Whole Genome Sequencing and Phenotypic Analysis of Antibiotic Resistance in Filifactor alocis Isolates
Abstract
1. Introduction
2. Results
2.1. Identification of Isolates and Sequencing Analysis
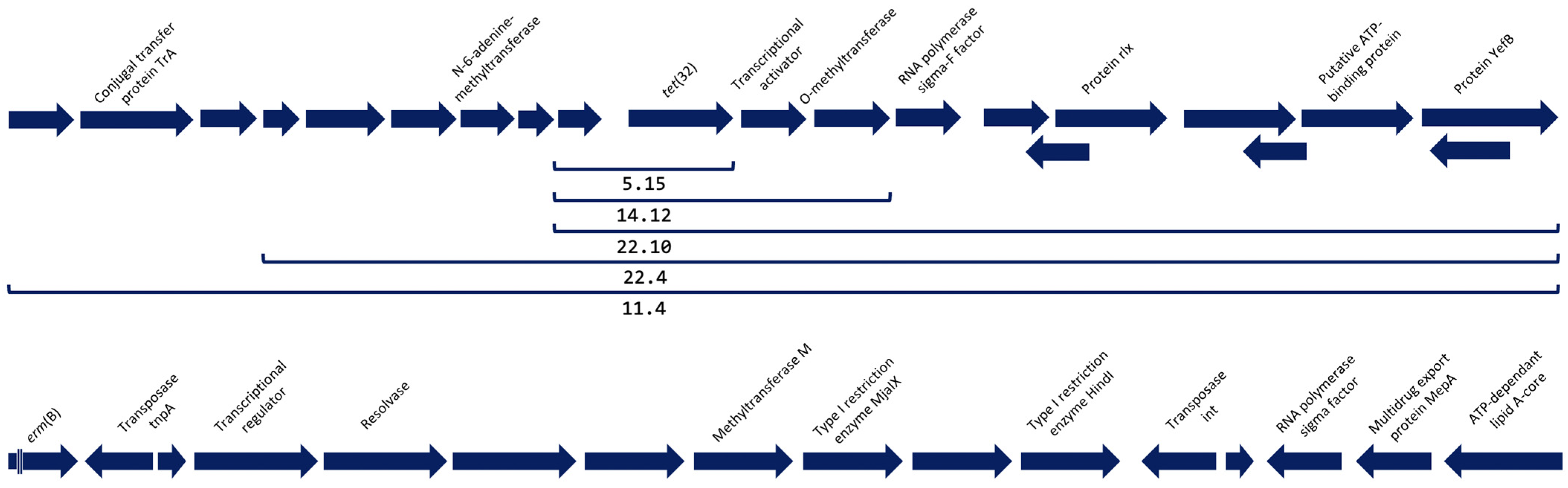
2.2. Detection and Analysis of Antimicrobial Resistance Genes
2.3. Antimicrobial Resistance Testing
3. Discussion
4. Material & Methods
4.1. Sample Collection and Culturing Conditions
4.2. DNA Isolation and Polymerase Chain Reaction-Based Characterization
4.3. Antibiotic Susceptibility Testing
4.4. Whole-Genome Sequencing
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jalava, J.; Eerola, E. Phylogenetic analysis of Fusobacterium alocis and Fusobacterium sulci based on 16S rRNA gene sequences: Proposal of Filifactor alocis (Cato, Moore and Moore) comb. nov. and Eubacterium sulci (Cato, Moore and Moore) comb. nov. Int. J. Syst. Evol. Microbiol. 1999, 49, 1375–1379. [Google Scholar] [CrossRef]
- Aja, E.; Mangar, M.; Fletcher, H.M.; Mishra, A. Filifactor alocis: Recent Insights and Advances. J. Dent. Res. 2021, 100, 790–797. [Google Scholar] [CrossRef]
- Cato, E.P.; Moore, L.V.H.; Moore, W.E.C. Fusobacterium alocis sp. nov. and Fusobacterium sulci sp. nov. from the Human Gingival Sulcus. Int. J. Syst. Evol. Microbiol. 1985, 35, 475–477. [Google Scholar] [CrossRef]
- Hutter, G.; Schlagenhauf, U.; Valenza, G.; Horn, M.; Burgemeister, S.; Claus, H.; Vogel, U. Molecular analysis of bacteria in periodontitis: Evaluation of clone libraries, novel phylotypes and putative pathogens. Microbiology 2003, 149, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Jr, J.F.S.; Rôças, I.N. Detection of Filifactor alocis in endodontic infections associated with different forms of periradicular diseases. Oral Microbiol. Immunol. 2003, 18, 263–265. [Google Scholar] [CrossRef]
- Gomes, B.P.; Jacinto, R.C.; Pinheiro, E.; Sousa, E.L.; Zaia, A.A.; Ferraz, C.; Souza-Filho, F.J. Molecular Analysis of Filifactor alocis, Tannerella forsythia, and Treponema denticola Associated with Primary Endodontic Infections and Failed Endodontic Treatment. J. Endod. 2006, 32, 937–940. [Google Scholar] [CrossRef]
- Kumar, P.S.; Leys, E.J.; Bryk, J.M.; Martinez, F.J.; Moeschberger, M.L.; Griffen, A.L. Changes in Periodontal Health Status Are Associated with Bacterial Community Shifts as Assessed by Quantitative 16S Cloning and Sequencing. J. Clin. Microbiol. 2006, 44, 3665–3673. [Google Scholar] [CrossRef]
- Chen, H.; Liu, Y.; Zhang, M.; Wang, G.; Qi, Z.; Bridgewater, L.; Zhao, L.; Tang, Z.; Pang, X. A Filifactor alocis-centered co-occurrence group associates with periodontitis across different oral habitats. Sci. Rep. 2015, 5, srep09053. [Google Scholar] [CrossRef]
- Griffen, A.L.; Beall, C.J.; Campbell, J.H.; Firestone, N.D.; Kumar, P.S.; Yang, Z.K.; Podar, M.; Leys, E.J. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 2011, 6, 1176–1185. [Google Scholar] [CrossRef]
- Moffatt, C.; Whitmore, S.; Griffen, A.; Leys, E.; Lamont, R. Filifactor alocis interactions with gingival epithelial cells. Mol. Oral Microbiol. 2011, 26, 365–373. [Google Scholar] [CrossRef]
