Abstract
There is scarce knowledge regarding the antimicrobial resistance profile of F. alocis. Therefore, the objective of this research was to assess antimicrobial resistance in recently obtained F. alocis clinical isolates and to identify the presence of antimicrobial resistance genes. Isolates were obtained from patients with periodontal or peri-implant diseases and confirmed by sequencing their 16S rRNA gene. Confirmed isolates had their genome sequenced by whole genome sequencing and their phenotypical resistance to nine antibiotics (amoxicillin clavulanate, amoxicillin, azithromycin, clindamycin, ciprofloxacin, doxycycline, minocycline, metronidazole, and tetracycline) tested by E-test strips. Antimicrobial resistance genes were detected in six of the eight isolates analyzed, of which five carried tet(32) and one erm(B). Overall, susceptibility to the nine antibiotics tested was high except for azithromycin in the isolate that carried erm(B). Moreover, susceptibility to tetracycline, doxycycline, and minocycline was lower in those isolates that carried tet(32). The genetic surroundings of the detected genes suggested their inclusion in mobile genetic elements that might be transferrable to other bacteria. These findings suggest that, despite showing high susceptibility to several antibiotics, F. alocis might obtain new antimicrobial resistance traits due to its acceptance of mobile genetic elements with antibiotic resistance genes in their genome.
1. Introduction
Filifactor alocis is an obligate anaerobic, non-sporeforming, Gram-positive rod [1,2]. This microorganism was first described in 1985 after being isolated from patients with periodontitis or gingivitis [3]. However, it was not until the early to mid-2000s, with the improvement of molecular identification techniques, when F. alocis was first acknowledged to be associated with oral diseases such as periodontitis or endodontic infections [4,5,6,7]. Later, the emergence of high-throughput sequencing allowed researchers to obtain a more realistic picture of the complexity of the oral microbiome and helped to confirm the strong association between F. alocis and periodontitis [8,9]. Moreover, F. alocis has been reported to employ several virulence strategies, including the invasion of epithelial cells, which induces the secretion of proinflammatory cytokines, subsequently triggering apoptosis [10,11]. The effects of F. alocis on the host cells also include the neutrophils, whose lifespan is extended through the inhibition of their apoptotic signaling while reducing their antimicrobial activities [2,12]. This elongates and worsens the inflammatory state of the periodontium, a situation in which F. alocis seems to thrive, due to the extended exposure of the periodontium to degradative enzymes secreted by neutrophils and the delayed initiation of tissue restorative mechanisms [12]. Other weapons in F. alocis’ arsenal include (1) the production of exotoxins, such as FtxA, which belongs to the RTX family and might help damage the surrounding cells by forming pores in the membranes of epithelial cells [13], (2) the ability to overcome oxidative stress thanks to enzymes such as superoxide reductase, which helps F. alocis to overcome the neutrophil response [14], and (3) the capability of mechanically adapting to the environment by forming pili-like projections [15,16]. To make things worse, and similar to other periodontopathogens such as Porphyromonas gingivalis, Fusobacterium nucleatum, and Aggregatibacter actinomycetemcomitans, F. alocis has also been detected in non-oral environments such as lung and brain abscesses [17,18,19,20,21].
Therefore, it seems that F. alocis might have an important role in the occurrence of several infectious diseases, especially periodontitis. Nevertheless, little is known of its antimicrobial resistance (AMR) properties [22]. Given that most infections still need antimicrobials to resolve successfully [23], it seems crucial to have updated and thorough information regarding the levels of AMR as well as the genetic determinants that might confer such resistance. Therefore, the phenotypic resistance levels of F. alocis isolates, obtained from subgingival samples of patients with different oral diseases, were tested. Moreover, their genomes were screened for AMR genes and mobile genetic elements (MGEs) that might spread these genes.
2. Results
2.1. Identification of Isolates and Sequencing Analysis
A total of eight isolates of F. alocis were obtained from eight different patients, two with periodontitis, one with peri-implant mucositis, and five with peri-implantitis. Isolates were identified by their colony morphology and then confirmed using a combination of three sets of primers designed to amplify a specific region of the 16S rRNA gene of F. alocis and the pilN and the ftxA genes. All isolates were positive for the 16S rRNA and pilN amplicons, and all but the 14.12, 22.1, and 22.4 isolates were positive for ftxA. Sequencing of their genome yielded a total of 155,032,230 reads, adding up to 23,409,866,730 base pairs sequenced (mean per isolate: 19,379,028.8 reads and 2,926,233,341 base pairs; standard deviation: ±3,458,607.2 reads and ±522,249,686.6 base pairs).
