Dissemination of Enterococcal Genetic Lineages: A One Health Perspective
Abstract
1. Enterococcus spp.—An Overview
2. Genetic Lineages
3. Enterococcal Fitness Factors, Virulence Traits, and Antimicrobial Resistance
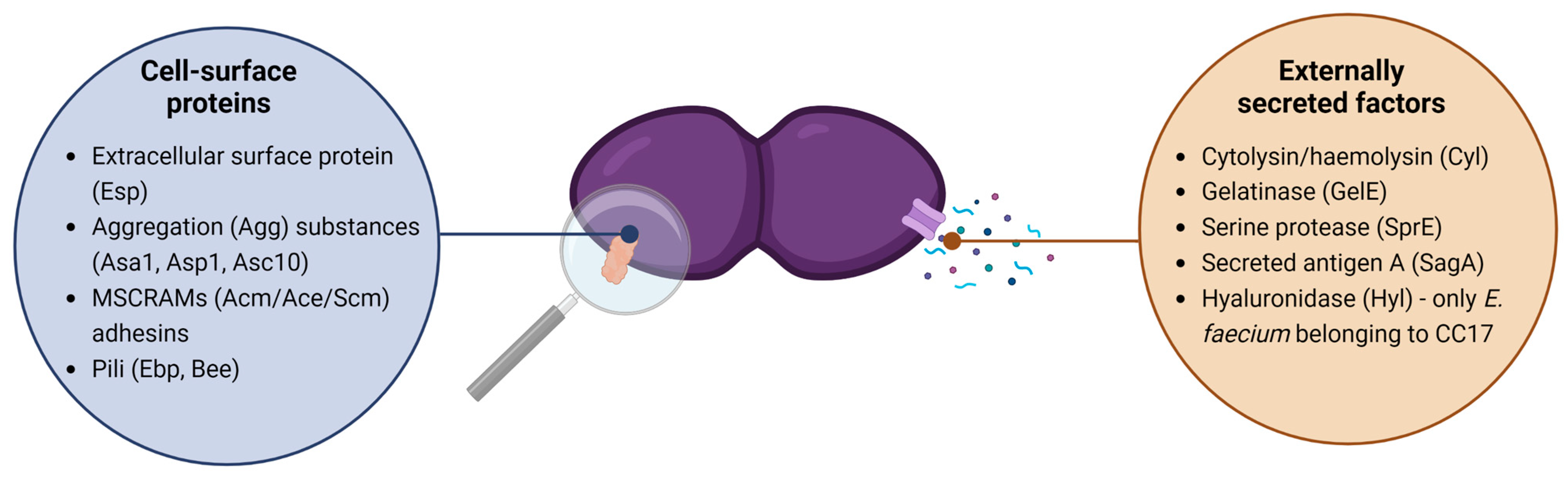
3.1. Fitness Factors
3.2. Virulence Traits
3.3. Antimicrobial Resistance and Associated Mechanisms
4. Enterococcal Genetic Traits: Dissemination in the One Health Continuum
4.1. Human Healthcare-Associated Genetic Traits
4.2. Dissemination of Genetic Traits across One Health Sectors
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Geraldes, C.; Tavares, L.; Gil, S.; Oliveira, M. Enterococcus Virulence and Resistant Traits Associated with Its Permanence in the Hospital Environment. Antibiotics 2022, 11, 857. [Google Scholar] [CrossRef]
- Lebreton, F.; Willems, R.J.L.; Gilmore, M.S. Enterococcus Diversity, Origins in Nature, and Gut Colonization. In Enterococci: From Commensals to Leading Causes of Drug Resistant Infection; Massachusetts Eye and Ear Infirmary: Boston, MA, USA, 2014; pp. 1–46. [Google Scholar]
- Lee, T.; Pang, S.; Abraham, S.; Coombs, G.W. Antimicrobial-Resistant CC17 Enterococcus faecium: The Past, the Present and the Future. J. Glob. Antimicrob. Resist. 2019, 16, 36–47. [Google Scholar] [CrossRef]
- Lebreton, F.; Manson, A.L.; Saavedra, J.T.; Straub, T.J.; Earl, A.M.; Gilmore, M.S. Tracing the Enterococci from Paleozoic Origins to the Hospital. Cell 2017, 169, 849–861. [Google Scholar] [CrossRef] [PubMed]
- Krawczyk, B.; Wityk, P.; Gałęcka, M.; Michalik, M. The Many Faces of Enterococcus spp.—Commensal, Probiotic and Opportunistic Pathogen. Microorganisms 2021, 9, 1900. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.W.; Cha, C.J. Antibiotic Resistome from the One-Health Perspective: Understanding and Controlling Antimicrobial Resistance Transmission. Exp. Mol. Med. 2021, 53, 301–309. [Google Scholar] [CrossRef] [PubMed]
- van Hal, S.J.; Willems, R.J.L.; Gouliouris, T.; Ballard, S.A.; Coque, T.M.; Hammerum, A.M.; Hegstad, K.; Pinholt, M.; Howden, B.P.; Malhotra-Kumar, S.; et al. The Interplay between Community and Hospital Enterococcus faecium Clones within Health-Care Settings: A Genomic Analysis. Lancet Microbe 2022, 3, e133–e141. [Google Scholar] [CrossRef] [PubMed]
- Semedo, T.; Almeida Santos, M.; Silva Lopes, M.F.; Figueiredo Marques, J.J.; Barreto Crespo, M.T.; Tenreiro, R. Virulence Factors in Food, Clinical and Reference Enterococci: A Common Trait in the Genus? Syst. Appl. Microbiol. 2003, 26, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Stępień-Pyśniak, D.; Bertelloni, F.; Dec, M.; Cagnoli, G.; Pietras-Ożga, D.; Urban-Chmiel, R.; Ebani, V.V. Characterization and Comparison of Enterococcus spp. Isolates from Feces of Healthy Dogs and Urine of Dogs with UTIs. Animals 2021, 11, 2845. [Google Scholar] [CrossRef]
- Hashem, Y.A.; Abdelrahman, K.A.; Aziz, R.K. Phenotype–Genotype Correlations and Distribution of Key Virulence Factors in Enterococcus faecalis Isolated from Patients with Urinary Tract Infections. Infect. Drug Resist. 2021, 14, 1713–1723. [Google Scholar] [CrossRef]
- Zhou, X.; Willems, R.J.L.; Friedrich, A.W.; Rossen, J.W.A.; Bathoorn, E. Enterococcus faecium: From Microbiological Insights to Practical Recommendations for Infection Control and Diagnostics. Antimicrob. Resist. Infect. Control. 2020, 9, 1–13. [Google Scholar] [CrossRef]
- Markwart, R.; Willrich, N.; Haller, S.; Noll, I.; Koppe, U.; Werner, G.; Eckmanns, T.; Reuss, A. The Rise in Vancomycin-Resistant Enterococcus faecium in Germany: Data from the German Antimicrobial Resistance Surveillance (ARS). Antimicrob. Resist. Infect. Control. 2019, 8, 1–11. [Google Scholar] [CrossRef]
- Denissen, J.; Reyneke, B.; Waso-Reyneke, M.; Havenga, B.; Barnard, T.; Khan, S.; Khan, W. Prevalence of ESKAPE Pathogens in the Environment: Antibiotic Resistance Status, Community-Acquired Infection and Risk to Human Health. Int. J. Hyg. Environ. Health 2022, 244, 1–17. [Google Scholar] [CrossRef]
- Rice, L.B. Federal Funding for the Study of Antimicrobial Resistance in Nosocomial Pathogens: No ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef] [PubMed]
- Brinkwirth, S.; Ayobami, O.; Eckmanns, T.; Markwart, R. Hospital-Acquired Infections Caused by Enterococci: A Systematic Review and Meta-Analysis, Who European Region, 1 January 2010 to 4 February 2020. Eurosurveillance 2021, 26, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Cattoir, V. The Multifaceted Lifestyle of Enterococci: Genetic Diversity, Ecology and Risks for Public Health. Curr. Opin. Microbiol. 2022, 65, 73–80. [Google Scholar] [CrossRef]
- Palmer, K.L.; Godfrey, P.; Griggs, A.; Kos, V.N.; Zucker, J.; Desjardins, C.; Cerqueira, G.; Gevers, D.; Walker, S.; Wortman, J.; et al. Comparative Genomics of Enterococci: Variation in Enterococcus faecalis, Clade Structure in E. Faecium, and Defining Characteristics of E. gallinarum and E. casseliflavus. mBio 2012, 3, 1–11. [Google Scholar] [CrossRef]
- Sanderson, H.; Gray, K.L.; Manuele, A.; Maguire, F.; Khan, A.; Liu, C.; Navanekere Rudrappa, C.; Nash, J.H.E.; Robertson, J.; Bessonov, K.; et al. Exploring the Mobilome and Resistome of Enterococcus faecium in a One Health Context across Two Continents. Microb. Genom. 2022, 8, 1–17. [Google Scholar] [CrossRef]