- Aruni, A.W.; Zhang, K.; Dou, Y.; Fletcher, H. Proteome Analysis of Coinfection of Epithelial Cells with Filifactor alocis and Porphyromonas gingivalis Shows Modulation of Pathogen and Host Regulatory Pathways. Infect. Immun. 2014, 82, 3261–3274. [Google Scholar] [CrossRef] [PubMed]
- Miralda, I.; Vashishta, A.; Rogers, M.N.; Lamont, R.J.; Uriarte, S.M. The emerging oral pathogen, Filifactor alocis, extends the functional lifespan of human neutrophils. Mol. Microbiol. 2022, 117, 1340–1351. [Google Scholar] [CrossRef]
- Bao, K.; Claesson, R.; Belibasakis, G.N.; Oscarsson, J. Extracellular Vesicle Subproteome Differences among Filifactor alocis Clinical Isolates. Microorganisms 2022, 10, 1826. [Google Scholar] [CrossRef]
- Mishra, A.; Aja, E.; Fletcher, H.M. Role of Superoxide Reductase FA796 in Oxidative Stress Resistance in Filifactor alocis. Sci. Rep. 2020, 10, 9178. [Google Scholar] [CrossRef] [PubMed]
- Aruni, A.W.; Roy, F.; Fletcher, H.M. Filifactor alocis Has Virulence Attributes That Can Enhance Its Persistence under Oxidative Stress Conditions and Mediate Invasion of Epithelial Cells by Porphyromonas gingivalis. Infect. Immun. 2011, 79, 3872–3886. [Google Scholar] [CrossRef] [PubMed]
- Aruni, A.W.; Roy, F.; Sandberg, L.; Fletcher, H.M. Proteome variation among Filifactor alocis strains. Proteomics 2012, 12, 3343–3364. [Google Scholar] [CrossRef]
- Schlafer, S.; Riep, B.; Griffen, A.L.; Petrich, A.; Hübner, J.; Berning, M.; Friedmann, A.; Göbel, U.B.; Moter, A. Filifactor alocis—involvement in periodontal biofilms. BMC Microbiol. 2010, 10, 66. [Google Scholar] [CrossRef]
- Gray, R.; Vidwans, M. Mixed anaerobic thoracic empyema: The first report of Filifactor alocis causing extra-oral disease. New Microbes New Infect. 2019, 29, 100528. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhu, Y.; Wan, H. Case report:Multiple abscesses caused by Porphyromonas gingivalis diagnosed by metagenomic next-generation sequencing. Front. Med. 2023, 9, 1089863. [Google Scholar] [CrossRef]
- Yusuf, K.; Sampath, V.; Umar, S. Bacterial Infections and Cancer: Exploring This Association And Its Implications for Cancer Patients. Int. J. Mol. Sci. 2023, 24, 3110. [Google Scholar] [CrossRef]
- Mesturino, M.A.; Bitetti, C.; Clemente, A.; Krzysztofiak, A.; Lancella, L.; Lombardi, R.; Cursi, L.; Boccuzzi, E.; Musolino, A.M.; Villani, A. Aggregatibacter actinomycetemcomitans infection in a 15-year-old boy with pulmonary empyema: A case report and review of literature. Ital. J. Pediatr. 2023, 49, 42. [Google Scholar] [CrossRef] [PubMed]
- Inubushi, J.; Liang, K. Update on minocycline in vitro activity against odontogenic bacteria. J. Infect. Chemother. 2020, 26, 1334–1337. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, M.I.; Römling, U.; Nadeem, F.; Bilal, H.M.; Zafar, M.; Jahan, H.; Ur-Rahman, A. Innovative Strategies to Overcome Antimicrobial Resistance and Tolerance. Microorganisms 2022, 11, 16. [Google Scholar] [CrossRef]
- European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint Tables for Interpretation of MICs and Zone Diameters, Version 13.0. EFSA Journal. Available online: http://www.eucast.org (accessed on 20 February 2023).
