Plasmid Costs Explain Plasmid Maintenance, Irrespective of the Nature of Compensatory Mutations
Abstract
1. Introduction
2. Results
2.1. Do Donors Benefit from Plasmids with Compensatory Mutations in Plasmids?
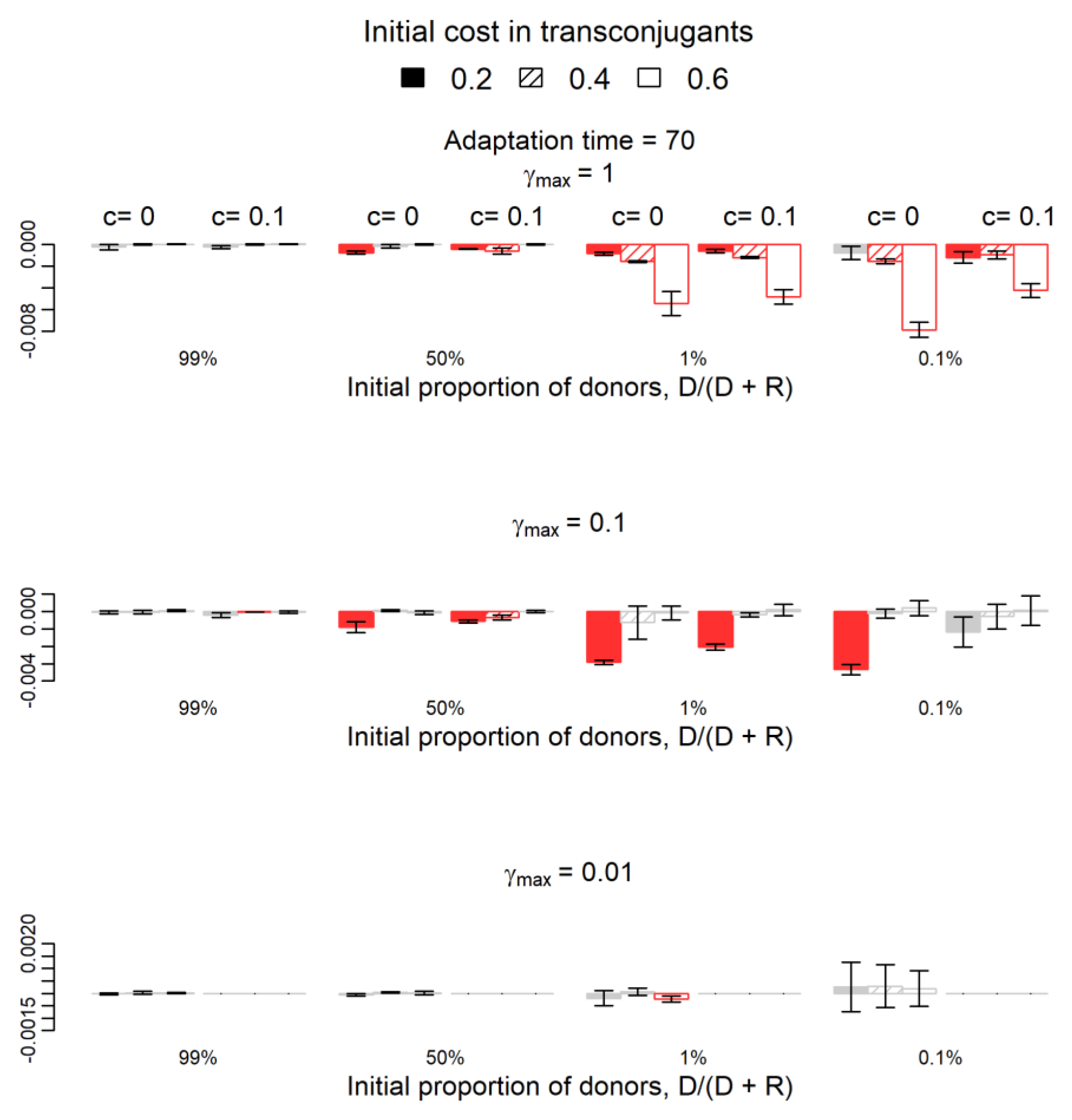
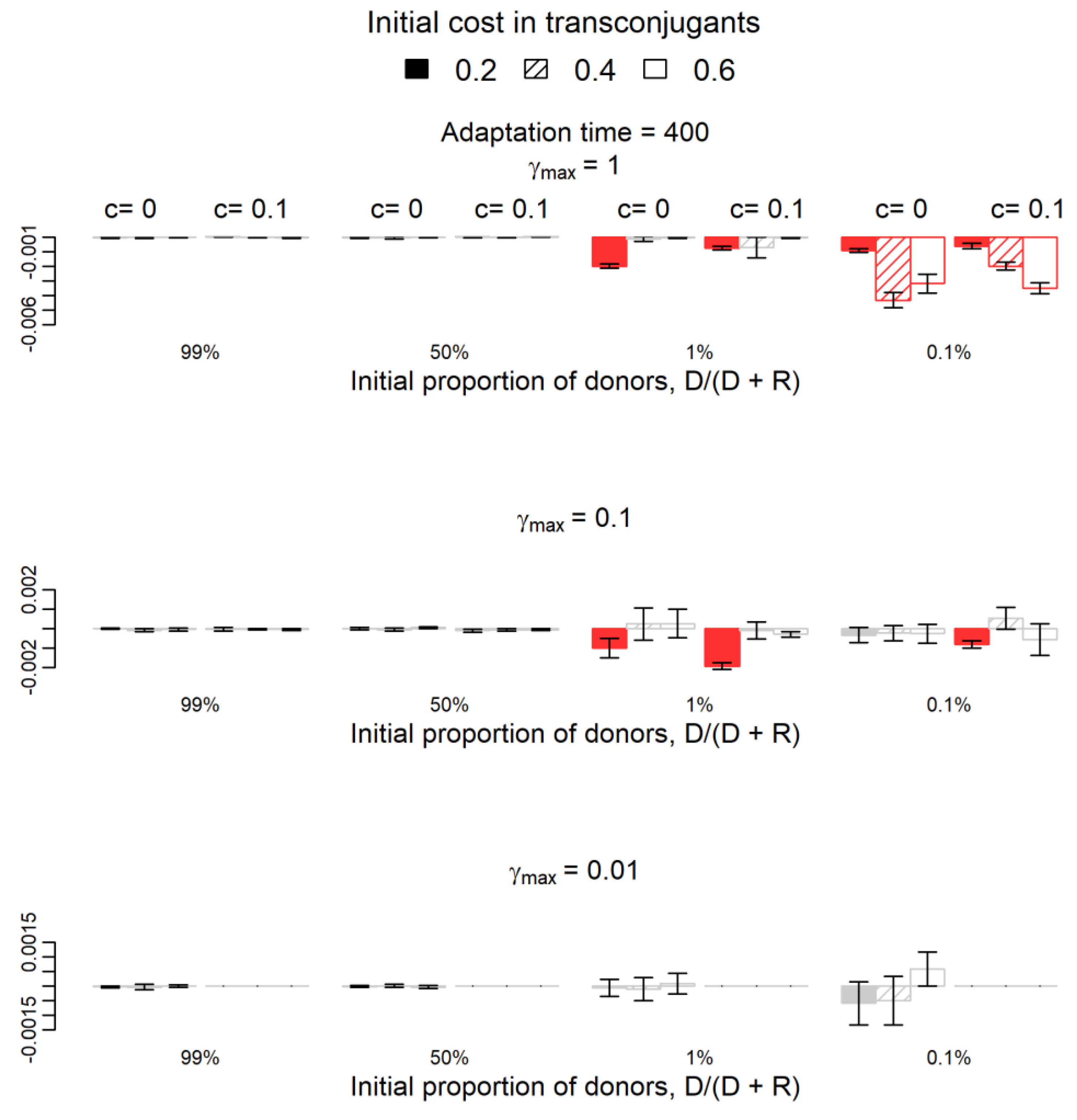
2.2. How Does Plasmid Fitness Compare When Mutations Occur in Plasmids versus Chromosomes?
3. Discussion
4. Materials and Methods
4.1. The Computational Model
4.1.1. Purpose
4.1.2. Entities, State Variables, and Scales
- Type, which ranges from 1 to 6—defines the type of bacteria, recipient (1), donor (2), transconjugant (3), adapted transconjugant that lost the plasmid (4), non-adapted transconjugant that lost the plasmid (5), or donor that lost the plasmid (6);
- Permanent plasmid fitness cost, which can be 0 or 0.1—this is the plasmid cost for donors and transconjugants carrying the plasmid already adapted;
- Initial plasmid fitness cost, which may be 0.2, 0.4, or 0.6—this is the plasmid cost for transconjugants carrying the non-adapted plasmid;
- Adaptation time, which can be 70 or 400—represents the number of times the plasmid is in a transconjugant bacterium that divides until its cost decreases;
- Local neighborhood, 3 × 3 space centered on the bacterium—these are the spaces into which the bacterium can duplicate or where another bacterium can be present to receive a plasmid through conjugation;
- Nutrient neighborhood, 7 × 7 space centered on the bacteria—allows defining the number of nutrients available, given by the proportion of empty spaces in these 49 spaces.
4.1.3. Process Overview and Scheduling
- We check if there is an empty space in the local neighborhood and if a random number is less than the conjugation rate—in that case, the submodel “bacterial_growth” is activated, and we update one of the empty spaces in the bacterium neighborhood, with the same characteristics as the original bacterium;
