Antimicrobial Resistance and Clonal Lineages of Escherichia coli from Food-Producing Animals
Abstract
1. Antibiotics Use in Food-Producing Animals
2. Food-Producing Animals as One of the Potential Sources of Food-Borne Pathogens: Escherichia coli
3. Clonal Lineages of Escherichia coli in Livestock
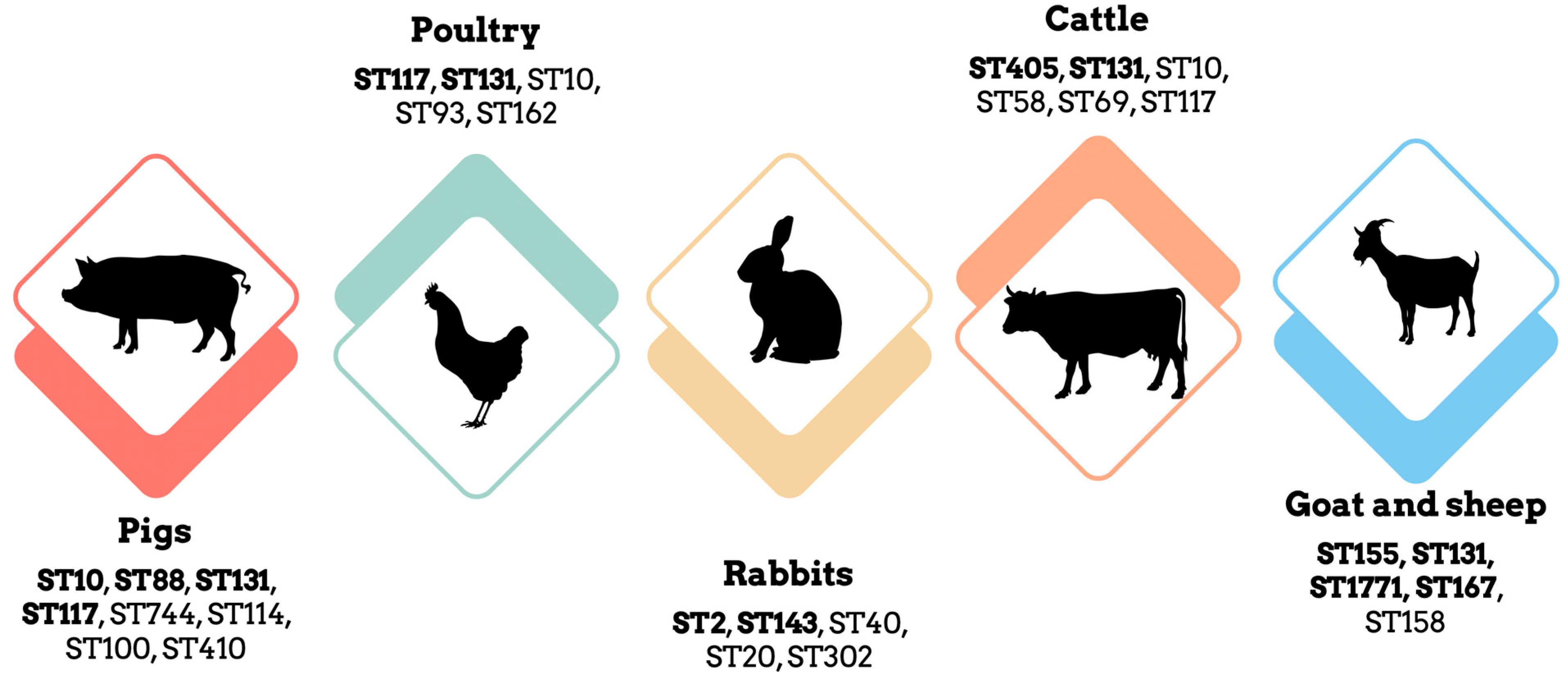
3.1. E. coli in Pigs: A Concerning Trend of Antimicrobial Resistance
3.2. E. coli in Poultry: Multi-Drug Resistant Strains and Potential Reservoirs
3.3. E. coli in Rabbits: High Levels of Resistance to β-Lactamases and Colistin
3.4. E. coli in Cattle: Potential Cross-Species Transmission
3.5. E. coli in Small Ruminants: Widespread Dissemination of Antibiotic Resistance
4. Methods
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lekagul, A.; Tangcharoensathien, V.; Yeung, S. Patterns of Antibiotic Use in Global Pig Production: A Systematic Review. Vet. Anim. Sci. 2019, 7, 100058. [Google Scholar] [CrossRef]
- Van Boeckel, T.P.; Brower, C.; Gilbert, M.; Grenfell, B.T.; Levin, S.A.; Robinson, T.P.; Teillant, A.; Laxminarayan, R. Global Trends in Antimicrobial Use in Food Animals. Proc. Natl. Acad. Sci. USA 2015, 112, 5649–5654. [Google Scholar] [CrossRef]
- Ma, W.; Guo, A.; Zhang, Y.; Wang, H.; Liu, Y.; Li, H. A Review on Astringency and Bitterness Perception of Tannins in Wine. Trends Food Sci. Technol. 2014, 40, 6–19. [Google Scholar] [CrossRef]
- Ceccarelli, D.; Hesp, A.; van der Goot, J.; Joosten, P.; Sarrazin, S.; Wagenaar, J.A.; Dewulf, J.; Mevius, D.J.; on behalf of the EFFORT consortium. Antimicrobial Resistance Prevalence in Commensal Escherichia coli from Broilers, Fattening Turkeys, Fattening Pigs and Veal Calves in European Countries and Association with Antimicrobial Usage at Country Level. J. Med. Microbiol. 2020, 69, 537–547. [Google Scholar] [CrossRef]
- Scott, H.M.; Acuff, G.; Bergeron, G.; Bourassa, M.W.; Gill, J.; Graham, D.W.; Kahn, L.H.; Morley, P.S.; Salois, M.J.; Simjee, S.; et al. Critically Important Antibiotics: Criteria and Approaches for Measuring and Reducing Their Use in Food Animal Agriculture. Ann. N. Y. Acad. Sci. 2019, 1441, 8–16. [Google Scholar] [CrossRef]
- Aidara-Kane, A.; Angulo, F.J.; Conly, J.M.; Minato, Y.; Silbergeld, E.K.; McEwen, S.A.; Collignon, P.J. World Health Organization (WHO) Guidelines on Use of Medically Important Antimicrobials in Food-Producing Animals. Antimicrob. Resist. Infect. Control 2018, 7, 7. [Google Scholar] [CrossRef]
- Patel, S.J.; Wellington, M.; Shah, R.M.; Ferreira, M.J. Antibiotic Stewardship in Food-Producing Animals: Challenges, Progress, and Opportunities. Clin. Ther. 2020, 42, 1649–1658. [Google Scholar] [CrossRef] [PubMed]
- Heredia, N.; García, S. Animals as Sources of Food-Borne Pathogens: A Review. Anim. Nutr. 2018, 4, 250–255. [Google Scholar] [CrossRef] [PubMed]
- Abdalla, S.E.; Abia, A.L.K.; Amoako, D.G.; Perrett, K.; Bester, L.A.; Essack, S.Y. From Farm-to-Fork: E. Coli from an Intensive Pig Production System in South Africa Shows High Resistance to Critically Important Antibiotics for Human and Animal Use. Antibiotics 2021, 10, 178. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Ahn, J. Emergence and Spread of Antibiotic-Resistant Foodborne Pathogens from Farm to Table. Food Sci. Biotechnol. 2022, 31, 1481–1499. [Google Scholar] [CrossRef]
- Vidovic, N.; Vidovic, S. Antimicrobial Resistance and Food Animals: Influence of Livestock Environment on the Emergence and Dissemination of Antimicrobial Resistance. Antibiotics 2020, 9, 52. [Google Scholar] [CrossRef] [PubMed]
- Hamid, H.; Zhao, L.H.; Ma, G.Y.; Li, W.X.; Shi, H.Q.; Zhang, J.Y.; Ji, C.; Ma, Q.G. Evaluation of the Overall Impact of Antibiotics Growth Promoters on Broiler Health and Productivity during the Medication and Withdrawal Period. Poult. Sci. 2019, 98, 3685–3694. [Google Scholar] [CrossRef]
- Food and Agriculture Organization of the United Nations. Monitoring and Surveillance of Antimicrobial Resistance in Bacteria from Healthy Food Animals Intended for Consumption; Regional Antimicrobial Resistance Monitoring and Surveillance Guidelines; Food & Agriculture Organization of the United Nations: Bangkok, Thailand, 2019; ISBN 978-92-5-131930-7. [Google Scholar]
- Ma, F.; Xu, S.; Tang, Z.; Li, Z.; Zhang, L. Use of Antimicrobials in Food Animals and Impact of Transmission of Antimicrobial Resistance on Humans. Biosaf. Health 2021, 3, 32–38. [Google Scholar] [CrossRef]
- Barton, M.D. Impact of Antibiotic Use in the Swine Industry. Curr. Opin. Microbiol. 2014, 19, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Apley, M.D.; Bush, E.J.; Morrison, R.B.; Singer, R.S.; Snelson, H. Use Estimates of In-Feed Antimicrobials in Swine Production in the United States. Foodborne Pathog. Dis. 2012, 9, 272–279. [Google Scholar] [CrossRef]
- Monger, X.C.; Gilbert, A.-A.; Saucier, L.; Vincent, A.T. Antibiotic Resistance: From Pig to Meat. Antibiotics 2021, 10, 1209. [Google Scholar] [CrossRef]