- Àlvarez, G.; Arredondo, A.; Isabal, S.; Teughels, W.; Laleman, I.; Contreras, M.J.; Isbej, L.; Huapaya, E.; Mendoza, G.; Mor, C.; et al. Association of nine pathobionts with periodontitis in four South American and European countries. J. Oral Microbiol. 2023, 15, 2188630. [Google Scholar] [CrossRef] [PubMed]
- Lafaurie, G.I.; Neuta, Y.; Ríos, R.; Pacheco-Montealegre, M.; Pianeta, R.; Castillo, D.M.; Herrera, D.; Reyes, J.; Diaz, L.; Castillo, Y.; et al. Differences in the subgingival microbiome according to stage of periodontitis: A comparison of two geographic regions. PLoS ONE 2022, 17, e0273523. [Google Scholar] [CrossRef]
- Thaker, M.; Spanogiannopoulos, P.; Wright, G.D. The tetracycline resistome. Cell. Mol. Life Sci. 2010, 67, 419–431. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, X.; Zong, Z. Genome sequence and virulence factors of a group G Streptococcus dysgalactiae subsp. equisimilis strain with a new element carrying erm(B). Sci. Rep. 2016, 6, 20389. [Google Scholar] [CrossRef]
- Arredondo, A.; Blanc, V.; Mor, C.; Nart, J.; León, R. Tetracycline and multidrug resistance in the oral microbiota: Differences between healthy subjects and patients with periodontitis in Spain. J. Oral Microbiol. 2021, 13, 1847431. [Google Scholar] [CrossRef]
- Warburton, P.; Roberts, A.P.; Allan, E.; Seville, L.; Lancaster, H.; Mullany, P. Characterization of tet (32) Genes from the Oral Metagenome. Antimicrob. Agents Chemother. 2009, 53, 273–276. [Google Scholar] [CrossRef]
- Speer, B.S.; Shoemaker, N.B.; Salyers, A.A. Bacterial Resistance to Tetracycline: Mechanisms, Transfer, and Clinical Significance. Clin. Microbiol. Rev. 1992, 5, 387–399. [Google Scholar] [CrossRef]
- Roberts, M.C. Resistance to Tetracycline, Macrolide-Lincosamide-Streptogramin, Trimethoprim, and Sulfonamide Drug Classes. Appl. Biochem. Biotechnol. Part B Mol. Biotechnol. 2002, 20, 261–284. [Google Scholar] [CrossRef] [PubMed]
- Weisblum, B. Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 1995, 39, 577–585. [Google Scholar] [CrossRef] [PubMed]
- Ciric, L.; Ellatif, M.; Sharma, P.; Patel, R.; Song, X.; Mullany, P.; Roberts, A.P. Tn916-like elements from human, oral, commensal streptococci possess a variety of antibiotic and antiseptic resistance genes. Int. J. Antimicrob. Agents 2012, 39, 360–361. [Google Scholar] [CrossRef] [PubMed]
- Arredondo, A.; Blanc, V.; Mor, C.; Nart, J.; León, R. Azithromycin and erythromycin susceptibility and macrolide resistance genes in Prevotella from patients with periodontal disease. Oral Dis. 2019, 25, 860–867. [Google Scholar] [CrossRef] [PubMed]
- Roberts, M.C. Location of the Various Genes Table. Available online: http://faculty.washington.edu/marilynr (accessed on 20 February 2023).
