J.S.; Valdez Alarcon, J.J.; Villegas, J.; Reyes-De la Cruz, H.; Campos-Garcia, J. Pathogenesis in Pseudomonas aeruginosa PAO1 Biofilm-Associated Is Dependent on the Pyoverdine and Pyocyanin Siderophores by Quorum Sensing Modulation. Microb. Ecol. 2023, 86, 727–741. [Google Scholar] [CrossRef]
- Costa, K.C.; Glasser, N.R.; Conway, S.J.; Newman, D.K. Pyocyanin degradation by a tautomerizing demethylase inhibits Pseudomonas aeruginosa biofilms. Science 2017, 355, 170–173. [Google Scholar] [CrossRef]
- Meirelles, L.A.; Newman, D.K. Both toxic and beneficial effects of pyocyanin contribute to the lifecycle of Pseudomonas aeruginosa. Mol. Microbiol. 2018, 110, 995–1010. [Google Scholar] [CrossRef] [PubMed]
- Quinn, R.J.; Barraza, I.; García-Diéguez, L.; Pajon, C.; Krausfeldt, L.E.; Ibrahim, K.; Enzinna, L.A.; Thorn, M.E.; Eldakar, O.T.; Craddock, T.J.; et al. Periodically disturbing the spatial structure of biofilms can affect the production of an essential virulence factor in Pseudomonas aeruginosa. Msystems 2021, 6, 10-1128. [Google Scholar] [CrossRef]
- Boles, B.R.; Thoendel, M.; Singh, P.K. Rhamnolipids mediate detachment of Pseudomonas aeruginosa from biofilms. Mol. Microbiol. 2005, 57, 1210–1223. [Google Scholar] [CrossRef]
- Bogiel, T.; Depka, D.; Rzepka, M.; Kwiecinska-Pirog, J.; Gospodarek-Komkowska, E. Prevalence of the Genes Associated with Biofilm and Toxins Synthesis amongst the Pseudomonas aeruginosa Clinical Strains. Antibiotics 2021, 10, 241. [Google Scholar] [CrossRef] [PubMed]
- Coggan, K.A.; Higgs, M.G.; Brutinel, E.D.; Marden, J.N.; Intile, P.J.; Winther-Larsen, H.C.; Koomey, M.; Yahr, T.L.; Wolfgang, M.C. Global Regulatory Pathways Converge To Control Expression of Pseudomonas aeruginosa Type IV Pili. mBio 2022, 13, e0369621. [Google Scholar] [CrossRef]
- Daboor, S.M.; Raudonis, R.; Cheng, Z. Characterizations of the viability and gene expression of dispersal cells from Pseudomonas aeruginosa biofilms released by alginate lyase and tobramycin. PLoS ONE 2021, 16, e0258950. [Google Scholar] [CrossRef] [PubMed]
- Sousa, S.A.; Feliciano, J.R.; Pita, T.; Soeiro, C.F.; Mendes, B.L.; Alves, L.G.; Leitao, J.H. Bacterial Nosocomial Infections: Multidrug Resistance as a Trigger for the Development of Novel Antimicrobials. Antibiotics 2021, 10, 942. [Google Scholar] [CrossRef] [PubMed]
- Castro, M.R.; Dias, G.M.; Salles, T.S.; Cabral, N.M.; Mariano, D.C.O.; Oliveira, H.L.; Abdelhay, E.; Binato, R.; Neves, B.C. Genome-wide analysis reveals a rhamnolipid-dependent modulation of flagellar genes in Pseudomonas aeruginosa PAO1. Curr. Genet. 2022, 68, 289–304. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, J.B.; Izano, E.A.; Gopal, P.; Karwacki, M.T.; Kim, S.; Bose, J.L.; Bayles, K.W.; Horswill, A.R. Low levels of beta-lactam antibiotics induce extracellular DNA release and biofilm formation in Staphylococcus aureus. mBio 2012, 3, e00198-12. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Melcon, C.; Capita, R.; Rodriguez-Jerez, J.J.; Martinez-Suarez, J.V.; Alonso-Calleja, C. Effect of Low Doses of Disinfectants on the Biofilm-Forming Ability of Listeria monocytogenes. Foodborne Pathog. Dis. 2019, 16, 262–268. [Google Scholar] [CrossRef]
- Oliveira, N.M.; Martinez-Garcia, E.; Xavier, J.; Durham, W.M.; Kolter, R.; Kim, W.; Foster, K.R. Biofilm Formation As a Response to Ecological Competition. PLoS Biol. 2015, 13, e1002191. [Google Scholar] [CrossRef]
- Kassis, I.; Ben-Zwi, M.; Petrou, P.; Halimi, M.; Karussis, D. Synergistic neuroprotective effects of Fingolimod and mesenchymal stem cells (MSC) in experimental autoimmune encephalomyelitis. Immunol. Lett. 2021, 233, 11–19. [Google Scholar] [CrossRef]