- Ricker, N.; Trachsel, J.; Colgan, P.; Jones, J.; Choi, J.; Lee, J.; Coetzee, J.F.; Howe, A.; Brockmeier, S.L.; Loving, C.L.; et al. Toward Antibiotic Stewardship: Route of Antibiotic Administration Impacts the Microbiota and Resistance Gene Diversity in Swine Feces. Front. Vet. Sci. 2020, 7, 255. [Google Scholar] [CrossRef] [PubMed]
- Zalewska, M.; Błażejewska, A.; Czapko, A.; Popowska, M. Antibiotics and Antibiotic Resistance Genes in Animal Manure—Consequences of Its Application in Agriculture. Front. Microbiol. 2021, 12, 610656. [Google Scholar] [CrossRef]
- Zhao, X.; Zhao, H.; Zhou, Z.; Miao, Y.; Li, R.; Yang, B.; Cao, C.; Xiao, S.; Wang, X.; Liu, H.; et al. Characterization of Extended-Spectrum β-Lactamase-Producing Escherichia coli Isolates That Cause Diarrhea in Sheep in Northwest China. Microbiol. Spectr. 2022, 10, e01595-22. [Google Scholar] [CrossRef]
- Ewers, C.; de Jong, A.; Prenger-Berninghoff, E.; El Garch, F.; Leidner, U.; Tiwari, S.K.; Semmler, T. Genomic Diversity and Virulence Potential of ESBL- and AmpC-β-Lactamase-Producing Escherichia coli Strains From Healthy Food Animals Across Europe. Front. Microbiol. 2021, 12, 626774. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, X.; Wan, F.; You, W.; Tan, X.; Sheng, Q.; Li, C.; Hu, Z.; Liu, G.; Zhao, H. Protective Effect of Resveratrol against Hydrogen Peroxide-Induced Oxidative Stress in Bovine Skeletal Muscle Cells. Meat Sci. 2022, 185, 108724. [Google Scholar] [CrossRef] [PubMed]
- Zil-e-Huma; Tareen, A.M.; Samad, A.; Mustafa, M.Z.; Maryam, M.; Rizwan, S.; Akbar, A. Immunogenic Protein Profiling of Pathogenic Escherichia coli Strains Isolated from Infants with Diarrhea in Quetta Balochistan. J. King Saud Univ. Sci. 2022, 34, 101883. [Google Scholar] [CrossRef]
- Arbab, S.; Ullah, H.; Wang, W.; Zhang, J. Antimicrobial Drug Resistance against Escherichia coli and Its Harmful Effect on Animal Health. Vet. Med. Sci. 2022, 8, 1780–1786. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Madec, J.-Y.; Lupo, A.; Schink, A.-K.; Kieffer, N.; Nordmann, P.; Schwarz, S. Antimicrobial Resistance in Escherichia coli. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef]
- Roth, N.; Käsbohrer, A.; Mayrhofer, S.; Zitz, U.; Hofacre, C.; Domig, K.J. The Application of Antibiotics in Broiler Production and the Resulting Antibiotic Resistance in Escherichia coli: A Global Overview. Poult. Sci. 2019, 98, 1791–1804. [Google Scholar] [CrossRef]
- Silva, N.; Igrejas, G.; Gonçalves, A.; Poeta, P. Commensal Gut Bacteria: Distribution of Enterococcus Species and Prevalence of Escherichia coli Phylogenetic Groups in Animals and Humans in Portugal. Ann. Microbiol. 2012, 62, 449–459. [Google Scholar] [CrossRef]
- Clermont, O.; Bonacorsi, S.; Bingen, E. Rapid and Simple Determination of the Escherichia coli Phylogenetic Group. Appl. Environ. Microbiol. 2000, 66, 4555–4558. [Google Scholar] [CrossRef]
- Carlos, C.; Pires, M.M.; Stoppe, N.C.; Hachich, E.M.; Sato, M.I.; Gomes, T.A.; Amaral, L.A.; Ottoboni, L.M. Escherichia coli Phylogenetic Group Determination and Its Application in the Identification of the Major Animal Source of Fecal Contamination. BMC Microbiol. 2010, 10, 161. [Google Scholar] [CrossRef]
- Lecointre, G.; Rachdi, L.; Darlu, P.; Denamur, E. Escherichia coli Molecular Phylogeny Using the Incongruence Length Difference Test. Mol. Biol. Evol. 1998, 15, 1685–1695. [Google Scholar] [CrossRef]
- Zhao, Q.; Hu, P.; Li, Q.; Zhang, S.; Li, H.; Chang, J.; Jiang, Q.; Zheng, Y.; Li, Y.; Liu, Z.; et al. Prevalence and Transmission Characteristics of Listeria Species from Ruminants in Farm and Slaughtering Environments in China. Emerg. Microbes Infect. 2021, 10, 356–364. [Google Scholar] [CrossRef]
- Ramos, S.; Silva, V.; Dapkevicius, M.d.L.E.; Caniça, M.; Tejedor-Junco, M.T.; Igrejas, G.; Poeta, P. Escherichia coli as Commensal and Pathogenic Bacteria among Food-Producing Animals: Health Implications of Extended Spectrum β-Lactamase (ESBL) Production. Animals 2020, 10, 2239. [Google Scholar] [CrossRef] [PubMed]
- Wibisono, F.J.; Sumiarto, B.; Untari, T.; Effendi, M.H.; Permatasari, D.A.; Witaningrum, M. CTX Gene of Extended Spectrum Beta-Lactamase (ESBL) Producing Escherichia coli on Broilers in Blitar, Indonesia. Syst. Rev. Pharm. 2020, 11, 396–403. [Google Scholar]
- Widodo, A.; Effendi, M.H.; Khairullah, A.R. Extended-Spectrum Beta-Lactamase (ESBL)-Producing Escherichia coli from Livestock. Syst. Rev. Pharm. 2020, 11, 382–392. [Google Scholar]
- Gruel, G.; Sellin, A.; Riveiro, H.; Pot, M.; Breurec, S.; Guyomard-Rabenirina, S.; Talarmin, A.; Ferdinand, S. Antimicrobial Use and Resistance in Escherichia coli from Healthy Food-Producing Animals in Guadeloupe. BMC Vet. Res. 2021, 17, 116. [Google Scholar] [CrossRef]
- Giufrè, M.; Mazzolini, E.; Cerquetti, M.; Brusaferro, S.; Accogli, M.; Agnoletti, F.; Agodi, A.; Alborali, G.L.; Arghittu, M.; Auxilia, F.; et al. Extended-Spectrum β-Lactamase-Producing Escherichia coli from Extraintestinal Infections in Humans and from Food-Producing Animals in Italy: A ‘One Health’ Study. Int. J. Antimicrob. Agents 2021, 58, 106433. [Google Scholar] [CrossRef]