- Tansirichaiya, S.; Rahman, A.; Roberts, A.P. The Transposon Registry. Mob. DNA 2019, 10, 40. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control (ECDC); European Food Safety Authority (EFSA); European Medicines Agency (EMA). ECDC/EFSA/EMA Second Joint Report on the Integrated Analysis of the Consumption of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Bacteria from Humans and Food-Producing Animals. EFSA J. 2017, 15, e04872. [Google Scholar] [CrossRef]
- Oscarsson, J.; Claesson, R.; Bao, K.; Brundin, M.; Belibasakis, G. Phylogenetic Analysis of Filifactor alocis Strains Isolated from Several Oral Infections Identified a Novel RTX Toxin, FtxA. Toxins 2020, 12, 687. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Feldgarden, M.; Brover, V.; Gonzalez-Escalona, N.; Frye, J.G.; Haendiges, J.; Haft, D.H.; Hoffmann, M.; Pettengill, J.B.; Prasad, A.B.; Tillman, G.E.; et al. AMRFinderPlus and the Reference Gene Catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci. Rep. 2021, 11, 12728. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
| Isolate | Gene | Class of Antimicrobial | Coverage of Sequence (%) | Identity of Sequence (%) |
|---|---|---|---|---|
| 5.15 | tet(32) | Tetracycline | 68.39 | 99.54 |
| 11.40 | tet(32) | Tetracycline | 100 | 100 |
| 14.12 | tet(32) | Tetracycline | 100 | 100 |
| 22.10 | tet(32) | Tetracycline | 100 | 100 |
| 22.40 | tet(32) | Tetracycline | 100 | 100 |
| 30.27 | erm(B) | Macrolide | 72.24 | 100 |
| Isolate | AMC | AMX | AZM | CDM | CIP | DOX | MIN | MTZ | TET |
|---|---|---|---|---|---|---|---|---|---|
| 5.15 | <0.016 | <0.016 | 1 | 0.047 | 0.008 | 0.047 | 0.032 | <0.016 | 0.25 |
| 8.25 | 0.016 | 0.023 | 0.032 | 0.016 | 0.023 | 0.023 | 0.016 | 0.016 | 0.016 |
| 11.40 | 0.032 | 0.023 | 0.047 | <0.016 | 0.032 | 0.125 | 0.094 | <0.016 | 0.75 |
| 14.12 | 0.25 | <0.016 | 0.032 | <0.016 | 0.012 | 0.5 | 0.125 | <0.016 | 0.75 |
| 22.10 | 0.016 | 0.064 | 0.047 | 0.016 | 0.023 | 0.25 | 0.19 | 0.016 | 1 |
| 22.40 | 0.023 | 0.032 | 0.032 | <0.016 | 0.016 | 0.25 | 0.032 | <0.016 | 0.75 |
| 30.27 | <0.016 | <0.016 | >256 | 1 | 0.125 | <0.016 | < 0.016 | <0.016 | 0.016 |
| 48B | 0.032 | 0.016 | 0.064 | <0.016 | 0.047 | 0.047 | < 0.016 | 0.016 | 0.016 |
| ATCC 35896 | 0.064 | 0.032 | 0.032 | <0.016 | 0.023 | 1 | 1 | 0.016 | 2 |
| Range | <0.016–0.094 | <0.016–0.25 | 0.032–>256 | <0.016–0.19 | 0.08–1 | <0.016–0.75 | <0.016–0.19 | <0.016–>256 | 0.016–1 |
| MIC50 | 0.016 | 0.023 | 0.047 | <0.016 | 0.023 | 0.047 | 0.032 | <0.016 | 0.25 |
| MIC90 | 0.032 | 0.064 | 1 | 0.047 | 0.047 | 0.25 | 0.125 | 0.016 | 0.75 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romero-Martínez, R.; Maher, A.; Àlvarez, G.; Figueiredo, R.; León, R.; Arredondo, A. Whole Genome Sequencing and Phenotypic Analysis of Antibiotic Resistance in Filifactor alocis Isolates. Antibiotics 2023, 12, 1059. https://doi.org/10.3390/antibiotics12061059
Romero-Martínez R, Maher A, Àlvarez G, Figueiredo R, León R, Arredondo A. Whole Genome Sequencing and Phenotypic Analysis of Antibiotic Resistance in Filifactor alocis Isolates. Antibiotics. 2023; 12(6):1059. https://doi.org/10.3390/antibiotics12061059
Chicago/Turabian StyleRomero-Martínez, Rosa, Anushiravan Maher, Gerard Àlvarez, Rui Figueiredo, Rubén León, and Alexandre Arredondo. 2023. "Whole Genome Sequencing and Phenotypic Analysis of Antibiotic Resistance in Filifactor alocis Isolates" Antibiotics 12, no. 6: 1059. https://doi.org/10.3390/antibiotics12061059
APA StyleRomero-Martínez, R., Maher, A., Àlvarez, G., Figueiredo, R., León, R., & Arredondo, A. (2023). Whole Genome Sequencing and Phenotypic Analysis of Antibiotic Resistance in Filifactor alocis Isolates. Antibiotics, 12(6), 1059. https://doi.org/10.3390/antibiotics12061059