- If the bacterium is a donor or a transconjugant, we check if there is a recipient bacterium in the local neighborhood, and if a random number is smaller than the conjugation rate—in that case, the submodel “conjugation” is activated and the recipient bacterium becomes a transconjugant with its characteristics;
- We verify if 95% of the total spaces are filled—in that case, we randomly eliminate bacteria until we have only 50% of the spaces filled.
4.1.4. Design Concepts
- Basic principles. This model is an adaptation of the model developed in [17]. The major difference is that we consider that the compensatory mutations that reduce the cost of bacteria carrying the plasmid occur in the plasmid rather than the chromosome. Therefore, after a compensatory mutation, the transconjugant bacteria will transfer the plasmid with the already reduced cost. Furthermore, each time the transconjugant bacteria transfers a plasmid, the adaptation time is also already reduced, being equal to that of the transconjugant bacterium that transferred the plasmid. This is because we consider that the plasmid has to be 70 or 400 times in transconjugant bacteria that divide;
- Emergence. The final densities of bacteria will depend on their growth throughout the simulations. In turn, this growth will vary depending on the type of bacteria that are in the nutrient neighborhood and consequently on the competition of those bacteria for resources. The rate of conjugation will also influence the final bacterial densities;
- Adaptation. The number of empty spaces in the nutrient neighborhood divided by the total number of spaces will define whether there is bacterial growth. If there is, the bacterium will grow to an empty position in the local neighborhood. Plasmids of transconjugant bacteria can adapt and reduce the cost to the bacteria carrying them. This adaptation depends on how many duplications the transconjugant bacteria carrying that plasmid already suffered;