- Palmer, K.L.; Gilmore, M.S. Multidrug-Resistant Enterococci Lack CRISPR-Cas. mBio 2010, 1, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Alduhaidhawi, A.H.M.; Alhuchaimi, S.N.; Al-Mayah, T.A.; Al-Ouqaili, M.T.S.; Alkafaas, S.S.; Muthupandian, S.; Saki, M. Prevalence of CRISPR-Cas Systems and Their Possible Association with Antibiotic Resistance in Enterococcus faecalis and Enterococcus faecium Collected from Hospital Wastewater. Infect. Drug Resist. 2022, 15, 1143–1154. [Google Scholar] [CrossRef] [PubMed]
- Galloway-Peña, J.; Roh, J.H.; Latorre, M.; Qin, X.; Murray, B.E. Genomic and SNP Analyses Demonstrate a Distant Separation of the Hospital and Community-Associated Clades of Enterococcus faecium. PLoS ONE 2012, 7, 1–10. [Google Scholar] [CrossRef]
- Lebreton, F.; van Schaik, W.; McGuire, A.M.; Godfrey, P.; Griggs, A.; Mazumdar, V.; Corander, J.; Cheng, L.; Saif, S.; Young, S.; et al. Emergence of Epidemic Multidrug-Resistant Enterococcus faecium from Animal and Commensal Strains. mBio 2013, 4, 1–10. [Google Scholar] [CrossRef]
- Raven, K.E.; Reuter, S.; Reynolds, R.; Brodrick, H.J.; Russell, J.E.; Török, M.E.; Parkhill, J.; Peacock, S.J. A Decade of Genomic History for Healthcare-Associated Enterococcus faecium in the United Kingdom and Ireland. Genome Res. 2016, 26, 1388–1396. [Google Scholar] [CrossRef] [PubMed]
- Leavis, H.L.; Bonten, M.J.; Willems, R.J. Identification of High-Risk Enterococcal Clonal Complexes: Global Dispersion and Antibiotic Resistance. Curr. Opin. Microbiol. 2006, 9, 454–460. [Google Scholar] [CrossRef]
- Gaca, A.O.; Lemos, J.A. Adaptation to Adversity: The Intermingling of Stress Tolerance and Pathogenesis in Enterococci. Microbiol. Mol. Biol. Rev. 2019, 83, 1–46. [Google Scholar] [CrossRef] [PubMed]
- Parga, A.; Manoil, D.; Brundin, M.; Otero, A.; Belibasakis, G.N. Gram-Negative Quorum Sensing Signalling Enhances Biofilm Formation and Virulence Traits in Gram-Positive Pathogen Enterococcus faecalis. J. Oral. Microbiol. 2023, 15, 1–13. [Google Scholar] [CrossRef]
- Fisher, K.; Phillips, C. The Ecology, Epidemiology and Virulence of Enterococcus. Microbiology 2009, 155, 1749–1757. [Google Scholar] [CrossRef] [PubMed]
- Segarra, R.A.; Booth, M.C.; Morales, D.A.; Huycke, M.M.; Gilmore, M.S. Molecular Characterization of the Enterococcus faecalis Cytolysin Activator. Infect. Immun. 1991, 59, 1239–1246. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Garbajosa, P.; Bonten, M.J.M.; Robinson, D.A.; Top, J.; Nallapareddy, S.R.; Torres, C.; Coque, T.M.; Cantón, R.; Baquero, F.; Murray, B.E.; et al. Multilocus Sequence Typing Scheme for Enterococcus faecalis Reveals Hospital-Adapted Genetic Complexes in a Background of High Rates of Recombination. J. Clin. Microbiol. 2006, 44, 2220–2228. [Google Scholar] [CrossRef]
- Gao, W.; Howden, B.P.; Stinear, T.P. Evolution of Virulence in Enterococcus faecium, a Hospital-Adapted Opportunistic Pathogen. Curr. Opin. Microbiol. 2018, 41, 76–82. [Google Scholar] [CrossRef]
- Teng, F.; Kawalec, M.; Weinstock, G.M.; Hryniewicz, W.; Murray, B.E. An Enterococcus faecium Secreted Antigen, SagA, Exhibits Broad-Spectrum Binding to Extracellular Matrix Proteins and Appears Essential for E. faecium Growth. Infect. Immun. 2003, 71, 5033–5041. [Google Scholar] [CrossRef]
- Paganelli, F.L.; de Been, M.; Braat, J.C.; Hoogenboezem, T.; Vink, C.; Bayjanov, J.; Rogers, M.R.C.; Huebner, J.; Bonten, M.J.M.; Willems, R.J.L.; et al. Distinct SagA from Hospital-Associated Clade A1 Enterococcus faecium Strains Contributes to Biofilm Formation. Appl. Environ. Microbiol. 2015, 81, 6873–6882. [Google Scholar] [CrossRef]
- Panesso, D.; Montealegre, M.C.; Rincán, S.; Mojica, M.F.; Rice, L.B.; Singh, K.V.; Murray, B.E.; Arias, C.A. The HylEfm Gene in PHylEfm of Enterococcus faecium Is Not Required in Pathogenesis of Murine Peritonitis. BMC Microbiol. 2011, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Tendolkar, P.M.; Baghdayan, A.S.; Gilmore, M.S.; Shankar, N. Enterococcal Surface Protein, Esp, Enhances Biofilm Formation by Enterococcus faecalis. Infect. Immun. 2004, 72, 6032–6039. [Google Scholar] [CrossRef] [PubMed]
- Heikens, E.; Bonten, M.J.M.; Willems, R.J.L. Enterococcal Surface Protein Esp Is Important for Biofilm Formation of Enterococcus faecium E1162. J. Bacteriol. 2007, 189, 8233–8240. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, S.-Z.; Zaheer, R.; Poulin-Laprade, D.; Scott, A.; Rehman, M.A.; Diarra, M.; Topp, E.; Van Domselaar, G.; Zovoilis, A.; McAllister, T.A. Comparative Genomic Analysis of Enterococci across Sectors of the One Health Continuum. Microorganisms 2023, 11, 727. [Google Scholar] [CrossRef]
- Willems, R.J.L.; Top, J.; Van Santen, M.; Robinson, D.A.; Coque, T.M.; Baquero, F.; Grundmann, H.; Bonten, M.J.M. Global Spread of Vancomycin-Resistant Enterococcus faecium from Distinct Nosocomial Genetic Complex. Emerg. Infect. Dis. 2005, 11, 821–828. [Google Scholar] [CrossRef]
- Zaheer, R.; Cook, S.R.; Barbieri, R.; Goji, N.; Cameron, A.; Petkau, A.; Polo, R.O.; Tymensen, L.; Stamm, C.; Song, J.; et al. Surveillance of Enterococcus spp. Reveals Distinct Species and Antimicrobial Resistance Diversity across a One-Health Continuum. Sci. Rep. 2020, 10, 1–16. [Google Scholar] [CrossRef]
- Top, J.; Willems, R.; Bonten, M. Emergence of CC17 Enterococcus faecium: From Commensal to Hospital-Adapted Pathogen. FEMS Immunol. Med. Microbiol. 2008, 52, 297–308. [Google Scholar] [CrossRef]
- WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed. Available online: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 25 May 2023).