2.2. Detection and Analysis of Antimicrobial Resistance Genes
Screening of the contigs revealed the presence of two ARM genes in six of the eight isolates (Table 1). The most prevalent AMR gene was tet(32) (5/8 isolates), which codes for a ribosomal protection protein (RPP) that confers resistance to tetracyclines, followed by erm(B) (1/8 isolates), a methyltransferase that confers resistance to macrolides. The genetic elements surrounding tet(32) were similar in all the isolates that carried this gene, suggesting the integration of tet(32) in an MGE (Figure 1). This MGE shares a high degree of homology with the MGE present in several bacterial species such as Streptococcus anginosus (accession number CP007573.1), Streptococcus oralis (accession number CP097843.1), or Clostridium scindens (accession number CP036170.1), among others (data not shown). The contig containing erm(B) did not comprise the full sequence of the gene, which was cut at the 5′ ends, covering 72.2% of its aminoacidic sequence with an identity of 100% with a previously published sequence (accession number WP_001038795.1). Nevertheless, a transposase was detected just downstream erm(B) and homology of this contig with other sequences suggests that this gene was also integrated in an MGE that has been detected in strains of S. anginosus (accession number CP069892.1), Streptococcus salivarius (accession number CP018189.1), Enterococcus saigonensis (accession number AP022822.1), and Enterococcus faecalis (accession number CP118057.1), among others (data not shown).

Table 1.
Antimicrobial resistance genes detected in the isolates. Reference sequences of tet(32) and erm(B) can be accessed with the accession numbers WP_003505402.1 and WP_001038795.1, respectively.
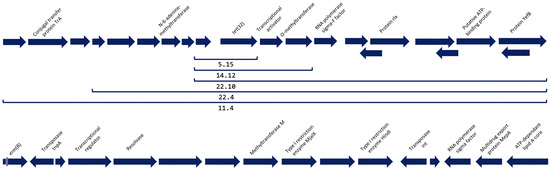
Figure 1.
Open Reading Frames of the surroundings of the antimicrobial resistance genes detected. At the top of the figure, the genetic surroundings of tet(32) and the length of the construction that was detected in each of the isolates indicated with a bracket. At the bottom of the figure, the genetic surroundings downstream gene erm(B). This gene was truncated in the contig and approximately 30% of its upstream sequence is missing, as well as its surroundings.
2.3. Antimicrobial Resistance Testing
Resistance to eight antibiotics pertaining to six different classes was tested using E-test strips. Susceptibility to these antibiotics was generally high (Table 2), with only one isolate (30.27) showing high levels of resistance, namely to azithromycin (AZM) (>256 µg/mL). Susceptibility or resistance to the antibiotics tested needs to be assessed with caution, since there are no established breakpoint concentrations for F. alocis in the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines [24]. Therefore, the MICs obtained must be analyzed by comparison with those of other anaerobic bacteria listed in the guidelines. According to this criterion, all isolates were susceptible to all the antibiotics tested except for isolate 30.27, which was resistant to clindamycin (CDM) and AZM.

Table 2.
Minimum inhibitory concentrations (µg/mL) of the tested F. alocis isolates. Antibiotic concentrations of the E-test strips ranged from 0.016 to 256 µg/mL in all antibiotics with the exception of ciprofloxacin, which ranged from 0.002 to 32 µg/mL. AMC: amoxicillin clavulanate, AMX: amoxicillin, AZM: azithromycin, CDM: clindamycin, CIP: ciprofloxacin, DOX: doxycycline, MIN: minocycline, MTZ: metronidazole, TET: tetracycline.
3. Discussion
This study aimed to determine the antimicrobial resistance profiles of F. alocis isolates obtained from subgingival and peri-implant samples from patients with periodontal or peri-implant diseases. Their genome was sequenced to screen for AMR genes, and their phenotypical AMR was tested with the E-test method.