- Interaction. Bacteria that have the plasmid (donors or transconjugants) can interact with bacteria that do not have the plasmid (recipients or segregants). This interaction happens when the bacteria are in the same local neighborhood. When there is conjugation from a donor or a non-adapted transconjugant, the bacteria receiving the plasmid have an associated cost. If the plasmid is transferred from a transconjugant that has already had an adaptation, the plasmid is already adapted and has no cost to the recipient bacterium. In addition to these interactions, the model takes into account mediated interactions since it considers that all bacteria compete for nutrients;
- Stochasticity. The model is initiated by the random distribution of bacteria across grid positions. Therefore, at the initial moment of each simulation, bacteria are always in different locations, and for this reason, the interactions between bacteria vary from simulation to simulation. Furthermore, in each cycle, the bacteria to be updated is randomly chosen. In this way, the number of times each bacterium is chosen can vary, making the model asynchronous. Additionally, in the bacterial growth and conjugation submodel, random numbers are obtained (see the “bacterial_growth” and “conjugation” submodels below), which confers stochasticity to the model. Finally, when we simulate bacterial death, we also randomly choose the bacteria that will be eliminated;
- Observation. The observation we take into account in this model is the densities of each type of bacteria whenever the grid reaches 95% capacity.
4.1.5. Initialization
4.1.6. Input Data
4.1.7. Submodels
- Bacterial growth
- 2.
- Conjugation
- 3.