- Ahmed, M.O.; Baptiste, K.E. Vancomycin-Resistant Enterococci: A Review of Antimicrobial Resistance Mechanisms and Perspectives of Human and Animal Health. Microb. Drug Resist. 2018, 24, 590–606. [Google Scholar] [CrossRef]
- Bender, J.K.; Cattoir, V.; Hegstad, K.; Sadowy, E.; Coque, T.M.; Westh, H.; Hammerum, A.M.; Schaffer, K.; Burns, K.; Murchan, S.; et al. Update on Prevalence and Mechanisms of Resistance to Linezolid, Tigecycline and Daptomycin in Enterococci in Europe: Towards a Common Nomenclature. Drug Resist. Updat. 2018, 40, 25–39. [Google Scholar] [CrossRef]
- Miller, W.R.; Munita, J.M.; Arias, C.A. Mechanisms of Antibiotic Resistance in Enterococci. Expert. Rev. Anti. Infect. 2014, 12, 1221–1236. [Google Scholar] [CrossRef]
- Park, I.J.; Wee, G.L.; Jong, H.S.; Kyung, W.L.; Gun, J.W. VanB Phenotype-VanA Genotype Enterococcus faecium with Heterogeneous Expression of Teicoplanin Resistance. J. Clin. Microbiol. 2008, 46, 3091–3093. [Google Scholar] [CrossRef] [PubMed]
- Sivertsen, A.; Pedersen, T.; Larssen, K.W.; Bergh, K.; Rønning, T.G.; Radtke, A.; Hegstad, K. A Silenced VanA Gene Cluster on a Transferable Plasmid Caused an Outbreak of Vancomycin-Variable Enterococci. Antimicrob. Agents Chemother. 2016, 60, 4119–4127. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.H.; Lee, Y.S.; Lee, S.Y.; Yoo, J.S.; Yoo, J.I.; Kim, H.S.; Kim, O.; Yu, J. yon Structure and Transfer of the VanA Cluster in VanA-Positive, Vancomycin-Susceptible Enterococcus faecium, and Its Revertant Mutant. Diagn. Microbiol. Infect. Dis. 2014, 80, 148–150. [Google Scholar] [CrossRef]
- Szakacs, T.A.; Kalan, L.; McConnell, M.J.; Eshaghi, A.; Shahinas, D.; McGeer, A.; Wright, G.D.; Low, D.E.; Patel, S.N. Outbreak of Vancomycin-Susceptible Enterococcus faecium Containing the Wild-Type VanA Gene. J. Clin. Microbiol. 2014, 52, 1682–1686. [Google Scholar] [CrossRef]
- Mutnick, A.H.; Enne, V.; Jones, R.N. Linezolid Resistance since 2001: SENTRY Antimicrobial Surveillance Program. Ann. Pharmacother. 2003, 37, 769–774. [Google Scholar] [CrossRef] [PubMed]
- One Health. Available online: https://www.who.int/news-room/fact-sheets/detail/one-health (accessed on 24 May 2023).
- Mackenzie, J.S.; Jeggo, M. The One Health Approach-Why Is It So Important? Trop. Med. Infect. Dis. 2019, 4, 88. [Google Scholar] [CrossRef]
- Guardabassi, L.; Butaye, P.; Dockrell, D.H.; Fitzgerald, J.R.; Kuijper, E.J. One Health: A Multifaceted Concept Combining Diverse Approaches to Prevent and Control Antimicrobial Resistance. Clin. Microbiol. Infect. 2020, 26, 1604–1605. [Google Scholar] [CrossRef]
- Semedo-Lemsaddek, T.; Pedroso, N.M.; Freire, D.; Nunes, T.; Tavares, L.; Verdade, L.M.; Oliveira, M. Otter Fecal Enterococci as General Indicators of Antimicrobial Resistance Dissemination in Aquatic Environments. Ecol. Indic. 2018, 85, 1113–1120. [Google Scholar] [CrossRef]
- Baquero, F.; Coque, T.M.; Martínez, J.L.; Aracil-Gisbert, S.; Lanza, V.F. Gene Transmission in the One Health Microbiosphere and the Channels of Antimicrobial Resistance. Front. Microbiol. 2019, 10, 1–10. [Google Scholar] [CrossRef]
- Gouliouris, T.; Coll, F.; Ludden, C.; Blane, B.; Raven, K.E.; Naydenova, P.; Crawley, C.; Török, M.E.; Enoch, D.A.; Brown, N.M.; et al. Quantifying Acquisition and Transmission of Enterococcus faecium Using Genomic Surveillance. Nat. Microbiol. 2021, 6, 103–111. [Google Scholar] [CrossRef]
- He, Q.; Hou, Q.; Wang, Y.; Li, J.; Li, W.; Kwok, L.-Y.; Sun, Z.; Zhang, H.; Zhong, Z. Comparative Genomic Analysis of Enterococcus faecalis: Insights into Their Environmental Adaptations. BMC Genom. 2018, 19, 527. [Google Scholar] [CrossRef] [PubMed]
- Guzman Prieto, A.M.; van Schaik, W.; Rogers, M.R.C.; Coque, T.M.; Baquero, F.; Corander, J.; Willems, R.J.L. Global Emergence and Dissemination of Enterococci as Nosocomial Pathogens: Attack of the Clones? Front. Microbiol. 2016, 7, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Suetens, C.; Latour, K.; Kärki, T.; Ricchizzi, E.; Kinross, P.; Moro, M.L.; Jans, B.; Hopkins, S.; Hansen, S.; Lyytikäinen, O.; et al. Prevalence of Healthcare-Associated Infections, Estimated Incidence and Composite Antimicrobial Resistance Index in Acute Care Hospitals and Long-Term Care Facilities: Results from Two European Point Prevalence Surveys, 2016 to 2017. Eurosurveillance 2018, 23, 1–15. [Google Scholar] [CrossRef]
- van Schaik, W.; Willems, R.J.L. Genome-Based Insights into the Evolution of Enterococci. Clin. Microbiol. Infect. 2010, 16, 527–532. [Google Scholar] [CrossRef]
- Nallapareddy, S.R.; Wenxiang, H.; Weinstock, G.M.; Murray, B.E. Molecular Characterization of a Widespread, Pathogenic, and Antibiotic Resistance-Receptive Enterococcus faecalis Lineage and Dissemination of Its Putative Pathogenicity Island. J. Bacteriol. 2005, 187, 5709–5718. [Google Scholar] [CrossRef] [PubMed]
- Willems, R.J.L.; Hanage, W.P.; Bessen, D.E.; Feil, E.J. Population Biology of Gram-Positive Pathogens: High-Risk Clones for Dissemination of Antibiotic Resistance. FEMS Microbiol. Rev. 2011, 35, 872–900. [Google Scholar] [CrossRef] [PubMed]
- McBride, S.M.; Fischetti, V.A.; LeBlanc, D.J.; Moellering, R.C.; Gilmore, M.S. Genetic Diversity among Enterococcus faecalis. PLoS ONE 2007, 2, 1–24. [Google Scholar] [CrossRef]
- Rao, C.; Dhawan, B.; Vishnubhatla, S.; Kapil, A.; Das, B.; Sood, S. Emergence of High-Risk Multidrug-Resistant Enterococcus faecalis CC2 (ST181) and CC87 (ST28) Causing Healthcare-Associated Infections in India. Infect. Genet. Evol. 2020, 85, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kuch, A.; Willems, R.J.L.; Werner, G.; Coque, T.M.; Hammerum, A.M.; Sundsfjord, A.; Klare, I.; Ruiz-Garbajosa, P.; Simonsen, G.S.; van Luit-Asbroek, M.; et al. Insight into Antimicrobial Susceptibility and Population Structure of Contemporary Human Enterococcus faecalis Isolates from Europe. J. Antimicrob. Chemother. 2012, 67, 551–558. [Google Scholar] [CrossRef]
- Kawalec, M.; Pietras, Z.; Daniłowicz, E.; Jakubczak, A.; Gniadkowski, M.; Hryniewicz, W.; Willems, R.J.L. Clonal Structure of Enterococcus faecalis Isolated from Polish Hospitals: Characterization of Epidemic Clones. J. Clin. Microbiol. 2007, 45, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Yan, L.; Yang, S.; Yang, D. Antimicrobial-Resistant Evolution and Global Spread of Enterococcus faecium Clonal Complex (CC) 17: Progressive Change from Gut Colonization to Hospital-Adapted Pathogen. China CDC Wkly. 2022, 4, 17–21. [Google Scholar] [CrossRef] [PubMed]