- Falgenhauer, L.; Imirzalioglu, C.; Oppong, K.; Akenten, C.W.; Hogan, B.; Krumkamp, R.; Poppert, S.; Levermann, V.; Schwengers, O.; Sarpong, N.; et al. Detection and Characterization of ESBL-Producing Escherichia coli from Humans and Poultry in Ghana. Front. Microbiol. 2019, 9, 3358. [Google Scholar] [CrossRef]
- De Koster, S.; Ringenier, M.; Xavier, B.B.; Lammens, C.; De Coninck, D.; De Bruyne, K.; Mensaert, K.; Kluytmans-van den Bergh, M.; Kluytmans, J.; Dewulf, J.; et al. Genetic Characterization of ESBL-Producing and Ciprofloxacin-Resistant Escherichia coli from Belgian Broilers and Pigs. Front. Microbiol. 2023, 14, 1150470. [Google Scholar] [CrossRef]
- Al-Mustapha, A.I.; Raufu, I.A.; Ogundijo, O.A.; Odetokun, I.A.; Tiwari, A.; Brouwer, M.S.M.; Adetunji, V.; Heikinheimo, A. Antibiotic Resistance Genes, Mobile Elements, Virulence Genes, and Phages in Cultivated ESBL-Producing Escherichia coli of Poultry Origin in Kwara State, North Central Nigeria. Int. J. Food Microbiol. 2023, 389, 110086. [Google Scholar] [CrossRef]
- Song, J.; Oh, S.-S.; Kim, J.; Park, S.; Shin, J. Clinically Relevant Extended-Spectrum β-Lactamase–Producing Escherichia coli Isolates from Food Animals in South Korea. Front. Microbiol. 2020, 11, 604. [Google Scholar] [CrossRef]
- Balázs, B.; Nagy, J.B.; Tóth, Z.; Nagy, F.; Károlyi, S.; Turcsányi, I.; Bistyák, A.; Kálmán, A.; Sárközi, R.; Kardos, G. Occurrence of Escherichia coli Producing Extended Spectrum β-Lactamases in Food-Producing Animals. Acta Vet. Hung. 2021, 69, 211–215. [Google Scholar] [CrossRef]
- Miltgen, G.; Martak, D.; Valot, B.; Kamus, L.; Garrigos, T.; Verchere, G.; Gbaguidi-Haore, H.; Ben Cimon, C.; Ramiandrisoa, M.; Picot, S.; et al. One Health Compartmental Analysis of ESBL-Producing Escherichia coli on Reunion Island Reveals Partitioning between Humans and Livestock. J. Antimicrob. Chemother. 2022, 77, 1254–1262. [Google Scholar] [CrossRef]
- Chai, M.H.; Sukiman, M.Z.; Jasmy, N.; Zulkifly, N.A.; Mohd Yusof, N.A.S.; Mohamad, N.M.; Ariffin, S.M.Z.; Ghazali, M.F. Molecular Detection and Antibiogram of ESBL-Producing and Carbapenem-Resistant Escherichia coli from Rabbit, Swine, and Poultry in Malaysia. Trop. Anim. Sci. J. 2022, 45, 16–23. [Google Scholar] [CrossRef]
- Sivaraman, G.K.; Rajan, V.; Vijayan, A.; Elangovan, R.; Prendiville, A.; Bachmann, T.T. Antibiotic Resistance Profiles and Molecular Characteristics of Extended-Spectrum Beta-Lactamase (ESBL)-Producing Escherichia coli and Klebsiella pneumoniae Isolated from Shrimp Aquaculture Farms in Kerala, India. Front. Microbiol. 2021, 12, 622891. [Google Scholar] [CrossRef] [PubMed]
- Chibuike, K.U.; Iroha, I.R.; Moses, I.B.; Chukwunwejim, C.R.; Peter, I.U.; Edemekong, C.I.; Ndugo, C.M.; Ngene, O.; Egbuna, N.R.; Okonkwo-Uzor, N.J. Phenotypic Screening of Multidrug-Resistant Escherichia coli from Water and Fish Collected from Different Fish Farms within Abakaliki Metropolis, Nigeria. Sci. Res. Essays 2021, 16, 15–19. [Google Scholar] [CrossRef]
- Sivaraman, G.K.; Sudha, S.; Muneeb, K.H.; Shome, B.; Holmes, M.; Cole, J. Molecular Assessment of Antimicrobial Resistance and Virulence in Multi Drug Resistant ESBL-Producing Escherichia coli and Klebsiella pneumoniae from Food Fishes, Assam, India. Microb. Pathog. 2020, 149, 104581. [Google Scholar] [CrossRef] [PubMed]
- Alegría, Á.; Arias-Temprano, M.; Fernández-Natal, I.; Rodríguez-Calleja, J.M.; García-López, M.-L.; Santos, J.A. Molecular Diversity of ESBL-Producing Escherichia coli from Foods of Animal Origin and Human Patients. IJERPH 2020, 17, 1312. [Google Scholar] [CrossRef] [PubMed]
- von Tippelskirch, P.; Gölz, G.; Projahn, M.; Daehre, K.; Friese, A.; Roesler, U.; Alter, T.; Orquera, S. Prevalence and Quantitative Analysis of ESBL and AmpC Beta-Lactamase Producing Enterobacteriaceae in Broiler Chicken during Slaughter in Germany. Int. J. Food Microbiol. 2018, 281, 82–89. [Google Scholar] [CrossRef]
- Alexandratos, N. World Agriculture towards 2030/2050: The 2012 Revision; ESA: Paris, France, 2012. [Google Scholar]
- Clemente, L.; Leão, C.; Moura, L.; Albuquerque, T.; Amaro, A. Prevalence and Characterization of ESBL/AmpC Producing Escherichia coli from Fresh Meat in Portugal. Antibiotics 2021, 10, 1333. [Google Scholar] [CrossRef]
- Kaesbohrer, A.; Bakran-Lebl, K.; Irrgang, A.; Fischer, J.; Kämpf, P.; Schiffmann, A.; Werckenthin, C.; Busch, M.; Kreienbrock, L.; Hille, K. Diversity in Prevalence and Characteristics of ESBL/PAmpC Producing Escherichia coli in Food in Germany. Vet. Microbiol. 2019, 233, 52–60. [Google Scholar] [CrossRef]
- Tseng, C.-H.; Liu, C.-W.; Liu, P.-Y. Extended-Spectrum β-Lactamases (ESBL) Producing Bacteria in Animals. Antibiotics 2023, 12, 661. [Google Scholar] [CrossRef]