- Fitness analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Redondo-Redondo-Salvo, S.; Fernández-López, R.; Ruiz, R.; Vielva, L.; de Toro, M.; Rocha, E.P.C.; Garcillán-Barcia, M.P.; de la Cruz, F. Pathways for Horizontal Gene Transfer in Bacteria Revealed by a Global Map of Their Plasmids. Nat. Commun. 2020, 11, 3602. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A. Resistance Plasmid Families in Enterobacteriaceae. Antimicrob. Agents Chemother. 2009, 53, 2227–2238. [Google Scholar] [CrossRef] [PubMed]
- Jordt, H.; Stalder, T.; Kosterlitz, O.; Ponciano, J.M.; Top, E.M.; Kerr, B. Coevolution of Host–Plasmid Pairs Facilitates the Emergence of Novel Multidrug Resistance. Nat. Ecol. Evol. 2020, 4, 863–869. [Google Scholar] [CrossRef] [PubMed]
- San Millan, A.; MacLean, R.C. Fitness Costs of Plasmids: A Limit to Plasmid Transmission. Microbiol. Spectr. 2017, 5, 5. [Google Scholar] [CrossRef]
- Winfield, M.D.; Groisman, E.A. Phenotypic Differences between Salmonella and Escherichia Coli Resulting from the Disparate Regulation of Homologous Genes. Proc. Natl. Acad. Sci. USA 2004, 101, 17162–17167. [Google Scholar] [CrossRef]
- Ochman, H.; Groisman, E.A. The Origin and Evolution of Species Differences in Escherichia Coli and Salmonella Typhimurium. In Molecular Ecology and Evolution: Approaches and Applications; Schierwater, B., Streit, B., Wagner, G.P., DeSalle, R., Eds.; Experientia Supplementum; Birkhäuser: Basel, Switzerland, 1994; pp. 479–493. ISBN 978-3-0348-7527-1. [Google Scholar]
- Frumkin, I.; Lajoie, M.J.; Gregg, C.J.; Hornung, G.; Church, G.M.; Pilpel, Y. Codon Usage of Highly Expressed Genes Affects Proteome-Wide Translation Efficiency. Proc. Natl. Acad. Sci. USA 2018, 115, E4940–E4949. [Google Scholar] [CrossRef]
- Rodríguez-Beltrán, J.; León-Sampedro, R.; Ramiro-Martínez, P.; de la Vega, C.; Baquero, F.; Levin, B.R.; San Millán, Á. Translational Demand Is Not a Major Source of Plasmid-Associated Fitness Costs. Philos. Trans. R. Soc. B Biol. Sci. 2021, 377, 20200463. [Google Scholar] [CrossRef]
- Gama, J.A.; Zilhao, R.; Dionisio, F. Multiple Plasmid Interference-Pledging Allegiance to My Enemy’s Enemy. Plasmid 2017, 93, 17–23. [Google Scholar] [CrossRef]
- Gama, J.A.; Zilhão, R.; Dionisio, F. Co-Resident Plasmids Travel Together. Plasmid 2017, 93, 24–29. [Google Scholar] [CrossRef]
- Gama, J.A.; Zilhão, R.; Dionisio, F. Conjugation Efficiency Depends on Intra and Intercellular Interactions between Distinct Plasmids: Plasmids Promote the Immigration of Other Plasmids but Repress Co-Colonizing Plasmids. Plasmid 2017, 93, 6–16. [Google Scholar] [CrossRef]
- Dionisio, F.; Zilhão, R.; Gama, J.A. Interactions between Plasmids and Other Mobile Genetic Elements Affect Their Transmission and Persistence. Plasmid 2019, 102, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Bergstrom, C.T.; Lipsitch, M.; Levin, B.R. Natural Selection, Infectious Transfer and the Existence Conditions for Bacterial Plasmids. Genetics 2000, 155, 1505–1519. [Google Scholar] [CrossRef] [PubMed]
- Wein, T.; Wang, Y.; Barz, M.; Stücker, F.T.; Hammerschmidt, K.; Dagan, T. Essential Gene Acquisition Destabilizes Plasmid Inheritance. PLoS Genet. 2021, 17, e1009656. [Google Scholar] [CrossRef]
- Datta, N.; Hughes, V.M. Plasmids of the Same Inc Groups in Enterobacteria before and after the Medical Use of Antibiotics. Nature 1983, 306, 616–617. [Google Scholar] [CrossRef] [PubMed]