- Lam, M.M.C.; Seemann, T.; Tobias, N.J.; Chen, H.; Haring, V.; Moore, R.J.; Ballard, S.; Grayson, L.M.; Johnson, P.D.R.; Howden, B.P.; et al. Comparative Analysis of the Complete Genome of an Epidemic Hospital Sequence Type 203 Clone of Vancomycin-Resistant Enterococcus faecium. BMC Genom. 2013, 14, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Buultjens, A.H.; Lam, M.M.C.; Ballard, S.; Monk, I.R.; Mahony, A.A.; Grabsch, E.A.; Grayson, M.L.; Pang, S.; Coombs, G.W.; Robinson, J.O.; et al. Evolutionary Origins of the Emergent ST796 Clone of Vancomycin Resistant Enterococcus faecium. PeerJ 2017, 2017, 1–22. [Google Scholar] [CrossRef]
- Leavis, H.L.; Willems, R.J.L.; Top, J.; Bonten, M.J.M. High-Level Ciprofloxacin Resistance from Point Mutations in GyrA and ParC Confined to Global Hospital-Adapted Clonal Lineage CC17 of Enterococcus faecium. J. Clin. Microbiol. 2006, 44, 1059–1064. [Google Scholar] [CrossRef] [PubMed]
- Galloway-Peña, J.R.; Nallapareddy, S.R.; Arias, C.A.; Eliopoulos, G.M.; Murray, B.E. Analysis of Clonality and Antibiotic Resistance among Early Clinical Isolates of Enterococcus faecium in the United States. J. Infect. Dis. 2009, 200, 1566–1573. [Google Scholar] [CrossRef]
- Mahony, A.A.; Buultjens, A.H.; Ballard, S.A.; Grabsch, E.A.; Xie, S.; Seemann, T.; Stuart, R.L.; Kotsanas, D.; Cheng, A.; Heffernan, H.; et al. Vancomycin-Resistant Enterococcus faecium Sequence Type 796—Rapid International Dissemination of a New Epidemic Clone. Antimicrob. Resist. Infect. Control. 2018, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Coombs, G.W.; Daley, D.A.; Lee, Y.T.; Pang, S. Australian Group on Antimicrobial Resistance (AGAR) Australian Enterococcal Sepsis Outcome Programme (AESOP) Annual Report 2016. Commun. Dis. Intell. 2018, 42, 1–12. [Google Scholar]
- Arias, C.A.; Murray, B.E. The Rise of the Enterococcus: Beyond Vancomycin Resistance. Nat. Rev. Microbiol. 2012, 10, 266–278. [Google Scholar] [CrossRef]
- Palmer, K.L.; van Schaik, W.; Willems, R.J.L.; Gilmore, M.S. Enterococcal Genomics. In Enterococci: From Commensals to Leading Causes of Drug Resistant Infection; Massachusetts Eye and Ear Infirmary: Boston, MA, USA, 2014; pp. 1–37. [Google Scholar]
- Arredondo-Alonso, S.; Top, J.; McNally, A.; Puranen, S.; Pesonen, M.; Pensar, J.; Marttinen, P.; Braat, J.C.; Rogers, M.R.C.; van Schaik, W.; et al. Plasmids Shaped the Recent Emergence of the Major Nosocomial Pathogen Enterococcus faecium. mBio 2020, 11, 1–17. [Google Scholar] [CrossRef]
- Solheim, M.; Brekke, M.C.; Snipen, L.G.; Willems, R.J.L.; Nes, I.F.; Brede, D.A. Comparative Genomic Analysis Reveals Significant Enrichment of Mobile Genetic Elements and Genes Encoding Surface Structure-Proteins in Hospital-Associated Clonal Complex 2 Enterococcus faecalis. BMC Microbiol. 2011, 11, 1–12. [Google Scholar] [CrossRef]
- Hasani, A.; Sharifi, Y.; Ghotaslou, R.; Naghili, B.; Hasani, A.; Aghazadeh, M.; Milani, M.; Bazmani, A. Molecular Screening of Virulence Genes in High-Level Gentamicin-Resistant Enterococcus faecalis and Enterococcus faecium Isolated from Clinical Specimens in Northwest Iran. Indian J. Med. Microbiol. 2012, 30, 175–181. [Google Scholar] [CrossRef]
- Zalipour, M.; Esfahani, B.N.; Halaji, M.; Azimian, A.; Havaei, S.A. Molecular Characterization of Vancomycin-Resistant Enterococcus faecalis among Inpatients at Iranian University Hospitals: Clonal Dissemination of ST6 and ST422. Infect. Drug Resist. 2019, 12, 3039–3047. [Google Scholar] [CrossRef] [PubMed]
- Wardal, E.; Żabicka, D.; Hryniewicz, W.; Sadowy, E. VanA-Enterococcus faecalis in Poland: Hospital Population Clonal Structure and VanA Mobilome. Eur. J. Clin. Microbiol. Infect. Dis. 2022, 41, 1245–1261. [Google Scholar] [CrossRef] [PubMed]
- Palmer, K.L.; Kos, V.N.; Gilmore, M.S. Horizontal Gene Transfer and the Genomics of Enterococcal Antibiotic Resistance. Curr. Opin. Microbiol. 2010, 13, 632–639. [Google Scholar] [CrossRef] [PubMed]
- Willems, R.J.L.; van Schaik, W. Transition of Enterococcus faecium from Commensal Organism to Nosocomial Pathogen. Future Microbiol. 2009, 4, 1125–1135. [Google Scholar] [CrossRef]
- Willems, R.J.L.; Top, J.; van Schaik, W.; Helen, L.; Bonten, M.; Sirén, J.; Hanage, W.P.; Corander, J.; Stephen, K.P. Restricted Gene Flow among Hospital Subpopulations of Enterococcus faecium. mBio 2012, 3, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Hammerum, A.M. Enterococci of Animal Origin and Their Significance for Public Health. Clin. Microbiol. Infect. 2012, 18, 619–625. [Google Scholar] [CrossRef]
- van Hal, S.J.; Willems, R.J.L.; Gouliouris, T.; Ballard, S.A.; Coque, T.M.; Hammerum, A.M.; Hegstad, K.; Westh, H.T.; Howden, B.P.; Malhotra-Kumar, S.; et al. The Global Dissemination of Hospital Clones of Enterococcus faecium. Genome Med. 2021, 13, 1–12. [Google Scholar] [CrossRef]
- van Hal, S.J.; Ip, C.L.C.; Ansari, M.A.; Wilson, D.J.; Espedido, B.A.; Jensen, S.O.; Bowden, R. Evolutionary Dynamics of Enterococcus faecium Reveals Complex Genomic Relationships between Isolates with Independent Emergence of Vancomycin Resistance. Microb. Genom. 2016, 2, 1–11. [Google Scholar] [CrossRef]
- Werner, G.; Neumann, B.; Weber, R.E.; Kresken, M.; Wendt, C.; Bender, J.K.; Becker, K.; Borgmann, S.; Diefenbach, A.; Hamprecht, A.; et al. Thirty Years of VRE in Germany—“Expect the Unexpected”: The View from the National Reference Centre for Staphylococci and Enterococci. Drug Resist. Updat. 2020, 53. [Google Scholar] [CrossRef] [PubMed]
- Fujiya, Y.; Harada, T.; Sugawara, Y.; Akeda, Y.; Yasuda, M.; Masumi, A.; Hayashi, J.; Tanimura, N.; Tsujimoto, Y.; Shibata, W.; et al. Transmission Dynamics of a Linear VanA-Plasmid during a Nosocomial Multiclonal Outbreak of Vancomycin-Resistant Enterococci in a Non-Endemic Area, Japan. Sci. Rep. 2021, 11, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Jiang, Y.; Guo, L.; Ye, L.; Ma, Y.; Luo, Y. Prevalence of Diverse Clones of Vancomycin-Resistant Enterococcus faecium ST78 in a Chinese Hospital. Microb. Drug Resist. 2016, 22, 294–300. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.X.; Li, T.; Ning, Y.Z.; Shao, D.H.; Liu, J.; Wang, S.Q.; Liang, G.W. Molecular Characterization of Resistance, Virulence and Clonality in Vancomycin-Resistant Enterococcus faecium and Enterococcus faecalis: A Hospital-Based Study in Beijing, China. Infect. Genet. Evol. 2015, 33, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Coombs, G.W.; Daley, D.A.; Yee, N.W.T.; Shoby, P.; Mowlaboccus, S. Australian Group on Antimicrobial Resistance (AGAR) Australian Enterococcal Sepsis Outcome Programme (AESOP) Annual Report 2020. Commun. Dis. Intell. 2022, 2018, 46. [Google Scholar] [CrossRef]
- Howden, B.P.; Holt, K.E.; Lam, M.M.C.; Seemann, T.; Ballard, S.; Coombs, G.W.; Tong, S.Y.C.; Grayson, M.L.; Johnson, P.D.R.; Stinear, T.P. Genomic Insights to Control the Emergence of Vancomycin-Resistant Enterococci. mBio 2013, 4, 1–9. [Google Scholar] [CrossRef]