- Riley, L.W. Pandemic Lineages of Extraintestinal Pathogenic Escherichia coli. Clin. Microbiol. Infect. 2014, 20, 380–390. [Google Scholar] [CrossRef]
- Storey, N.; Cawthraw, S.; Turner, O.; Rambaldi, M.; Lemma, F.; Horton, R.; Randall, L.; Duggett, N.A.; AbuOun, M.; Martelli, F.; et al. Use of Genomics to Explore AMR Persistence in an Outdoor Pig Farm with Low Antimicrobial Usage. Microb. Genom. 2022, 8, 000782. [Google Scholar] [CrossRef]
- Soncini, J.G.M.; Cerdeira, L.; Sano, E.; Koga, V.L.; Tizura, A.T.; Tano, Z.N.; Nakazato, G.; Kobayashi, R.K.T.; Aires, C.A.M.; Lincopan, N.; et al. Genomic Insights of High-Risk Clones of ESBL-Producing Escherichia coli Isolated from Community Infections and Commercial Meat in Southern Brazil. Sci. Rep. 2022, 12, 9354. [Google Scholar] [CrossRef]
- Massella, E.; Giacometti, F.; Bonilauri, P.; Reid, C.J.; Djordjevic, S.P.; Merialdi, G.; Bacci, C.; Fiorentini, L.; Massi, P.; Bardasi, L.; et al. Antimicrobial Resistance Profile and ExPEC Virulence Potential in Commensal Escherichia coli of Multiple Sources. Antibiotics 2021, 10, 351. [Google Scholar] [CrossRef]
- Adefioye, O.J.; Weinreich, J.; Rödiger, S.; Schierack, P.; Olowe, O.A. Phylogenetic Characterization and Multilocus Sequence Typing of Extended-Spectrum Beta Lactamase-Producing Escherichia coli from Food-Producing Animals, Beef, and Humans in Southwest Nigeria. Microb. Drug Resist. 2021, 27, 111–120. [Google Scholar] [CrossRef]
- Founou, L.L.; Founou, R.C.; Allam, M.; Ismail, A.; Essack, S.Y. Genome Analysis of ESBL-Producing Escherichia coli Isolated from Pigs. Pathogens 2022, 11, 776. [Google Scholar] [CrossRef]
- Fournier, C.; Nordmann, P.; Pittet, O.; Poirel, L. Does an Antibiotic Stewardship Applied in a Pig Farm Lead to Low ESBL Prevalence? Antibiotics 2021, 10, 574. [Google Scholar] [CrossRef] [PubMed]
- Bernreiter-Hofer, T.; Schwarz, L.; Müller, E.; Cabal-Rosel, A.; Korus, M.; Misic, D.; Frankenfeld, K.; Abraham, K.; Grünzweil, O.; Weiss, A.; et al. The Pheno- and Genotypic Characterization of Porcine Escherichia coli Isolates. Microorganisms 2021, 9, 1676. [Google Scholar] [CrossRef] [PubMed]
- Szmolka, A.; Wami, H.; Dobrindt, U. Comparative Genomics of Emerging Lineages and Mobile Resistomes of Contemporary Broiler Strains of Salmonella Infantis and Escherichia coli. Front. Microbiol. 2021, 12, 642125. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Kim, Y.A.; Seo, Y.H.; Lee, H.; Lee, K. Prevalence and Molecular Epidemiology of Extended-Spectrum-β-Lactamase (ESBL)-Producing Escherichia coli from Multiple Sectors of Poultry Industry in Korea. Antibiotics 2021, 10, 1050. [Google Scholar] [CrossRef]
- Zhao, X.; Yang, J.; Ju, Z.; Chang, W.; Sun, S. Molecular Characterization of Antimicrobial Resistance in Escherichia coli from Rabbit Farms in Tai’an, China. BioMed Res. Int. 2018, 2018, 8607647. [Google Scholar] [CrossRef]
- Freitas-Silva, J.; Inácio, Â.S.; Mourão, J.; Antunes, P.; Mendes, Â.; de Carvalho, A.P.; Vasconcelos, V.; Peixe, L.; da Costa, P.M. Occurrence of mcr-1 in Escherichia coli from Rabbits of Intensive Farming. Vet. Microbiol. 2018, 227, 78–81. [Google Scholar] [CrossRef]
- Massé, J.; Vanier, G.; Fairbrother, J.M.; de Lagarde, M.; Arsenault, J.; Francoz, D.; Dufour, S.; Archambault, M. Description of Antimicrobial-Resistant Escherichia coli and Their Dissemination Mechanisms on Dairy Farms. Vet. Sci. 2023, 10, 242. [Google Scholar] [CrossRef]
- Smoglica, C.; Barco, L.; Angelucci, S.; Orsini, M.; Marsilio, F.; Antonucci, A.; Di Francesco, C.E. Whole Genome Sequencing of Escherichia coli and Enterococcus spp. in Wildlife-Livestock Interface: A Pilot Study. J. Glob. Antimicrob. Resist. 2023, 32, 118–121. [Google Scholar] [CrossRef] [PubMed]
- AbuOun, M.; O’Connor, H.M.; Stubberfield, E.J.; Nunez-Garcia, J.; Sayers, E.; Crook, D.W.; Smith, R.P.; Anjum, M.F. Characterizing Antimicrobial Resistant Escherichia coli and Associated Risk Factors in a Cross-Sectional Study of Pig Farms in Great Britain. Front. Microbiol. 2020, 11, 861. [Google Scholar] [CrossRef] [PubMed]
- AbuOun, M.; Jones, H.; Stubberfield, E.; Gilson, D.; Shaw, L.P.; Hubbard, A.T.M.; Chau, K.K.; Sebra, R.; Peto, T.E.A.; Crook, D.W.; et al. A Genomic Epidemiological Study Shows That Prevalence of Antimicrobial Resistance in Enterobacterales Is Associated with the Livestock Host, as Well as Antimicrobial Usage. Microb. Genom. 2021, 7, 000630. [Google Scholar] [CrossRef] [PubMed]
- Duggett, N.; AbuOun, M.; Randall, L.; Horton, R.; Lemma, F.; Rogers, J.; Crook, D.; Teale, C.; Anjum, M.F. The Importance of Using Whole Genome Sequencing and Extended Spectrum Beta-lactamase Selective Media When Monitoring Antimicrobial Resistance. Sci. Rep. 2020, 10, 19880. [Google Scholar] [CrossRef]
- Guenther, S.; Aschenbrenner, K.; Stamm, I.; Bethe, A.; Semmler, T.; Stubbe, A.; Stubbe, M.; Batsajkhan, N.; Glupczynski, Y.; Wieler, L.H.; et al. Comparable High Rates of Extended-Spectrum-Beta-Lactamase-Producing Escherichia coli n Birds of Prey from Germany and Mongolia. PLoS ONE 2012, 7, e53039. [Google Scholar] [CrossRef]