- Hughes, V.M.; Datta, N. Conjugative Plasmids in Bacteria of the “pre-Antibiotic” Era. Nature 1983, 302, 725–726. [Google Scholar] [CrossRef] [PubMed]
- Domingues, C.P.F.; Rebelo, J.S.; Monteiro, F.; Nogueira, T.; Dionisio, F. Harmful Behaviour through Plasmid Transfer: A Successful Evolutionary Strategy of Bacteria Harbouring Conjugative Plasmids. Philos. Trans. R. Soc. B Biol. Sci. 2022, 377, 20200473. [Google Scholar] [CrossRef] [PubMed]
- Zwanzig, M.; Harrison, E.; Brockhurst, M.A.; Hall, J.P.J.; Berendonk, T.U.; Berger, U. Mobile Compensatory Mutations Promote Plasmid Survival. mSystems 2019, 4, e00186-18. [Google Scholar] [CrossRef]
- Zhong, X.; Droesch, J.; Fox, R.; Top, E.M.; Krone, S.M. On the Meaning and Estimation of Plasmid Transfer Rates for Surface-Associated and Well-Mixed Bacterial Populations. J. Theor. Biol. 2012, 294, 144–152. [Google Scholar] [CrossRef]
- Fox, R.E.; Zhong, X.; Krone, S.M.; Top, E.M. Spatial Structure and Nutrients Promote Invasion of IncP-1 Plasmids in Bacterial Populations. ISME J. 2008, 2, 1024–1039. [Google Scholar] [CrossRef]
- Krone, S.M.; Lu, R.; Fox, R.; Suzuki, H.; Top, E.M. Modelling the Spatial Dynamics of Plasmid Transfer and Persistence. Microbiology 2007, 153, 2803–2816. [Google Scholar] [CrossRef]
- Rebelo, J.S.; Domingues, C.P.F.; Nogueira, T.; Dionisio, F. Plasmids Increase the Competitive Ability of Plasmid-Bearing Cells Even When Transconjugants Are Poor Donors, as Shown by Computer Simulations. Microorganisms 2023, 11, 1238. [Google Scholar] [CrossRef]
- West, S.A.; Griffin, A.S.; Gardner, A.; Diggle, S.P. Social Evolution Theory for Microorganisms. Nat. Rev. Microbiol. 2006, 4, 597–607. [Google Scholar] [CrossRef]
- West, S.A.; Diggle, S.P.; Buckling, A.; Gardner, A.; Griffin, A.S. The Social Lives of Microbes. Annu. Rev. Ecol. Evol. Syst. 2007, 38, 53–77. [Google Scholar] [CrossRef]
- Dionisio, F. Selfish and Spiteful Behaviour through Parasites and Pathogens. Evol. Ecol. Res. 2007, 9, 1199–1210. [Google Scholar]
- Queller, D.C. Genetic Relatedness in Viscous Populations. Evol. Ecol. 1994, 8, 70–73. [Google Scholar] [CrossRef]
- Bouma, J.E.; Lenski, R.E. Evolution of a Bacteria Plasmid Association. Nature 1988, 335, 351–352. [Google Scholar] [CrossRef] [PubMed]
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Cassini, A.; Hogberg, L.D.; Plachouras, D.; Quattrocchi, A.; Hoxha, A.; Simonsen, G.S.; Colomb-Cotinat, M.; Kretzschmar, M.E.; Devleesschauwer, B.; Cecchini, M.; et al. Attributable Deaths and Disability-Adjusted Life-Years Caused by Infections with Antibiotic-Resistant Bacteria in the EU and the European Economic Area in 2015: A Population-Level Modelling Analysis. Lancet Infect. Dis. 2019, 19, 56–66. [Google Scholar] [CrossRef]
- Dionisio, F.; Baquero, F.; Fuertes, M. Psychological and Cultural Factors Influencing Antibiotic Prescription. Trends Microbiol. 2023. [Google Scholar] [CrossRef]
- Calvo-Villamañán, A.; San Millán, Á.; Carrilero, L. Tackling AMR from a Multidisciplinary Perspective: A Primer from Education and Psychology. Int. Microbiol. 2022, 26, 1–9. [Google Scholar] [CrossRef]
- André, J.-B.; Godelle, B. Multicellular Organization in Bacteria as a Target for Drug Therapy. Ecol. Lett. 2005, 8, 800–810. [Google Scholar] [CrossRef]
- Pepper, J.W. Drugs That Target Pathogen Public Goods Are Robust against Evolved Drug Resistance. Evol. Appl. 2012, 5, 757–761. [Google Scholar] [CrossRef] [PubMed]