- Bravo, G.A.; Cedeno, R.R.; Casadevall, M.P.; Ramio-Torrenta, L. Sphingosine-1-Phosphate (S1P) and S1P Signaling Pathway Modulators, from Current Insights to Future Perspectives. Cells 2022, 11, 2058. [Google Scholar] [CrossRef] [PubMed]
- Pournajaf, S.; Valian, N.; Mohaghegh Shalmani, L.; Khodabakhsh, P.; Jorjani, M.; Dargahi, L. Fingolimod increases oligodendrocytes markers expression in epidermal neural crest stem cells. Eur. J. Pharmacol. 2020, 885, 173502. [Google Scholar] [CrossRef]
- De Kleijn, K.M.A.; Martens, G.J.M. Molecular Effects of FDA-Approved Multiple Sclerosis Drugs on Glial Cells and Neurons of the Central Nervous System. Int. J. Mol. Sci. 2020, 21, 4229. [Google Scholar] [CrossRef]
- Balasubramanian, S.; Thej, C.; Venugopal, P.; Priya, N.; Zakaria, Z.; Sundarraj, S.; Majumdar, A.S. Higher Propensity of Wharton’s Jelly Derived Mesenchymal Stromal Cells towards Neuronal Lineage in Comparison to Those Derived from Adipose and Bone Marrow: Increased Neuronal Differentiation Propensity of Wharton’s Jelly Derived MSCs. Cell Biol. Int. 2013, 37, 507–515. [Google Scholar] [CrossRef]
- Brichta, D.M.; Azad, K.N.; Ralli, P.; O’Donovan, G.A. Pseudomonas aeruginosa dihydroorotases: A tale of three pyrCs. Arch. Microbiol. 2004, 182, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Elshikh, M.; Ahmed, S.; Funston, S.; Dunlop, P.; McGaw, M.; Marchant, R.; Banat, I.M. Resazurin-based 96-well plate microdilution method for the determination of minimum inhibitory concentration of biosurfactants. Biotechnol. Lett. 2016, 38, 1015–1019. [Google Scholar] [CrossRef]
- Vidal-Aroca, F.; Meng, A.; Minz, T.; Page, M.G.; Dreier, J. Use of resazurin to detect mefloquine as an efflux-pump inhibitor in Pseudomonas aeruginosa and Escherichia coli. J. Microbiol. Methods 2009, 79, 232–237. [Google Scholar] [CrossRef]
- Lalitha, M. Manual on antimicrobial susceptibility testing. Perform. Stand. Antimicrob. Test. Twelfth Inf. Suppl. 2004, 56238, 454–456. [Google Scholar]
- Aljarbou, F.; Niazy, A.A.; Lambarte, R.N.A.; Mothana, R.A.; Binrayes, A.; Al-Obaida, M.; Alamri, H.M. Efficacy of Salvadora persica root extract as an endodontic irrigant–an in-vitro evaluation. J. Herb. Med. 2022, 34, 100564. [Google Scholar] [CrossRef]
- Alfawaz, A.; Alzahrani, K.; Niazy, A.; Alghamadi, H.; Lambarte, R.; Beagan, A.; Alfhaid, L.; Alotaibi, K.; Alswieleh, A. Smart nanocarrier based on poly (oligo (ethylene glycol) methyl ether acrylate) terminated pH-responsive polymer brushes grafted mesoporous silica nanoparticles. Appl. Sci. 2022, 12, 3688. [Google Scholar] [CrossRef]
- Yang, L.; Wang, L.; Wang, M.; Bajinka, O.; Wu, G.; Qin, L.; Tan, Y. Oligoribonuclease mediates high adaptability of P. aeruginosa through metabolic conversion. BMC Microbiol. 2024, 24, 25. [Google Scholar] [CrossRef]
- Niazy, A.A.; Lambarte, R.N.A.; Alghamdi, H.S. De novo pyrimidine synthesis pathway inhibition reduces motility virulence of Pseudomonas aeruginosa despite complementation. J. King Saud Univ. Sci. 2022, 34, 102040. [Google Scholar] [CrossRef]
- Wang, Q.; Sun, F.J.; Liu, Y.; Xiong, L.R.; Xie, L.L.; Xia, P.Y. Enhancement of biofilm formation by subinhibitory concentrations of macrolides in icaADBC-positive and -negative clinical isolates of Staphylococcus epidermidis. Antimicrob. Agents Chemother. 2010, 54, 2707–2711. [Google Scholar] [CrossRef]
- Patel, H.; Gajjar, D. Cell adhesion and twitching motility influence strong biofilm formation in Pseudomonas aeruginosa. Biofouling 2022, 38, 235–249. [Google Scholar] [CrossRef] [PubMed]
- Rasband, W.S. ImageJ; US National Institutes of Health: Bethesda, MD, USA, 2011.