- Cormier, A.; Zhang, P.L.C.; Chalmers, G.; Weese, J.S.; Deckert, A.; Mulvey, M.; McAllister, T.; Boerlin, P. Diversity of CTX-M-Positive Escherichia coli Recovered from Animals in Canada. Vet. Microbiol. 2019, 231, 71–75. [Google Scholar] [CrossRef]
- Xia, L.; Liu, Y.; Xia, S.; Kudinha, T.; Xiao, S.; Zhong, N.; Ren, G.; Zhuo, C. Prevalence of ST1193 Clone and IncI1/ST16 Plasmid in Escherichia coli Isolates Carrying blaCTX-M-55 Gene from Urinary Tract Infections Patients in China. Sci. Rep. 2017, 7, 44866. [Google Scholar] [CrossRef]
- Clermont, O.; Dixit, O.V.A.; Vangchhia, B.; Condamine, B.; Dion, S.; Bridier-Nahmias, A.; Denamur, E.; Gordon, D. Characterization and Rapid Identification of Phylogroup G in Escherichia coli, a Lineage with High Virulence and Antibiotic Resistance Potential. Environ. Microbiol. 2019, 21, 3107–3117. [Google Scholar] [CrossRef] [PubMed]
- Rafaï, C.; Frank, T.; Manirakiza, A.; Gaudeuille, A.; Mbecko, J.-R.; Nghario, L.; Serdouma, E.; Tekpa, B.; Garin, B.; Breurec, S. Dissemination of IncF-Type Plasmids in Multiresistant CTX-M-15-Producing Enterobacteriaceae Isolates from Surgical-Site Infections in Bangui, Central African Republic. BMC Microbiol. 2015, 15, 15. [Google Scholar] [CrossRef] [PubMed]
- Founou, L.L.; Founou, R.C.; Essack, S.Y. Antimicrobial Resistance in the Farm-to-Plate Continuum: More than a Food Safety Issue. Future Sci. OA 2021, 7, FSO692. [Google Scholar] [CrossRef]
- Reid, C.J.; Blau, K.; Jechalke, S.; Smalla, K.; Djordjevic, S.P. Whole Genome Sequencing of Escherichia coli from Store-Bought Produce. Front. Microbiol. 2020, 10, 3050. [Google Scholar] [CrossRef] [PubMed]
- Manges, A.R. Escherichia coli and Urinary Tract Infections: The Role of Poultry-Meat. Clin. Microbiol. Infect. 2016, 22, 122–129. [Google Scholar] [CrossRef]
- García, V.; García-Meniño, I.; Mora, A.; Flament-Simon, S.C.; Díaz-Jiménez, D.; Blanco, J.E.; Alonso, M.P.; Blanco, J. Co-Occurrence of mcr-1, mcr-4 and mcr-5 Genes in Multidrug-Resistant ST10 Enterotoxigenic and Shiga Toxin-Producing Escherichia coli in Spain (2006-2017). Int. J. Antimicrob. Agents 2018, 52, 104–108. [Google Scholar] [CrossRef]
- Cuong, N.; Padungtod, P.; Thwaites, G.; Carrique-Mas, J. Antimicrobial Usage in Animal Production: A Review of the Literature with a Focus on Low- and Middle-Income Countries. Antibiotics 2018, 7, 75. [Google Scholar] [CrossRef]
- Zhou, Z.; Alikhan, N.-F.; Mohamed, K.; Fan, Y.; The Agama Study Group; Achtman, M. The EnteroBase User’s Guide, with Case Studies on Salmonella Transmissions, Yersinia pestis Phylogeny, and Escherichia coli Genomic Diversity. Genome Res. 2020, 30, 138–152. [Google Scholar] [CrossRef]
- Liu, X.; Liu, H.; Wang, L.; Peng, Q.; Li, Y.; Zhou, H.; Li, Q. Molecular Characterization of Extended-Spectrum β-Lactamase-Producing Multidrug Resistant Escherichia coli from Swine in Northwest China. Front. Microbiol. 2018, 9, 1756. [Google Scholar] [CrossRef]
- Birgy, A.; Madhi, F.; Jung, C.; Levy, C.; Cointe, A.; Bidet, P.; Hobson, C.A.; Bechet, S.; Sobral, E.; Vuthien, H.; et al. Diversity and Trends in Population Structure of ESBL-Producing Enterobacteriaceae in Febrile Urinary Tract Infections in Children in France from 2014 to 2017. J. Antimicrob. Chemother. 2020, 75, 96–105. [Google Scholar] [CrossRef]
- Li, G.; Cai, W.; Hussein, A.; Wannemuehler, Y.M.; Logue, C.M.; Nolan, L.K. Proteome Response of an Extraintestinal Pathogenic Escherichia coli Strain with Zoonotic Potential to Human and Chicken Sera. J. Proteom. 2012, 75, 4853–4862. [Google Scholar] [CrossRef] [PubMed]
- Tansawai, U.; Walsh, T.R.; Niumsup, P.R. Extended Spectrum SS-Lactamase-Producing Escherichia coli among Backyard Poultry Farms, Farmers, and Environments in Thailand. Poult. Sci. 2019, 98, 2622–2631. [Google Scholar] [CrossRef] [PubMed]
- Aarestrup, F.M.; Wegener, H.C.; Collignon, P. Resistance in Bacteria of the Food Chain: Epidemiology and Control Strategies. Expert Rev. Anti-Infect. Ther. 2008, 6, 733–750. [Google Scholar] [CrossRef] [PubMed]
- Agnoletti, F.; Brunetta, R.; Bano, L.; Drigo, I.; Mazzolini, E. Longitudinal Study on Antimicrobial Consumption and Resistance in Rabbit Farming. Int. J. Antimicrob. Agents 2018, 51, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Cunha, S.; Mendes, Â.; Rego, D.; Meireles, D.; Fernandes, R.; Carvalho, A.; Costa, P.M. da Effect of Competitive Exclusion in Rabbits Using an Autochthonous Probiotic. World Rabbit Sci. 2017, 25, 123. [Google Scholar] [CrossRef]
- Gelbíčová, T.; Baráková, A.; Florianová, M.; Jamborová, I.; Zelendová, M.; Pospíšilová, L.; Koláčková, I.; Karpíšková, R. Dissemination and Comparison of Genetic Determinants of mcr-Mediated Colistin Resistance in Enterobacteriaceae via Retailed Raw Meat Products. Front. Microbiol. 2019, 10, 2824. [Google Scholar] [CrossRef]