- O’Toole, R.F.; Leong, K.W.C.; Cumming, V.; van Hal, S.J. Vancomycin-Resistant Enterococcus faecium and the Emergence of New Sequence Types Associated with Hospital Infection. Res. Microbiol. 2023, 174, 104046. [Google Scholar] [CrossRef] [PubMed]
- Wassilew, N.; Seth-Smith, H.M.B.; Rolli, E.; Fietze, Y.; Casanova, C.; Führer, U.; Egli, A.; Marschall, J.; Buetti, N. Outbreak of Vancomycin-Resistant Enterococcus faecium Clone ST796, Switzerland, December 2017 to April 2018. Eurosurveillance 2018, 23, 1800351. [Google Scholar] [CrossRef]
- Piezzi, V.; Wassilew, N.; Atkinson, A.; D’Incau, S.; Kaspar, T.; Seth-Smith, H.M.B.; Casanova, C.; Bittel, P.; Jent, P.; Sommerstein, R.; et al. Nosocomial Outbreak of Vancomycin-Resistant Enterococcus faecium (VRE) ST796, Switzerland, 2017 to 2020. Eurosurveillance 2022, 27, 1–11. [Google Scholar] [CrossRef]
- De Oliveira, E.S.; Freitas, A.R.; Peixe, L.; Novais, C.; Melo, M.C. Silent Clonal Spread of Vancomycin-Resistant Enterococcus faecalis ST6 and ST525 Colonizing Patients at Hospital Admission in Natal, Brazil. Infect. Control. Hosp. Epidemiol. 2020, 41, 485–487. [Google Scholar] [CrossRef]
- Pinholt, M.; Bayliss, S.C.; Gumpert, H.; Worning, P.; Jensen, V.V.S.; Pedersen, M.; Feil, E.J.; Westh, H. WGS of 1058 Enterococcus faecium from Copenhagen, Denmark, Reveals Rapid Clonal Expansion of Vancomycin-Resistant Clone ST80 Combined with Widespread Dissemination of a VanA-Containing Plasmid and Acquisition of a Heterogeneous Accessory Genome. J. Antimicrob. Chemother. 2019, 74, 1776–1785. [Google Scholar] [CrossRef]
- Hammerum, A.M.; Justesen, U.S.; Pinholt, M.; Roer, L.; Kaya, H.; Worning, P.; Nygaard, S.; Kemp, M.; Clausen, M.E.; Nielsen, K.L.; et al. Surveillance of Vancomycin-Resistant Enterococci Reveals Shift in Dominating Clones and National Spread of a Vancomycin-Variable VanA Enterococcus faecium ST1421-CT1134 Clone, Denmark, 2015 to March 2019. Eurosurveillance 2019, 24, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Ekwanzala, M.D.; Dewar, J.B.; Kamika, I.; Momba, M.N.B. Comparative Genomics of Vancomycin-Resistant Enterococcus spp. Revealed Common Resistome Determinants from Hospital Wastewater to Aquatic Environments. Sci. Total Environ. 2020, 719, 1–11. [Google Scholar] [CrossRef]
- Zischka, M.; Künne, C.T.; Blom, J.; Wobser, D.; Sakinç, T.; Schmidt-Hohagen, K.; Dabrowski, P.W.; Nitsche, A.; Hübner, J.; Hain, T.; et al. Comprehensive Molecular, Genomic and Phenotypic Analysis of a Major Clone of Enterococcus faecalis MLST ST40. BMC Genom. 2015, 16, 1–20. [Google Scholar] [CrossRef]
- Novais, C.; Freitas, A.R.; Silveira, E.; Antunes, P.; Silva, R.; Coque, T.M.; Peixe, L. Spread of Multidrug-Resistant Enterococcus to Animals and Humans: An Underestimated Role for the Pig Farm Environment. J. Antimicrob. Chemother. 2013, 68, 2746–2754. [Google Scholar] [CrossRef]
- Choi, J.M.; Woo, G.J. Molecular Characterization of High-Level Gentamicin-Resistant Enterococcus faecalis from Chicken Meat in Korea. Int. J. Food Microbiol. 2013, 165, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yang, L.; Huang, X.; Wen, Y.; Zhao, Q.; Huang, X.; Xia, J.; Huang, Y.; Cao, S.; Du, S.; et al. Molecular Characterization of Antimicrobial Resistance and Virulence Factors of Enterococcus faecalis from Ducks at Slaughterhouses. Poult. Sci. 2022, 101, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Sanderson, H.; Ortega-Polo, R.; Zaheer, R.; Goji, N.; Amoako, K.K.; Brown, R.S.; Majury, A.; Liss, S.N.; McAllister, T.A. Comparative Genomics of Multidrug-Resistant Enterococcus spp. Isolated from Wastewater Treatment Plants. BMC Microbiol. 2020, 20, 1–17. [Google Scholar] [CrossRef]
- Wist, V.; Morach, M.; Schneeberger, M.; Cernela, N.; Stevens, M.J.A.; Zurfluh, K.; Stephan, R.; Nüesch-Inderbinen, M. Phenotypic and Genotypic Traits of Vancomycin-Resistant Enterococci from Healthy Food-Producing Animals. Microorganisms 2020, 8, 261. [Google Scholar] [CrossRef]
- Beukers, A.G.; Zaheer, R.; Goji, N.; Amoako, K.K.; Chaves, A.V.; Ward, M.P.; McAllister, T.A. Comparative Genomics of Enterococcus spp. Isolated from Bovine Feces. BMC Microbiol. 2017, 17. [Google Scholar] [CrossRef]
- Fatoba, D.O.; Amoako, D.G.; Akebe, A.L.K.; Ismail, A.; Essack, S.Y. Genomic Analysis of Antibiotic-Resistant Enterococcus spp. Reveals Novel Enterococci Strains and the Spread of Plasmid-Borne Tet(M), Tet(L) and Erm(B) Genes from Chicken Litter to Agricultural Soil in South Africa. J. Environ. Manag. 2022, 302, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Freitas, A.R.; Coque, T.M.; Novais, C.; Hammerum, A.M.; Lester, C.H.; Zervos, M.J.; Donabedian, S.; Jensen, L.B.; Francia, M.V.; Baquero, F.; et al. Human and Swine Hosts Share Vancomycin-Resistant Enterococcus faecium CC17 and CC5 and Enterococcus faecalis CC2 Clonal Clusters Harboring Tn1546 on Indistinguishable Plasmids. J. Clin. Microbiol. 2011, 49, 925–931. [Google Scholar] [CrossRef] [PubMed]
- Sadowy, E.; Luczkiewicz, A. Drug-Resistant and Hospital-Associated Enterococcus faecium from Wastewater, Riverine Estuary and Anthropogenically Impacted Marine Catchment Basin. BMC Microbiol. 2014, 14, 1–15. [Google Scholar] [CrossRef]
- Getachew, Y.; Hassan, L.; Zakaria, Z.; Abdul Aziz, S. Genetic Variability of Vancomycin-Resistant Enterococcus faecium and Enterococcus faecalis Isolates from Humans, Chickens, and Pigs in Malaysia. Appl. Environ. Microbiol. 2013, 79, 4528–4533. [Google Scholar] [CrossRef] [PubMed]
- Trościańczyk, A.; Nowakiewicz, A.; Osińska, M.; Łagowski, D.; Gnat, S.; Chudzik-Rząd, B. Comparative Characteristics of Sequence Types, Genotypes and Virulence of Multidrug-Resistant Enterococcus faecium Isolated from Various Hosts in Eastern Poland. Spread of Clonal Complex 17 in Humans and Animals. Res. Microbiol. 2022, 173, 1–10. [Google Scholar] [CrossRef]
- Finisterra, L.; Duarte, B.; Peixe, L.; Novais, C.; Freitas, A.R. Industrial Dog Food Is a Vehicle of Multidrug-Resistant Enterococci Carrying Virulence Genes Often Linked to Human Infections. Int. J. Food Microbiol. 2021, 358, 1–9. [Google Scholar] [CrossRef] [PubMed]
- López, M.; Sáenz, Y.; Rojo-Bezares, B.; Martínez, S.; del Campo, R.; Ruiz-Larrea, F.; Zarazaga, M.; Torres, C. Detection of VanA and VanB2-Containing Enterococci from Food Samples in Spain, Including Enterococcus faecium Strains of CC17 and the New Singleton ST425. Int. J. Food Microbiol. 2009, 133, 172–178. [Google Scholar] [CrossRef]