The significance of the role of F. alocis in periodontal and peri-implant diseases is increasingly being emphasized, as a growing number of studies utilizing molecular approaches are establishing its association with these diseases [25,26]. The fastidious conditions for growing this species have been a barrier to conduct studies based on its growth in isolation such as Whole Genome Sequencing or antimicrobial susceptibility analyses. In fact, only a study from 1985 [3] described the susceptibility of 20 F. alocis isolates to breakpoint concentrations of five antibiotics (chloramphenicol [12 µg/mL], clindamycin [1.6 µg/mL], erythromycin [3 µg/mL], tetracycline [6 µg/mL], and penicillin [2 U/mL]), observing that those isolates were susceptible to all breakpoint concentrations except for one isolate which was resistant to penicillin. Nevertheless, the aging of such data, the geographical constraints of the study, and the fact that the identity of those isolates remains to be confirmed with molecular approaches, call for new reliable data.
The results of this study show the high susceptibility of F. alocis to the most used antimicrobials in the clinical practice, with some exceptions such as isolate 30.27, which showed reduced susceptibility to CDM and extremely high tolerance to AZM; the ATCC 35896 strain, whose reduced susceptibility to tetracyclines has already been reported [22]. Resistance to tetracycline (TET) can be mediated through tetracycline resistance genes that code for RPPs, efflux pumps or inactivation enzymes [27]. Tolerance to this class of antibiotics by the ATCC 35896 strain has been linked to the presence of the tet(M) gene, which codes for an RPP and is enclosed in an MGE [28]. However, none of the isolates in this study carried tet(M). Instead, five of them carried tet(32) (Table 1), which was integrated into another MGE that shared a highly homologous 12-kb region with strains of S. anginosus, S. oralis, and C. scindens, among others, which also contain tet(32) in their genome. Moreover, the presence of this gene in the isolates seemed to increase their tolerance to minocycline (MIN), doxycycline (DOX), and even more to TET when comparing with the isolates that did not carry it (5.15, 48B, and 30.27), despite not reaching the levels conferred by tet(M) in the ATCC 35896 strain (Table 2). tet(32) codes for an RPP that binds to a ribosome that is being altered by a molecule of TET, or one of its derivatives, and breaks the union to the detriment of a GTP molecule [27]. Both TetM and Tet32 use this same mechanism, which suggests that the different levels of resistance of the isolates that carry one gene or the other may be due to something else, such as for example, different levels of gene expression or the impact that genetic surroundings might have on these genes. In a recent study, the genes tet(M) and tet(32) were the two most detected TET resistance genes among subgingival isolates obtained from patients with periodontitis [29]. However, to our knowledge, this is the first report of tet(32) in bacteria from the genus Filifactor. The high prevalence in the oral environment of these genes, which are often embedded in MGEs, such as conjugative transposons [30,31], might be an indicator of their ease of spread, which does not seem to bend under phylogenetic barriers and could render the use of tetracyclines useless in the oral environment.
erm(B) codes for a methyltransferase that methylates an adenine in the domain where macrolides and lincosamides bind to the 23S rRNA region of the major subunit of the bacterial ribosome, preventing such binding and thus conferring resistance [32,33]. Therefore, the presence of this gene might have provided isolate 30.27 with a very high tolerance to AZM (>256 µg/mL) and reduced its susceptibility to CDM (1 µg/mL). The present study is, to our knowledge, the first report of a macrolide resistance gene in F. alocis. As observed in TET resistance genes, erm(B) shows a high prevalence in oral bacteria and can be detected in conjugative transposons [29,34]. In fact, in a study from 2019, the authors also detected erm(B) for the first time in bacteria from the genus Prevotella [35], which until that moment were only known to carry erm(A), erm(C), erm(F) and erm(G) [36], highlighting the transferrable properties of these genes through MGEs that carry them. Interestingly, a previous study pointed out the high resemblance of a 45-kb region of the Streptococcus dysgalactiae subsp. equisimilis containing erm(B), with the region of F. alocis ATCC 35,896 that contains tet(M), suggesting that a swap between tet(M) and erm(B) took place at some time [28]. However, the surroundings of erm(B) in isolate 30.27 showed little resemblance with either F. alocis ATCC 35896 or the strain of S. dysgalactiae of the mentioned study. Given its close proximity to the tnpA and int transposases, also present in conjugative transposons such as Tn3, Tn916, Tn1721, Tn1545 and Tn6261 among many others [37], and the high homology of the 9-kb downstream of the gene erm(B) with a region of S. anginosus, S. salivarius, E. saigonensis and, to a lesser extent, E. faecalis, it is highly probable that this gene is being carried within an MGE.