- Zhang, Y.-L.; Huang, F.-Y.; Gan, L.-L.; Yu, X.; Cai, D.-J.; Fang, J.; Zhong, Z.; Guo, H.; Xie, Y.; Yi, J.; et al. High Prevalence of BlaCTX-M and BlaSHV among ESBL Producing E. coli Isolates from Beef Cattle in China’s Sichuan-Chongqing Circle. Sci. Rep. 2021, 11, 13725. [Google Scholar] [CrossRef]
- Millar, N.; Aenishaenslin, C.; Lardé, H.; Roy, J.; Fourichon, C.; Francoz, D.; Paradis, M.; Dufour, S. Evidence of a Decrease in Sales of Antimicrobials of Very High Importance for Humans in Dairy Herds after a New Regulation Restricting Their Use in Quebec, Canada. Zoonoses Public Health 2022, 69, 370–381. [Google Scholar] [CrossRef]
- Dantas Palmeira, J.; Haenni, M.; Madec, J.-Y.; Ferreira, H.M.N. First Global Report of Plasmid-Mediated mcr-1 and Extended-Spectrum Beta-Lactamase-Producing Escherichia coli from Sheep in Portugal. Antibiotics 2021, 10, 1403. [Google Scholar] [CrossRef]
- Atlaw, N.A.; Keelara, S.; Correa, M.; Foster, D.; Gebreyes, W.; Aidara-Kane, A.; Harden, L.; Thakur, S.; Fedorka-Cray, P.J. Evidence of Sheep and Abattoir Environment as Important Reservoirs of Multidrug Resistant Salmonella and Extended-Spectrum Beta-Lactamase Escherichia coli. Int. J. Food Microbiol. 2022, 363, 109516. [Google Scholar] [CrossRef]
- Eger, E.; Domke, M.; Heiden, S.E.; Paditz, M.; Balau, V.; Huxdorff, C.; Zimmermann, D.; Homeier-Bachmann, T.; Schaufler, K. Highly Virulent and Multidrug-Resistant Escherichia coli Sequence Type 58 from a Sausage in Germany. Antibiotics 2022, 11, 1006. [Google Scholar] [CrossRef] [PubMed]
- Skurnik, D.; Clermont, O.; Guillard, T.; Launay, A.; Danilchanka, O.; Pons, S.; Diancourt, L.; Lebreton, F.; Kadlec, K.; Roux, D.; et al. Emergence of Antimicrobial-Resistant Escherichia coli of Animal Origin Spreading in Humans. Mol. Biol. Evol. 2016, 33, 898–914. [Google Scholar] [CrossRef] [PubMed]
- Mihaila, L.; Wyplosz, B.; Clermont, O.; Garry, L.; Hipeaux, M.C.; Vittecoq, D.; Dussaix, E.; Denamur, E.; Branger, C. Probable Intrafamily Transmission of a Highly Virulent CTX-M-3-Producing Escherichia coli Belonging to the Emerging Phylogenetic Subgroup D2 O102-ST405 Clone. J. Antimicrob. Chemother. 2010, 65, 1537–1539. [Google Scholar] [CrossRef] [PubMed]

| Samples | Location | Data of Isolation Samples | Food-Producing Animals | Total Samples Collected | Method Used for ESBL Detected | Number of Samples Detected with ESBL—E. coli Positive | Prevalence ESBL—E. coli Positive (%) | Reference |
|---|---|---|---|---|---|---|---|---|
| Fecal | Guadeloupe | Poultry | 216 | Double-disk synergy test | 6 | 14.7 | [35] | |
| 2018–2019 | Pigs | 9 | 7.3 | |||||
| Pigs | 11 | 35.3 | ||||||
| Fecal/cacecal intestinal content | Italy | 2016–2017 | Pigs | 445 | Enrichment broth+CTX/double-disk synergy test | 120 | 27 | [36] |
| Cattle | 120 | 27 | ||||||
| Poultry | 194 | 43.6 | ||||||
| Fecal | Ghana | 2015 | Poultry | 140 | Enrichment broth+CTX/double-disk synergy test | 41 | 29 | [37] |
| Fecal | Belgium | 2017–2018 | Pigs | 798 | Double-disk synergy test | 37 | 48 | [38] |
| Broilers | 45 | 58.4 | ||||||
| Fecal | Hungary | ND | Poultry | 124 | Double-disk synergy test | 39 | 34.2 | [41] |
| Porcine | 100 | 72 | 72 | |||||
| Food (Farms) | Spain | ND | Chicken meat | 10 | Chromagar ESBL | 2 | 15.4 | [47] |
| Goat milk | 68 | 3 | 23 | |||||
| Fresh cheese | 20 | 3 | 23 | |||||
| Ewe’s milk | 10 | 5 | 38.5 | |||||
| Caecal | Nigeria | 2019 | Spent layers | 50 | Double-disk synergy test | 4 | 8 | [39] |
| Broilers | 304 | 33 | 10.8 | |||||
| Boot/rectal (cattle) | Reunion island | 2015–2018 | Rabbits | 39 | Double-disk synergy | 0 | 0 | [42] |
| Cattle | 124 | 2 | 8.3 | |||||
| Sheep and Goats | 50 | 3 | 18.5 | |||||
| Pig | 177 | 33 | 50 | |||||
| Poultry | 176 | 50 | 70 | |||||
| Fecal | Malaysia | ND | Rabbits | 100 | PCR | 0 | 0 | [43] |
| Swine | 100 | 0 | 0 | |||||
| Poultry | 200 | 11 | 5.5 | |||||
| Fecal | South Korea | 2018 | Chickens | 32 | Chromagar ESBL+ Double-disk synergy test | 4 | 7 | [40] |
| Pigs | 59 | 41 | 69.5 | |||||
| Cattle | 34 | 32 | 94.1 | |||||
| Food (meat) | Portugal | 2016–2017 | Pork | 220 | Double-disk synergy test | 23 | 10.5 | [48] |
| Beef | 220 | 26 | 11.8 | |||||
| Broiler | 198 | 60 | 30.3 | |||||
| Fish samples-site | India | 2019 | Fish | 66 | Double-disk synergy test | 54 | 81.8 | [46] |
| Shrimp, water, and sediment | India | 2018–2020 | Aquaculture farms | 261 | BD PhoenixTM M50 automated system | 14 | 5.4 | [44] |
| Fish | Nigeria | ND | Fish farms | 90 | Double-disk synergy test | 54 | 60 | [45] |
| Animal | Samples | Location | Data of Isolation Samples | Most Prevalent Phylogroup | Clonal Lineages (MLST) | AMR Phenotypes | AMR Genotypes | MDR (%) | References |
|---|---|---|---|---|---|---|---|---|---|
| Pigs | Faecal | United Kingdon | 2017–2018 | ND | ST44, ST88, ST10, ST744, ST58, ST117, ST48, ST2721 | Aminoglycosides, β-lactams, fluoroquinolones, sulphonamides, tetracyclines | aadA5, strA, strBaph (6)-Ib, blaTEM-1b, mphA, sul, sul2, tetAB, tetB, dfrA5 | 7 | [54] |