- Oravcová, V.; Peixe, L.; Coque, T.M.; Novais, C.; Francia, M.V.; Literák, I.; Freitas, A.R. Wild Corvid Birds Colonized with Vancomycin-Resistant Enterococcus faecium of Human Origin Harbor Epidemic VanA Plasmids. Environ. Int. 2018, 118, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Biggel, M.; Nüesch-Inderbinen, M.; Raschle, S.; Stevens, M.J.A.; Stephan, R. Spread of Vancomycin-Resistant Enterococcus faecium ST133 in the Aquatic Environment in Switzerland. J. Glob. Antimicrob. Resist. 2021, 27, 31–36. [Google Scholar] [CrossRef]
- Belloso Daza, M.V.; Milani, G.; Cortimiglia, C.; Pietta, E.; Bassi, D.; Cocconcelli, P.S. Genomic Insights of Enterococcus faecium UC7251, a Multi-Drug Resistant Strain From Ready-to-Eat Food, Highlight the Risk of Antimicrobial Resistance in the Food Chain. Front. Microbiol. 2022, 13, 1–15. [Google Scholar] [CrossRef]
- Ghosh, A.; KuKanich, K.; Brown, C.E.; Zurek, L. Resident Cats in Small Animal Veterinary Hospitals Carry Multi-Drug Resistant Enterococci and Are Likely Involved in Cross-Contamination of the Hospital Environment. Front. Microbiol. 2012, 3, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Rousham, E.K.; Unicomb, L.; Islam, M.A. Human, Animal and Environmental Contributors to Antibiotic Resistance in Low-Resource Settings: Integrating Behavioural, Epidemiological and One Health Approaches. Proc. R. Soc. B Biol. Sci. 2018, 285, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Guardabassi, L.; Schwarz, S.; Lloyd, D.H. Pet Animals as Reservoirs of Antimicrobial-Resistant Bacteria. J. Antimicrob. Chemother. 2004, 54, 321–332. [Google Scholar] [CrossRef] [PubMed]
- Pomba, C.; Rantala, M.; Greko, C.; Baptiste, K.E.; Catry, B.; van Duijkeren, E.; Mateus, A.; Moreno, M.A.; Pyörälä, S.; Ružauskas, M.; et al. Public Health Risk of Antimicrobial Resistance Transfer from Companion Animals. J. Antimicrob. Chemother. 2017, 72, 957–968. [Google Scholar] [CrossRef]
- Freitas, A.R.; Tedim, A.P.; Almeida-santos, A.C.; Duarte, B.; Elghaieb, H.; Abbassi, M.S.; Hassen, A.; Novais, C. High-Resolution Genotyping Unveils Identical Ampicillin-Resistant Enterococcus faecium Strains in Different Sources and Countries: A One Health Approach. Microorganisms 2022, 10, 632. [Google Scholar] [CrossRef]
- Leite-Martins, L.; Meireles, D.; Bessa, L.J.; Mendes, Â.; de Matos, A.J.; Martins da Costa, P. Spread of Multidrug-Resistant Enterococcus faecalis Within the Household Setting. Microb. Drug Resist. 2014, 20, 501–507. [Google Scholar] [CrossRef]
- Ghosh, A.; Dowd, S.E.; Zurek, L. Dogs Leaving the ICU Carry a Very Large Multi-Drug Resistant Enterococcal Population with Capacity for Biofilm Formation and Horizontal Gene Transfer. PLoS ONE 2011, 6, 1–14. [Google Scholar] [CrossRef]
- Damborg, P.; Sørensen, A.H.; Guardabassi, L. Monitoring of Antimicrobial Resistance in Healthy Dogs: First Report of Canine Ampicillin-Resistant Enterococcus faecium Clonal Complex 17. Vet. Microbiol. 2008, 132, 190–196. [Google Scholar] [CrossRef]
- KuKanich, K.S.; Ghosh, A.; Skarbek, J.V.; Lothamer, K.M.; Zurek, L. Surveillance of Bacterial Contamination in Small Animal Veterinary Hospitals with Special Focus on Antimicrobial Resistance and Virulence Traits of Enterococci. J. Am. Vet. Med. Assoc. 2012, 240, 437–445. [Google Scholar] [CrossRef]
- Jernberg, C.; Löfmark, S.; Edlund, C.; Jansson, J.K. Long-Term Ecological Impacts of Antibiotic Administration on the Human Intestinal Microbiota. ISME J. 2007, 1, 56–66. [Google Scholar] [CrossRef]
- Dethlefsen, L.; Huse, S.; Sogin, M.L.; Relman, D.A. The Pervasive Effects of an Antibiotic on the Human Gut Microbiota, as Revealed by Deep 16S RRNA Sequencing. PLoS Biol. 2008, 6, e280. [Google Scholar] [CrossRef] [PubMed]
- Jernberg, C.; Löfmark, S.; Edlund, C.; Jansson, J.K. Long-Term Impacts of Antibiotic Exposure on the Human Intestinal Microbiota. Microbiology 2010, 156, 3216–3223. [Google Scholar] [CrossRef] [PubMed]
- Antonopoulos, D.A.; Huse, S.M.; Morrison, H.G.; Schmidt, T.M.; Sogin, M.L.; Young, V.B. Reproducible Community Dynamics of the Gastrointestinal Microbiota Following Antibiotic Perturbation. Infect. Immun. 2009, 77, 2367–2375. [Google Scholar] [CrossRef]
- Neumann, B.; Prior, K.; Bender, J.K.; Harmsen, D.; Klare, I.; Fuchs, S.; Bethe, A.; Zühlke, D.; Göhler, A.; Schwarz, S.; et al. A Core Genome Multilocus Sequence Typing Scheme for Enterococcus faecalis. J. Clin. Microbiol. 2019, 57, 1–33. [Google Scholar] [CrossRef] [PubMed]
- Barros, J.; Andrade, M.; Radhouani, H.; López, M.; Igrejas, G.; Poeta, P.; Torres, C. Detection of VanA-Containing Enterococcus Species in Faecal Microbiota of Gilthead Seabream (Sparus aurata). Microbes Environ. 2012, 27, 509–511. [Google Scholar] [CrossRef]
- Almeida, L.M.; Lebreton, F.; Gaca, A.; Bispo, P.M.; Saavedra, J.T.; Calumby, R.N.; Grillo, L.M.; Nascimento, T.G.; Filsner, P.H.; Moreno, A.M.; et al. Transferable Resistance Gene OptrA in Enterococcus faecalis from Swine in Brazil. Antimicrob. Agents Chemother. 2020, 64, 1–23. [Google Scholar] [CrossRef]
- Almeida, L.M.; Gaca, A.; Bispo, P.M.; Lebreton, F.; Saavedra, J.T.; Silva, R.A.; Basílio-Júnior, I.D.; Zorzi, F.M.; Filsner, P.H.; Moreno, A.M.; et al. Coexistence of the Oxazolidinone Resistance–Associated Genes Cfr and OptrA in Enterococcus faecalis From a Healthy Piglet in Brazil. Front. Public Health 2020, 8, 1–8. [Google Scholar] [CrossRef]
- Torres, C.; Alonso, C.A.; Ruiz-Ripa, L.; León-Sampedro, R.; Del Campo, R.; Coque, T.M. Antimicrobial Resistance in Enterococcus Spp. of Animal Origin. Microbiol. Spectr. 2018, 6, 1–41. [Google Scholar] [CrossRef] [PubMed]
- Prichula, J.; Van Tyne, D.; Schwartzman, J.; Sant’Anna, F.H.; Pereira, R.I.; da Cunha, G.R.; Tavares, M.; Lebreton, F.; Frazzon, J.; D’Azevedo, P.A.; et al. Enterococci from Wild Magellanic Penguins (Spheniscus magellanicus) as an Indicator of Marine Ecosystem Health and Human Impact. Appl. Environ. Microbiol. 2020, 86, 1–48. [Google Scholar] [CrossRef]
- Li, W.; Wang, A. Genomic Islands Mediate Environmental Adaptation and the Spread of Antibiotic Resistance in Multiresistant Enterococci—Evidence from Genomic Sequences. BMC Microbiol. 2021, 21, 1–10. [Google Scholar] [CrossRef]
- Boyd, D.A.; Lévesque, S.; Picard, A.C.; Golding, G.R. Vancomycin-Resistant Enterococcus faecium Harbouring VanN in Canada: A Case and Complete Sequence of PEfm12493 Harbouring the VanN Operon. J. Antimicrob. Chemother. 2015, 70, 2163–2165. [Google Scholar] [CrossRef]
- Braga, T.M.; Pomba, C.; Lopes, M.F.S. High-Level Vancomycin Resistant Enterococcus faecium Related to Humans and Pigs Found in Dust from Pig Breeding Facilities. Vet. Microbiol. 2013, 161, 344–349. [Google Scholar] [CrossRef] [PubMed]