This study is not exempt from limitations. For instance, due to the fastidious culture, identification, and isolation involved, the number of isolates was not high enough to extract conclusions of AMR in F. alocis, even more given the variability observed in the eight isolates of the study and despite working with a similar or higher number of isolates than other publications [13,22]. Moreover, full assemblies of the genomes using long-read amplicon sequencing in conjunction with shotgun sequencing might help to better understand those regions that were truncated due to methodological limitations. On the other hand, the reduced sample size did not allow to make comparisons between the isolates found in the different pathologies (periodontitis, peri-implant mucositis, or peri-implantitis). Further studies are needed to assess the contribution of the AMR genes detected to the phenotypic resistance observed in the study isolates.
AMR in bacteria that have a potential role in infectious diseases should be strictly monitored [38]. Although its role in periodontal disease is still largely unknown, evidence that F. alocis is strongly associated with periodontitis is growing [17,25,26]. This study shows that F. alocis isolated from either subgingival or peri-implant samples is still highly susceptible to the most common antibiotics. However, AMR genes embedded in MGEs were observed in almost all isolates, and one of them showed a very high tolerance to AZM, which highlights the need to keep a close eye on the AMR profiles in these microorganisms if antibiotics are to be kept as a treatment option.
4. Material & Methods
4.1. Sample Collection and Culturing Conditions
Patients diagnosed with periodontitis, peri-implant mucositis or peri-implantitis attending the Dental Hospital of the University of Barcelona, were asked to volunteer for 2 studies that aimed at determining the presence of Filifactor alocis in periodontitis and peri-implant diseases. The study protocols were approved by the local Ethics Committee (protocols ref. 15/2022 and 36/2022). All volunteers signed an Institutional review board-approved informed consent form. Subgingival and peri-implant samples were obtained by placing sterile paper points for 30 s in subgingival pockets. Then, paper points were pooled in a vial containing either 1.5 mL of cold sterilized reduced transport medium (RTF) without ethylenediaminetetraacetic acid or modified Amies medium containing 20.9 g/L Amies transport with charcoal (CondalabTM, Madrid, Spain), cysteine (0.012 g/L), tryptose (0.5 g/L) and peptone (0.5 g/L). Both mediums were incubated for 24 h in anaerobic conditions (10% H2, 10% CO2 and 80% N2) at 37 °C prior to the sample collection. Samples were sent to the Dentaid Research Center (Cerdanyola del Vallès, Spain) to be processed.
Samples in Amies medium were resuspended in 400 µL of phosphate-buffered saline (PBS), transferred to a new 1.5 mL sterile centrifuge tube, and vortexed for 1 min. Samples in RTF were directly vortexed and 400 µL were transferred to a new 1.5 mL sterile centrifuge tube. Then, serial dilutions were made and plated on blood agar plates containing 40 g/L Oxoid Nutrient Broth No. 2, 5% horse blood, hemin (5 mg/L) and menadione (1 mg/L) and were incubated under anaerobic conditions at 37 °C for 10 days.
Colonies suspected of being F. alocis were isolated, replated in blood agar plates, and incubated in anaerobic conditions for 7 days. Then, they were recovered under a stereo microscope to ensure correct identification. Translucent flattened colonies with a wide halo and small round-shaped center were considered to be F. alocis, isolated on new plates and cryo-conserved at −80 °C. In order to extract their DNA and to perform the antimicrobial susceptibility tests, 5–10 colonies were inoculated in liquid cultures with BHI-GA medium containing 37 g/L Brain Heart Infusion (Becton Dickinson), yeast extract (1 g/L), sodium bicarbonate (2 g/L), L-cysteine (1 g/L), L-arginine (2 g/L), and noble agar (1 g/L). Liquid cultures were grown under anaerobic conditions at 37 °C for at least 3 days.