| Meat | Brazil | 2016–2019 | ND | ST410, ST117 | Aminoglycosides, β-lactams, trimethoprim, phenicols, tetracyclines, macrolides, sulfonamides, quinolones, lincosamides, fosfomycin | strA and strB, aadA1, dfrA17, dfrA1, tet(A), tet(B), sul1, sul2, of blaCTX-M-55, blaCTX-M-15, blaCTX-M-2, blaCMY-2 | ND | [55] | |
| Feacal | Italy | 2010–2018 | A (32%), B1 (32%),C (32%), E(4%) | ST10, ST641, ST3744,ST575,ST100, ST20, ST206, ST871, ST410, ST7093, ST88 | Streptomycin, chloramphenicol, sulfisoxazole, trimethoprim/sulfamethoxazole, tetracycline, nalidixic acid; enrofloxacin, colistin | blaTEM-1b, aadA1, strA, strB, sul1, dfrA1 tetA, tetB | 24 | [56] | |
| Feacal | Nigeria | 2015–2016 | D, B2, B1, A | ST131, ST2348 | Clindamycin, penicillin, ceftazidime, tobramycin, cefazolin, enrofloxacin, levofloxacin, sulfamethoxazole/trimethoprim, kanamycin, cefuroxime, piperacillin/tazobactam, ampicillin, cefalexin, streptomycin, doxycycline, neomycin, spectinomycin, amoxicillin/clavulanate, sulfamethoxazole, ampicillin/sulbactam, cefotaxime, ticarcillin, ciprofloxacin, trimethoprim, tetracycline | blaCTX-M-15, blaCTX-M-1, blaCTX-2, blaCTX-M-9 | 42.1 | [57] | |
| Nasal and rectal | Cameroon and South Africa | 2016 | A (45%), B1 (28%) and C (18%), D (9%) | ST88, ST2144, ST10, ST69, ST226, ST944, ST4450, ST44 | ampicillin, cefuroxime, cefuroxime acetyl, cefotaxime, ceftazidime, cefepime, trimethoprim–sulfamethoxazole | blaCTX-M-15, blaTEM-1B, blaTEM-141, blaTEM-206, aph(3″)-Ib, aph(6)-1d, aadA5, aadA1, qnrS1, aac(6′)Ib-cr, oqxAB, gyrA, sul2, sul1, dfrA17, dfrA14 | 18.18 | [58] | |
| Rectal | Switzerland | ND | A (57.9%), B (34.3%), C (2.6%), B1 (31.5%), B2 (2.6%) | ST10, ST34, ST744 | Tetracycline, sulfonamides, gentamicin, tobramycin; kanamycin | blaCTX-M-1 | ND | [59] | |
| gut-associated | Austria | ND | A (50.98%), B1 (25.48%), C (8.81%), D (5.87%), B2 (3.91%), F (1.95%), E (0.97%), G (0.97%) | ST10, ST100, ST354, ST131, ST6404,ST636,ST1112,ST107,ST760, ST744, ST641, ST117, ST101, ST56, ST42, ST23, ST12008, ST12009, ST12010 | Ampicillin, tetracycline, piperacillin, sulfamethoxazole–trimethoprim, cefotaxime, chloramphenicol, ceftazidime, cefepime, gentamicin, fluoroquinolone, aztreonam, tobramycin, and fosfomycin, colistin | blaCTX, blaCMY-2, blaTEM-1, blaCTX M-1, mcr-1.1, gyrA, parC | 36.27 | [60] | |
| Poultry | Faecal, caecum and bone marrow | Hungary | 2016–2018 | A (27%) B1 (37%) | ST10, ST93, ST117, ST162, ST155, ST8702, ST10088 | Aminoglycosides, β-lactams, fluoroquinolone, sulphonamides, tetracyclines, trimethoprim. | blaTEM-1, tet (A), aadA1, aph (3″)-Ib, aph (6)-Id, sul2 | ND | [61] |
| Meat | Brazil | 2016–2019 | ND | ST38, ST131, ST354, ST1196, ST117 | Aminoglycosides, β-lactams, trimethoprim, phenicols, tetracyclines, macrolides, sulfonamides, quinolones, lincosamides, fosfomycin | strA and strB, aadA1, dfrA17, dfrA, tet(A), tet(B), sul1, sul2 | ND | [55] | |
| Food | Italy | 2010–2018 | A (8%), B1 (24%), B2 (4%), C (32%), E (8%) F (20%) | ST23, ST101, ST359, ST131, ST117, ST744, ST57, ST48, ST162, ST10, ST155, ST2614, ST297, ST93, ST69, ST1286 | Gentamicin, Streptomycin, chloramphenicol, sulfisoxazole, trimethoprim/sulfamethoxazole, tetracycline, nalidixic acid; enrofloxacin | blaTEM-1b, aadA1, strA, strB, sul1, dfrA1 tetA, tetB | 64 | [56] | |
| Feacal | Nigeria | 2015–2016 | B2, B1, A | ST131, ST156, ST167, ST410, ST1056 | Clindamycin, penicillin, ceftazidime, tobramycin, cefazolin, enrofloxacin, levofloxacin, sulfamethoxazole/trimethoprim, kanamycin, cefuroxime, piperacillin/tazobactam, ampicillin, cefalexin, streptomycin, doxycycline, neomycin, spectinomycin, amoxicillin/clavulanate, sulfamethoxazole, ampicillin/sulbactam, cefotaxime, ticarcillin, ciprofloxacin, trimethoprim, and tetracycline | blaCTX-M-15, blaCTX-M-1, blaCTX-2, blaCTX-M-9 | 38.9 | [57] | |
| Chicken meat and environment | Korea | 2019 | ND | ST93, ST131, ST48, ST57, ST69, ST88, ST115, ST117, ST162, ST297, ST362, ST457, ST770, ST919, ST1011, ST143, ST1485, ST163, ST165, ST2179, ST2334, ST278, ST2792, ST328, ST3941,ST455, ST6779 | nalidixic acid, ciprofloxacin, tetracycline, chloramphenicol, cotrimoxazole, gentamicin | bla, str, aad, aac, aph, mph, aac(6′)Ib-cr, Qnr, dfr, sul, tet, cat, fos, ARR-3, Inu,CTX-M-55, CTX-M-14, CTX-M-, CTX-M-15, CTX-M-65, CTX-M-27 and CTX-M-61 | ND | [62] | |
| Rabbits | Food | Italy | 2010–2018 | B1 (86.96%), B2 (4.35%), E (8%), D (8.70%) | ST40, ST20, ST1611, ST297, ST533, ST20, ST129, ST706, ST906, ST351, ST501, ST224, ST111, ST539, ST1431, ST491, ST1727 | Gentamicin, Streptomycin, chloramphenicol, sulfisoxazole, trimethoprim/sulfamethoxazole, tetracycline, nalidixic acid; enrofloxacin, colistin | blaTEM-1b, aadA1, strA, strB, sul1, dfrA1 tetA, tetB | 95.65 | [56] |
| Feacal | China | 2016 | ND | ST302, ST468, ST370, ST87, ST314, ST370, ST636, ST2, ST24, ST88, ST353, ST370, ST461, ST731, ST73 | Tetracycline, ampicillin, chloramphenicol, ciprofloxacin gentamicin, nalidixic acid, trimethoprim/sulfamethoxazole | blaTEM, blaCTX-M, sul2, tetB qnrS, aac(6)-Ib-cr | 50.9 | [63] | |