- Alsubait, S.; Albader, S.; Alajlan, N.; Alkhunaini, N.; Niazy, A.; Almahdy, A. Comparison of the antibacterial activity of calcium silicate- and epoxy resin-based endodontic sealers against Enterococcus faecalis biofilms: A confocal laser-scanning microscopy analysis. Odontology 2019, 107, 513–520. [Google Scholar] [CrossRef]
- Lekbach, Y.; Dong, Y.; Li, Z.; Xu, D.; El Abed, S.; Yi, Y.; Li, L.; Koraichi, S.I.; Sun, T.; Wang, F. Catechin hydrate as an eco-friendly biocorrosion inhibitor for 304L stainless steel with dual-action antibacterial properties against Pseudomonas aeruginosa biofilm. Corros. Sci. 2019, 157, 98–108. [Google Scholar] [CrossRef]
- Niazy, A.; Hughes, L.E. Accumulation of pyrimidine intermediate orotate decreases virulence factor production in Pseudomonas aeruginosa. Curr. Microbiol. 2015, 71, 229–234. [Google Scholar] [CrossRef]
- King, E.O.; Ward, M.K.; Raney, D.E. Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 1954, 44, 301–307. [Google Scholar] [CrossRef]
- Hosseinidoust, Z.; Tufenkji, N.; van de Ven, T.G. Predation in homogeneous and heterogeneous phage environments affects virulence determinants of Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2013, 79, 2862–2871. [Google Scholar] [CrossRef] [PubMed]
- Geng, Y.F.; Yang, C.; Zhang, Y.; Tao, S.N.; Mei, J.; Zhang, X.C.; Sun, Y.J.; Zhao, B.T. An innovative role for luteolin as a natural quorum sensing inhibitor in Pseudomonas aeruginosa. Life Sci. 2021, 274, 119325. [Google Scholar] [CrossRef] [PubMed]





| Genes | Primer Sequence (5′-3′) |
|---|---|
| rhlA | F: GGC GAT CGG CCA TCT G R: AGC GAA GCC ATG TGC TGA T |
| rhlB | F: GCC TGT CGG CGT TTC ATG R: CAT CCC CCT CCC TAT GAC AA |
| pilA | F: TGC GCG TTC GGA AGG T R: CGA CTC TTC AAC AGT GGT CTT CA |
| pilI | F: GCA CTG CAA CCC TTC ATT CAT R: CGC ATG CGG GCT GAA C |
| fliC | F: CAG TGC CAA GGA CGA TGC T R: AAC GTT CAG ACC GCT GAT CTG |
| fliD | F: TGG CTG GCA CCT ACC AGA TC R: GGC CTG GAG GGC AAT CTT |
| algR | F: CCT CGG CCA GCA ATG G R: CGG ATA TCC AGC AGG ACG AT |
| 16S rRNA | F: GCGCAACCCTTGTCCTTAGTT R: TGT CACCGGCAGTCTCCTTAG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niazy, A.A.; Lambarte, R.N.A.; Sumague, T.S.; Vigilla, M.G.B.; Bin Shwish, N.M.; Kamalan, R.; Daeab, E.K.; Aljehani, N.M. FTY720 Reduces the Biomass of Biofilms in Pseudomonas aeruginosa in a Dose-Dependent Manner. Antibiotics 2024, 13, 621. https://doi.org/10.3390/antibiotics13070621
Niazy AA, Lambarte RNA, Sumague TS, Vigilla MGB, Bin Shwish NM, Kamalan R, Daeab EK, Aljehani NM. FTY720 Reduces the Biomass of Biofilms in Pseudomonas aeruginosa in a Dose-Dependent Manner. Antibiotics. 2024; 13(7):621. https://doi.org/10.3390/antibiotics13070621
Chicago/Turabian StyleNiazy, Abdurahman A., Rhodanne Nicole A. Lambarte, Terrence S. Sumague, Mary Grace B. Vigilla, Najla M. Bin Shwish, Ranan Kamalan, Eid Khulaif Daeab, and Nami M. Aljehani. 2024. "FTY720 Reduces the Biomass of Biofilms in Pseudomonas aeruginosa in a Dose-Dependent Manner" Antibiotics 13, no. 7: 621. https://doi.org/10.3390/antibiotics13070621
APA StyleNiazy, A. A., Lambarte, R. N. A., Sumague, T. S., Vigilla, M. G. B., Bin Shwish, N. M., Kamalan, R., Daeab, E. K., & Aljehani, N. M. (2024). FTY720 Reduces the Biomass of Biofilms in Pseudomonas aeruginosa in a Dose-Dependent Manner. Antibiotics, 13(7), 621. https://doi.org/10.3390/antibiotics13070621