- Obeng, A.S.; Rickard, H.; Ndi, O.; Sexton, M.; Barton, M. Comparison of Antimicrobial Resistance Patterns in Enterococci from Intensive and Free Range Chickens in Australia. Avian. Pathol. 2013, 42, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Mencía-Ares, O.; Borowiak, M.; Argüello, H.; Cobo-Díaz, J.F.; Malorny, B.; Álvarez-Ordóñez, A.; Carvajal, A.; Deneke, C. Genomic Insights into the Mobilome and Resistome of Sentinel Microorganisms Originating from Farms of Two Different Swine Production Systems. Microbiol. Spectr. 2022, 10, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Bastião Rocha, P.A.; Monteiro Marques, J.M.; Barreto, A.S.; Semedo-Lemsaddek, T. Enterococcus spp. from Azeitão and Nisa PDO-Cheeses: Surveillance for Antimicrobial Drug Resistance. LWT 2021, 154, 112622. [Google Scholar] [CrossRef]
- Ben Said, L.; Hamdaoui, M.; Klibi, A.; Ben Slama, K.; Torres, C.; Klibi, N. Diversity of Species and Antibiotic Resistance in Enterococci Isolated from Seafood in Tunisia. Ann. Microbiol. 2017, 67, 135–141. [Google Scholar] [CrossRef]
- Pesavento, G.; Calonico, C.; Ducci, B.; Magnanini, A.; Lo Nostro, A. Prevalence and Antibiotic Resistance of Enterococcus spp. Isolated from Retail Cheese, Ready-to-Eat Salads, Ham, and Raw Meat. Food Microbiol. 2014, 41, 1–7. [Google Scholar] [CrossRef]
- Gouliouris, T.; Raven, K.E.; Ludden, C.; Blane, B.; Corander, J.; Horner, C.S.; Hernandez-Garcia, J.; Wood, P.; Hadjirin, N.F.; Radakovic, M.; et al. Genomic Surveillance of Enterococcus faecium Reveals Limited Sharing of Strains and Resistance Genes between Livestock and Humans in the United Kingdom. mBio 2018, 9, 1–15. [Google Scholar] [CrossRef]
- Elghaieb, H.; Freitas, A.R.; Abbassi, M.S.; Novais, C.; Zouari, M.; Hassen, A.; Peixe, L. Dispersal of Linezolid-Resistant Enterococci Carrying PoxtA or OptrA in Retail Meat and Food-Producing Animals from Tunisia. J. Antimicrob. Chemother. 2019, 74, 2865–2869. [Google Scholar] [CrossRef]
- Ni, J.; Long, X.; Wang, M. Transmission of Linezolid-Resistant Enterococcus Isolates Carrying OptrA and PoxtA Genes in Slaughterhouses. Front. Sustain. Food Syst. 2023, 7, 1–12. [Google Scholar] [CrossRef]
- Byappanahalli, M.N.; Nevers, M.B.; Korajkic, A.; Staley, Z.R.; Harwood, V.J. Enterococci in the Environment. Microbiol. Mol. Biol. Rev. 2012, 76, 685–706. [Google Scholar] [CrossRef]
- dos Santos, L.D.R.; Furlan, J.P.R.; Gallo, I.F.L.; Ramos, M.S.; Savazzi, E.A.; Stehling, E.G. Occurrence of Multidrug-Resistant Enterococcus faecium Isolated from Environmental Samples. Lett. Appl. Microbiol. 2021, 73, 237–246. [Google Scholar] [CrossRef]
- Monteiro, S.; Santos, R. Incidence of Enterococci Resistant to Clinically Relevant Antibiotics in Environmental Waters and in Reclaimed Waters Used for Irrigation. J. Water Health 2020, 18, 911–924. [Google Scholar] [CrossRef] [PubMed]
- Gouliouris, T.; Raven, K.E.; Moradigaravand, D.; Ludden, C.; Coll, F.; Blane, B.; Naydenova, P.; Horner, C.; Brown, N.M.; Corander, J.; et al. Detection of Vancomycin-Resistant Enterococcus faecium Hospital-Adapted Lineages in Municipal Wastewater Treatment Plants Indicates Widespread Distribution and Release into the Environment. Genome Res. 2019, 29, 626–634. [Google Scholar] [CrossRef]
- Cherak, Z.; Bendjama, E.; Moussi, A.; Benbouza, A.; Grainat, N.; Rolain, J.M.; Loucif, L. First Detection of VanA Positive Enterococcus faecium Clonal Complex 17 in Hospital Wastewater in Algeria: An Epidemiological Report. New Microbes New Infect. 2022, 47, 100977. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Liu, R.; Sun, Y.; Yang, X.; Yao, J. Tracing Enterococci Persistence along a Pork Production Chain from Feed to Food in China. Anim. Nutr. 2022, 9, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Rathnayake, I.; Hargreaves, M.; Huygens, F. SNP Diversity of Enterococcus faecalis and Enterococcus faecium in a South East Queensland Waterway, Australia, and Associated Antibiotic Resistance Gene Profiles. BMC Microbiol. 2011, 11, 1–13. [Google Scholar] [CrossRef]
- Noh, E.B.; Kim, Y.B.; Seo, K.W.; Son, S.H.; Ha, J.S.; Lee, Y.J. Antimicrobial Resistance Monitoring of Commensal Enterococcus faecalis in Broiler Breeders. Poult. Sci. 2020, 99, 2675–2683. [Google Scholar] [CrossRef]

| Antibiotic Class | Cellular Target | Species (Mostly Present) | Type of Resistance | Mechanism of Resistance | Associated Antimicrobial Resistance Genes |
|---|---|---|---|---|---|
| β-lactams | Peptidoglycan synthesis | All enterococci | Intrinsic | Low affinity of penicillin-binding proteins (PBP) | pbp5/pbp4 |
| E. faecium, E. hirae, E. faecalis | Acquired (ampicillin high-level) | Overproduction or PBP alterations lead to lower affinity; β-lactamase | - | ||
| Aminoglycosides | Protein synthesis (30 s) | All enterococci | Intrinsic (low-level) | Poor antibiotic uptake | - |
| E. faecium | Intrinsic (moderate-level) | Modification of the antibiotic molecule | aac | ||
| E. faecium | Intrinsic | Target modification with rRNA methyltransferase | efmM | ||
| E. faecalis, E. faecium, E. gallinarum, E. casseliflavus | Acquired (high-level) | Modification of the antibiotic molecule | aph, ant, aac-aph | ||
| E. faecium, E. faecalis | Acquired | Target modification (point mutations) | - | ||
| Ansamycins | DNA replication | E. faecium, E. faecalis | Acquired | Mutations in the gene that encodes the β-subunit of RNA polymerase | rpoB |
| Glycopeptides | Cell wall synthesis | E. gallinarum, E. casseliflavus | Intrinsic (low-level) | Production of D-Ala-D-Lac/D-Ala-D-Ser terminus peptidoglycan precursors | van operons (vanA–vanM) |
| E. faecium, E. faecalis | Acquired (high-level) | Precursor modification | |||
| Fluoroquinolones | DNA replication | E. faecium, E. faecalis | Acquired | Target-site modification (gene mutation in DNA gyrase and topoisomerase IV) | gyrA, pacC |
| Tetracyclines | Protein synthesis (30 s) | E. faecium, E. faecalis | Intrinsic/acquired | Target-site modification | poxtA |
| - | Antibiotic efflux | tet (K), tet (L) | |||
| - | Target-site protection | tet (M), tet (O), tet (S) | |||
| Macrolides | Protein synthesis (50 s) | Most enterococci | Acquired | Ribosomal methylation | ermA, ermB, ermC, ermS, ermV |
| Oxazolidinones | Protein synthesis (23 s) | E. faecium, E. faecalis | Intrinsic/acquired | Target-site protection | poxtA, optrA |
| E. faecium | Acquired | Target-site modification (point mutations) | - | ||
| Phenicols (Chloramphenicol) | Protein synthesis | E. faecium, E. faecalis | Acquired | CAT-encoding enzymes | catA, catB (mostly) |
| StreptoGramins | Protein synthesis (early/late stage) | E. faecalis, E. avium, E. gallinarum, E. casseliflavus | Intrinsic | Antibiotic efflux | - |