| Intestinal content | Portugal | ND | A, B1 | ST206, ST1589, ST1431, ST2, ST4 | Colistin | mcr-1 | ND | [64] | |
| Cattle | Food | Italy | 2010–2018 | A (32%), B1 (36%) B2 (8%), C (16%), D (4%), E (4%) | ST1510, ST398, ST10, ST583, ST1303, ST58, ST155, ST69, ST278, ST1091, ST731, ST2328, ST216, ST1125 | Gentamicin, Streptomycin, chloramphenicol, sulfisoxazole, trimethoprim/sulfamethoxazole, tetracycline, nalidixic acid; enrofloxacin, colistin, ceftiofur, ceftazidime | blaTEM-1b, aadA1, strA, strB, sul1, dfrA1 tetA, tetB | 24 | [56] |
| Beef | Nigeria | 2015–2016 | D, B2, B1, A | ST58, ST131, ST405 | Clindamycin, penicillin, ceftazidime, tobramycin, cefazolin, enrofloxacin, levofloxacin, sulfamethoxazole/trimethoprim, kanamycin, cefuroxime, piperacillin/tazobactam, ampicillin, cefalexin, streptomycin, doxycycline, neomycin, spectinomycin, amoxicillin/clavulanate, sulfamethoxazole, ampicillin/sulbactam, cefotaxime, ticarcillin, ciprofloxacin, trimethoprim, and tetracycline | blaCTX-M-15, blaCTX-M-1, blaCTX-2, blaCTX-M-9 | 22.6 | [57] | |
| Feacal | 2015–2016 | D, B2, B1, A | ST131, ST405 | 33.3 | |||||
| Beef cattle/Cloacal | China | 2016 | A (50%), B1 (34%), B2 (10.22%), D (5.6%) | ST398, ST7130, ST297, ST48, ST4977, ST202 | β-lactam, penicillin derivatives and third generation cephalosporins | blaCTX-M, blaTEM, blaSHV | ND | [63] | |
| Dairy farms | Feacal | Canada | ND | A (33.7%), E (4.6%), D (5.8%), F (3.4%), G (4.6%), C (18.6%), B1(29%) | ST10, ST88, ST744, ST4981, ST2500, ST34, ST48, ST11813, ST5708, ST408, ST540, ST1204, ST219, ST3018, ST2449, ST38, ST69, ST967, ST648, ST1163, ST657, ST117, ST783, ST21, ST4559, ST162, ST2522, ST172, ST345, ST297, ST155, ST683 | Ampicillin, ceftriaxone, sulfisoxazole, ceftiofur, tetracycline, ciprofloxacin, danofloxacin, enrofloxacin, and nalidixic acid, azithromycin, gentamicin | sul2, strA/strB, tet(A), aph(3′)-1a, blaCTX-M, blaCMY-2, ampC, qnrS1, blaCTX-M-15, blaCTX-M-1, gyrA, parC, parE | ND | [65] |
| Goats | Feacal | Nigeria | 2015–2016 | B2, B1, A | ST131, ST155, ST167, ST406, ST1771 | Clindamycin, penicillin, ceftazidime, tobramycin, cefazolin, enrofloxacin, levofloxacin, sulfamethoxazole/trimethoprim, kanamycin, cefuroxime, piperacillin/tazobactam, ampicillin, cefalexin, streptomycin, doxycycline, neomycin, spectinomycin, amoxicillin/clavulanate, sulfamethoxazole, ampicillin/sulbactam, cefotaxime, ticarcillin, ciprofloxacin, trimethoprim, and tetracycline | blaCTX-M-15, blaCTX-M-1, blaCTX-2, blaCTX-M-9, blaCTX-M-11 | 50 | [57] |
| Feacal | Italy | 2019 | ND | ST675 | Colistin, tetracycline | KpnE, KpnF, acrD, baeR, baeS, cpxA, tolC, soxS, soxR, marA, ampC, ampC 1, acrA, acrB, acrE, acrF, acrR, acrS, CRP, emrE, evgA, evgS, gadX, gadW | ND | [66] | |
| Sheeps | Feacal | Nigeria | 2015–2016 | B1 | ST58, ST131, ST155, ST156, ST167, ST405, ST406, ST1056, ST1771, ST2348 | Clindamycin, penicillin, ceftazidime, tobramycin, cefazolin, enrofloxacin, levofloxacin, sulfamethoxazole/trimethoprim, kanamycin, cefuroxime, piperacillin/tazobactam, ampicillin, cefalexin, streptomycin, doxycycline, neomycin, spectinomycin, amoxicillin/clavulanate, sulfamethoxazole, ampicillin/sulbactam, cefotaxime, ticarcillin, ciprofloxacin, trimethoprim, and tetracycline | ND | 5.5 | [57] |
| Cloacal swabbing | China | 2019–2020 | B1(70.1%), B2 (1.5%), C (20.9%), E (1.5%), F (1.5%) | ST10, ST23, ST58, ST162, ST167, ST361, ST602,ST1137 | Ceftazidime, ceftiofur, ceftriaxone, cefixime (third generation), cefepime (fourth generation), sulfisoxazole, florfenicol, tetracyclines, mequindox, enrofloxacin, ampicillin, spectinomycin, gentamicin, colistin | blaCTX-M, blaTEM, blaOXA. blaSHV, blaCMY, blaKPC | 6.7 | [20] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva, A.; Silva, V.; Pereira, J.E.; Maltez, L.; Igrejas, G.; Valentão, P.; Falco, V.; Poeta, P. Antimicrobial Resistance and Clonal Lineages of Escherichia coli from Food-Producing Animals. Antibiotics 2023, 12, 1061. https://doi.org/10.3390/antibiotics12061061
Silva A, Silva V, Pereira JE, Maltez L, Igrejas G, Valentão P, Falco V, Poeta P. Antimicrobial Resistance and Clonal Lineages of Escherichia coli from Food-Producing Animals. Antibiotics. 2023; 12(6):1061. https://doi.org/10.3390/antibiotics12061061
Chicago/Turabian StyleSilva, Adriana, Vanessa Silva, José Eduardo Pereira, Luís Maltez, Gilberto Igrejas, Patrícia Valentão, Virgílio Falco, and Patrícia Poeta. 2023. "Antimicrobial Resistance and Clonal Lineages of Escherichia coli from Food-Producing Animals" Antibiotics 12, no. 6: 1061. https://doi.org/10.3390/antibiotics12061061
APA StyleSilva, A., Silva, V., Pereira, J. E., Maltez, L., Igrejas, G., Valentão, P., Falco, V., & Poeta, P. (2023). Antimicrobial Resistance and Clonal Lineages of Escherichia coli from Food-Producing Animals. Antibiotics, 12(6), 1061. https://doi.org/10.3390/antibiotics12061061