4.2. DNA Isolation and Polymerase Chain Reaction-Based Characterization
DNA extractions were performed using the QIAamp DNA Mini Kit (Qiagen, Hildem Germany) according to the manufacturer’s instructions. DNA was quantified using a NanoDrop 2000C UV-vis Spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). PCR reactions were made with 3 different pairs of oligonucleotides. First, DNA amplicons of the 16S rRNA gene of F. alocis were obtained as described [5]. Then, a second reaction was performed for pilN, a specific gene of F. alocis, with forward primer (pilN_F: 5′-GCTCAGCAAACATGCGATTG-3′) and reverse primer (pilN_R: 5′-GAAGGCTATGATTTGATTGTTTCC-3′) to amplify a 156-bp-length fragment. Finally, a third reaction was conducted for the ftxA gene to amplify a 798-bp-length fragment as described [39]. A total of 0.5 µM was the final concentration used for all primers. Between 30 and 100 nanograms of DNA were used, together with 1× PCR buffer, 1× dNTPs solution, 2.5 mM of MgCl2, and 1 unit of Taq polymerase (Takara TaqTM DNA Polymerase). PCR cycling conditions were 10 min at 94 °C, followed by 30 cycles of 94 °C for 30 s, 60 °C for 30 s, and 72 °C for 40 s, followed by a final extension of 10 min at 72 °C, performed in a T3000 Thermocycler (Biometra, Göttingen, Germany). PCR products were assessed by electrophoresis in 2% agarose gels with SybrSafeTM DNA Gel Stain (Life Technologies, Carisbad, CA, USA).
4.3. Antibiotic Susceptibility Testing
To determine antibiotic susceptibility, strain ATCC 35896 and the F. alocis clinical isolates were plated on blood agar, E-test strips (BioMérieuxTM, Marcy-l’Étoile, France) were added and then plates were incubated at 37 °C in anaerobic conditions. E-test strips contained a concentration gradient of amoxicillin, amoxicillin with clavulanic acid, MIN, DOX, TET, metronidazole, AZM, CDM, or ciprofloxacin. The minimum inhibitory concentration of all the antibiotics was measured as instructed by the manufacturer. Streptococcus pneumoniae ATCC 49619 and Escherichia coli DSM 1576 were used as quality control strains.
4.4. Whole-Genome Sequencing
DNA from the isolates (5.15, 8.25, 11.40, 14.12, 22.10, 22.40, 30.27, and 48B) was sent to Macrogen, Inc. (Seoul, South Korea) for Whole-Genome Sequencing using the Nextera DNA XT Library and the NovaSeq6000 platform (Illumina, San Diego, CA, USA). Quality of the reads was assessed using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 1 March 2023)) and trimmed using Cutadapt [40]. Trimmed reads were assembled using Bowtie2 and using the F. alocis ATCC 35896 assembly available at Genbank (accession number GCA_000163895.2) as a reference genome. Genomes were screened for AMR genes using AMRFinderPlus [41] fed with contigs assembled with SPAdes 3.15.4 [42]. DNA regions of interest were further analyzed using de NCBI ORF finder (https://www.ncbi.nlm.nih.gov/orffinder/ (accessed on 5 March 2023)) and BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 5 March 2023)).
Author Contributions
A.A. and R.L. designed the study. A.M. and R.F. provided the samples and the characterization of the patients. R.R.-M. carried out the experiments and wrote the manuscript with the corrections and supervision of A.A., G.À. and R.L. A.A. performed the bioinformatic and statistical analyses. All authors did a critical review of the manuscript and agreed on the submission. All authors have read and agreed to the published version of the manuscript.
Funding
This study was partially funded by a research agreement between the University of Barcelona and Dentaid S.L. (Càtedra UB-Dentaid).
Institutional Review Board Statement
The protocol complied with the Helsinki Declaration guidelines and was submitted to and approved by the Ethics Committee for Clinical Research (CEIC) of the Dental Hospital of the University of Barcelona (register number 488, protocol ref. 36/2022), and all the patients signed an informed consent form before enrollment.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Data might be shared upon reasonable request.
Acknowledgments
We would like to thank Ann Bangle for the English revision of the manuscript.
Conflicts of Interest
The authors have no conflict of interest to declare.