- Mei, H.; Arbeithuber, B.; Cremona, M.A.; DeGiorgio, M.; Nekrutenko, A. A High-Resolution View of Adaptive Event Dynamics in a Plasmid. Genome Biol. Evol. 2019, 11, 3022–3034. [Google Scholar] [CrossRef] [PubMed]
- Grimm, V.; Railsback, S.F.; Vincenot, C.E.; Berger, U.; Gallagher, C.; DeAngelis, D.L.; Edmonds, B.; Ge, J.; Giske, J.; Groeneveld, J.; et al. The ODD Protocol for Describing Agent-Based and Other Simulation Models: A Second Update to Improve Clarity, Replication, and Structural Realism. J. Artif. Soc. Soc. Simul. 2020, 23, 7. [Google Scholar] [CrossRef]
- Grimm, V.; Berger, U.; Bastiansen, F.; Eliassen, S.; Ginot, V.; Giske, J.; Goss-Custard, J.; Grand, T.; Heinz, S.K.; Huse, G.; et al. A Standard Protocol for Describing Individual-Based and Agent-Based Models. Ecol. Model. 2006, 198, 115–126. [Google Scholar] [CrossRef]
- Railsback, S.F.; Grimm, V. Agent-Based and Individual-Based Modeling; Princeton University Press: Princeton, NJ, USA, 2019; ISBN 978-0-691-19082-2. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2015. [Google Scholar]
| Entities | Parameter Range | Description |
|---|---|---|
| grid_edge | 1000 | Maximum valeu for x and y coordinates |
| maximum_proportion_full_grid | 0.95 | Maximum proportion of bacteria that the grid can contain |
| remaining_proportion_grid | 0.5 | Proportion of bacteria remaining in the grid when bacteria are randomly removed when the grid reaches 95% capacity |
| number_plasmid_free_bacteria | 9990, 9900, 5000, 100 | Initial number of bacteria not carrying plasmid |
| donor_bacteria | 10, 100, 5000, 9900 | Initial number of bacteria that carry plasmid |
| maximum_growth_rate (ψmax) | 1 | Maximum bacterial growth rate |
| maximum_conjugation_rate (γmax) | 1 | Maximum bacterial conjugation rate |
| theta (θ) | 0.8 | Value of theta (bacterial growth) |
| theta_1 (𝜃1) | 0.2 | Value of theta 1 (conjugation) |
| theta_2 (𝜃2) | 0.3 | Value of theta 2 (conjugation) |
| initial_plasmid_cost (b) | 0.2, 0.4, 0.6 | Cost that bacterium has when receiving the plasmid |
| permanent_plasmid_cost (c) | 0, 0.1 | Cost associated with the presence of the plasmid in donors and adapted transconjugants |
| adaptation_time | 70, 400 | Number of duplications that bacteria need until the initial plasmid cost changes to permanent plasmid cost |
| segregation_probability | 0.001 | Probability of a bacterium losing the plasmid at the moment of its duplication |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rebelo, J.S.; Domingues, C.P.F.; Dionisio, F. Plasmid Costs Explain Plasmid Maintenance, Irrespective of the Nature of Compensatory Mutations. Antibiotics 2023, 12, 841. https://doi.org/10.3390/antibiotics12050841
Rebelo JS, Domingues CPF, Dionisio F. Plasmid Costs Explain Plasmid Maintenance, Irrespective of the Nature of Compensatory Mutations. Antibiotics. 2023; 12(5):841. https://doi.org/10.3390/antibiotics12050841
Chicago/Turabian StyleRebelo, João S., Célia P. F. Domingues, and Francisco Dionisio. 2023. "Plasmid Costs Explain Plasmid Maintenance, Irrespective of the Nature of Compensatory Mutations" Antibiotics 12, no. 5: 841. https://doi.org/10.3390/antibiotics12050841
APA StyleRebelo, J. S., Domingues, C. P. F., & Dionisio, F. (2023). Plasmid Costs Explain Plasmid Maintenance, Irrespective of the Nature of Compensatory Mutations. Antibiotics, 12(5), 841. https://doi.org/10.3390/antibiotics12050841