| Species | Source | Location (Date) | Clade | Clonal Lineages | Virulence Genes | Antimicrobial Resistance Genes | Ref. | |
|---|---|---|---|---|---|---|---|---|
| MLST Sequence Type | Clonal Complex | |||||||
| E. faecalis | Septicemias and endocarditis | Worldwide | NA | ST2 | CC2 | gelE | blaZ, aac6′-aph2”, vanB | [61,62,63] |
| ST6 | esp, gelE | blaZ, aac6′-aph2”, ermB, vanA, vanB | [61,63] | |||||
| Healthcare-associated infections | Worldwide (first in Argentina the USA) | NA | ST9 | CC9 | esp, gelE | blaZ, aac6′-aph2”, ermB | [29,61,64] | |
| ST106 | gelE | aac6′-aph2” | [61] | |||||
| Bacteremia | Poland | NA | ST28 | CC87 | asaI, ace esp, gelEcyl, hyl | aac6′-aph2”, vanA | [61,62,64] | |
| E. faecium | Healthcare-associated infections | Worldwide (first in the USA) | Clade A | ST17 | CC17 | acm, esp, hyl, fms | vanA, vanB, dfrG, tetM, msrC, ermB, aac6′-aph2″ | [37,65,66,67,68,69] |
| ST18 | acm, esp, hyl | - | [65,66,67,68,69] | |||||
| ST78 | - | - | [65,68] | |||||
| ST203 | acm, esp | vanB | [65,66,67,68,70] | |||||
| ST796 | acm, esp | vanA, vanB, dfrG, tetM, msrC, ermB, aac6′-aph2″ | [65,66,67,68,70] | |||||
| ST1421 | - | vanA | [71] | |||||
| Species | Clonal Lineages | Virulence Genes | Antimicrobial Resistance Genes | Source | Location | Ref. | |
|---|---|---|---|---|---|---|---|
| Clonal Complex | MLST Sequence Type | ||||||
| E. faecalis | CC2 | ST40 | GelE | tetM | Cheese | Poland | [98] |
| ST6 | ND | vanA, tetM, aac6′-Ie-aph2″-Ia | Water in swine facility | Portugal | [99] | ||
| CC21 | ST21 | ND | tetM, tetL, aac6′-Ie-aph2″-Ia | Air in facility; liquid manure | |||
| ST202 | gelE, efaA, ace | aac6’–aph2”-Ia | Poultry meat | Korea | [100] | ||
| CC82 | ST170 | gdh, gyd, pstS, gki, aroE, xpt, yqiL | ND | Duck’s cecum | China | [101] | |
| CC93 | ST93 | ||||||
| CC192 | ST192 | ||||||
| CC314 | ST314 | ||||||
| CC593 | ST593 | ||||||
| CC903 | ST903 | ||||||
| CC476 | ST116 | Duck’s skin | |||||
| - | ST16, ST21, ST26, ST84, ST138/501, ST207, ST209, ST277, ST326, ST672, ST674, ST715 | tuf, hyl, ebpA, bopD, fss1 | ermB | WWTP | Canada | [102] | |
| ST29 | ND | dfrE, emeA, efrA, efrB, lsaA, vanA | Cattle feces | Switzerland | [103] | ||
| - | ST242 | efaA, ace, ebpA, ebpB, epbC, gelE, fsrB | - | Bovine feces | Canada | [104] | |
| - | ST271 | ND | lsaA, tetM, tetL, dfrE, emeA | Unamended soil | South Africa | [105] | |
| - | ST1004, ST1006 | ND | dfrE, lsaA, emeA | Litter-amended soil | |||
| E. faecium | CC5 | ST5 | ND | vanA, tetM, ermB | Swine feces in slaughterhouse | Denmark | [106] |
| ST150 | ND | tetM, ermB | Adult swine feces | Portugal | [99] | ||
| ST185 | ND | vanA, tetM, tetL, ermB | Soil, solid manure, and adult swine diarrheal feces | ||||
| ND | vanA, tetM, ermB | Swine feces in slaughterhouse | Denmark | [106] | |||
| ST66 | fms5, fms17, fms21 | tetM, tetL | WWTP | Poland | [107] | ||
| CC9 | ST433 | ND | tetM, tetL | Air from swine facility | Portugal | [99] | |
| ST437 | ND | tetM, tetL, ermB | Adult swine feed | Portugal | [99] | ||
| ST29, ST57 | aptA, ddl, gdh, purK, gyd, pstS, adk | ND | Poultry | Malaysia | [108] | ||
| CC17 | ST10 | ND | aac6′-Ii, ant6-Ia, aph3′- III, ermB, lnuG, eatA, tetM, tetL, dfrG, efmA | Litter-amended soil | South Africa | [105] | |
| efaA, ccf/6 | ND | Poultry | Poland | [109] | |||
| ST17 | ptsD, sgrA, IS16, orf1481, esp | ND | Dog food | Portugal | [110] | ||
| ST18 | tuf, aga, efaA, sgrA, uppS, lisR, acm, esp, scm, bsh, tip/ropA, bopD, eno, rfbA-1 | ND | WWTP | Canada | [102] | ||
| ST203 | aptA, ddl, gdh, purK, gyd, pstS, adk | - | Poultry | Malaysia | [108] | ||
| ST78 | ND | vanA, ermB, ant6-Ia, aac6′-Ie-aph2″-Ia, aph3′-IIIa | Rabbit meat | Spain | [111] | ||
| ST78 | fms20, fms14, ebpA, fms16 | ND | WWTP | Czech Republic | [112] | ||
| ST132 | ND | vanA, aac6′-Ie-aph2″-Ia | Water in swine facilities | Portugal | [99] | ||
| ST431 | ND | tetM, tetL, ermB | Swine facility dust | ||||
| ST386 | esp/intA, fms5, fms17, fms19, fms21 | ant6’-la | WWTP | Poland | [107] | ||
| CC18 | ST273 | fms20, fms14, ebpA, fms16 | ND | WWTP | Czech Republic | [112] | |
| CC22 | ST21 | fms17, fms19, fms21 | ant6’-la, tetM), tetL | WWTP | Poland | [107] | |
| ST32 | ND | tetM, tetL, tetS | Antiseptic, drinking water in a swine facility | Portugal | [99] | ||
| ST55 | aptA, ddl, gdh, purK, gyd, pstS, adk | ND | Poultry and swine | Malaysia | [108] | ||
| CC94 | ST40 | tuf, aga, efaA, sgrA, upp, lisR, acm, esp, scm, bsh, tip/ropA, bopD, eno, rfbA-1 | ND | WWTP | Canada | [102] | |
| ST361 | fms5, fms17, fms19, fms21 | ant6’-la, tetL | WWTP | Poland | [107] | ||
| ST1754 | ND | aac6′-Ii, msrC, eatA | Poultry litter | South Africa | [105] | ||
| - | ST1700 | ND | aac6′-Ii, ant6-Ia, ant9- Ia, ermB, lnuB, lsaE, msrC, cat, tetL, tetM, eatA | ||||
| - | ST1752, ST1756 | ND | aac6′-Ii, ermB, msrC, tetL, tetM, eatA, efmA | ||||
| - | ST1753, ST1755 | aac6′-Ii, ermB, msr C, tetL, tetM, eatA, efmA | Litter-amended soil | ||||
| - | ST214, ST955 | efaA, acm | ermB, msrC, aac6′-Ii, tetL, tetM, tetO | Bovine feces | Canada | [104] | |
| - | ST133 | ND | aac60-Ii, eatAv, cadA, cadC, copZ, czrA, merA, merR, tetW/N/W, vanA, zosA | Swine feces | Switzerland | [103] | |
| - | ND | aac6′-Ii, eatAv, efrA, msrC, tetM, vanA | Environmental water | Switzerland | [113] | ||
| - | ST425 | ND | vanA, erm (B), tet (M) | Poultry meat | Spain | [111] | |
| - | ST13 | ND | aac60-Ii, aadK, eatAv, vanA | Poultry feces | Switzerland | [103] | |
| CC117 | ST117 | efaA, sagA, malR, swpA, swpB, swpC | ant6-la, ant1, aph, lnuB, isaE, tetL, satA, erm_1, erm_2, aad6-la | Fermented dry sausage | Italy | [114] | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Monteiro Marques, J.; Coelho, M.; Santana, A.R.; Pinto, D.; Semedo-Lemsaddek, T. Dissemination of Enterococcal Genetic Lineages: A One Health Perspective. Antibiotics 2023, 12, 1140. https://doi.org/10.3390/antibiotics12071140
Monteiro Marques J, Coelho M, Santana AR, Pinto D, Semedo-Lemsaddek T. Dissemination of Enterococcal Genetic Lineages: A One Health Perspective. Antibiotics. 2023; 12(7):1140. https://doi.org/10.3390/antibiotics12071140
Chicago/Turabian StyleMonteiro Marques, Joana, Mariana Coelho, Andressa Rodrigues Santana, Daniel Pinto, and Teresa Semedo-Lemsaddek. 2023. "Dissemination of Enterococcal Genetic Lineages: A One Health Perspective" Antibiotics 12, no. 7: 1140. https://doi.org/10.3390/antibiotics12071140
APA StyleMonteiro Marques, J., Coelho, M., Santana, A. R., Pinto, D., & Semedo-Lemsaddek, T. (2023). Dissemination of Enterococcal Genetic Lineages: A One Health Perspective. Antibiotics, 12(7), 1140. https://doi.org/10.3390/antibiotics12071140