References
- Jalava, J.; Eerola, E. Phylogenetic analysis of Fusobacterium alocis and Fusobacterium sulci based on 16S rRNA gene sequences: Proposal of Filifactor alocis (Cato, Moore and Moore) comb. nov. and Eubacterium sulci (Cato, Moore and Moore) comb. nov. Int. J. Syst. Evol. Microbiol. 1999, 49, 1375–1379. [Google Scholar] [CrossRef]
- Aja, E.; Mangar, M.; Fletcher, H.M.; Mishra, A. Filifactor alocis: Recent Insights and Advances. J. Dent. Res. 2021, 100, 790–797. [Google Scholar] [CrossRef]
- Cato, E.P.; Moore, L.V.H.; Moore, W.E.C. Fusobacterium alocis sp. nov. and Fusobacterium sulci sp. nov. from the Human Gingival Sulcus. Int. J. Syst. Evol. Microbiol. 1985, 35, 475–477. [Google Scholar] [CrossRef]
- Hutter, G.; Schlagenhauf, U.; Valenza, G.; Horn, M.; Burgemeister, S.; Claus, H.; Vogel, U. Molecular analysis of bacteria in periodontitis: Evaluation of clone libraries, novel phylotypes and putative pathogens. Microbiology 2003, 149, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Jr, J.F.S.; Rôças, I.N. Detection of Filifactor alocis in endodontic infections associated with different forms of periradicular diseases. Oral Microbiol. Immunol. 2003, 18, 263–265. [Google Scholar] [CrossRef]
- Gomes, B.P.; Jacinto, R.C.; Pinheiro, E.; Sousa, E.L.; Zaia, A.A.; Ferraz, C.; Souza-Filho, F.J. Molecular Analysis of Filifactor alocis, Tannerella forsythia, and Treponema denticola Associated with Primary Endodontic Infections and Failed Endodontic Treatment. J. Endod. 2006, 32, 937–940. [Google Scholar] [CrossRef]
- Kumar, P.S.; Leys, E.J.; Bryk, J.M.; Martinez, F.J.; Moeschberger, M.L.; Griffen, A.L. Changes in Periodontal Health Status Are Associated with Bacterial Community Shifts as Assessed by Quantitative 16S Cloning and Sequencing. J. Clin. Microbiol. 2006, 44, 3665–3673. [Google Scholar] [CrossRef]
- Chen, H.; Liu, Y.; Zhang, M.; Wang, G.; Qi, Z.; Bridgewater, L.; Zhao, L.; Tang, Z.; Pang, X. A Filifactor alocis-centered co-occurrence group associates with periodontitis across different oral habitats. Sci. Rep. 2015, 5, srep09053. [Google Scholar] [CrossRef]
- Griffen, A.L.; Beall, C.J.; Campbell, J.H.; Firestone, N.D.; Kumar, P.S.; Yang, Z.K.; Podar, M.; Leys, E.J. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 2011, 6, 1176–1185. [Google Scholar] [CrossRef]
- Moffatt, C.; Whitmore, S.; Griffen, A.; Leys, E.; Lamont, R. Filifactor alocis interactions with gingival epithelial cells. Mol. Oral Microbiol. 2011, 26, 365–373. [Google Scholar] [CrossRef]
- Aruni, A.W.; Zhang, K.; Dou, Y.; Fletcher, H. Proteome Analysis of Coinfection of Epithelial Cells with Filifactor alocis and Porphyromonas gingivalis Shows Modulation of Pathogen and Host Regulatory Pathways. Infect. Immun. 2014, 82, 3261–3274. [Google Scholar] [CrossRef] [PubMed]
- Miralda, I.; Vashishta, A.; Rogers, M.N.; Lamont, R.J.; Uriarte, S.M. The emerging oral pathogen, Filifactor alocis, extends the functional lifespan of human neutrophils. Mol. Microbiol. 2022, 117, 1340–1351. [Google Scholar] [CrossRef]
- Bao, K.; Claesson, R.; Belibasakis, G.N.; Oscarsson, J. Extracellular Vesicle Subproteome Differences among Filifactor alocis Clinical Isolates. Microorganisms 2022, 10, 1826. [Google Scholar] [CrossRef]
- Mishra, A.; Aja, E.; Fletcher, H.M. Role of Superoxide Reductase FA796 in Oxidative Stress Resistance in Filifactor alocis. Sci. Rep. 2020, 10, 9178. [Google Scholar] [CrossRef] [PubMed]
- Aruni, A.W.; Roy, F.; Fletcher, H.M. Filifactor alocis Has Virulence Attributes That Can Enhance Its Persistence under Oxidative Stress Conditions and Mediate Invasion of Epithelial Cells by Porphyromonas gingivalis. Infect. Immun. 2011, 79, 3872–3886. [Google Scholar] [CrossRef] [PubMed]
- Aruni, A.W.; Roy, F.; Sandberg, L.; Fletcher, H.M. Proteome variation among Filifactor alocis strains. Proteomics 2012, 12, 3343–3364. [Google Scholar] [CrossRef]
- Schlafer, S.; Riep, B.; Griffen, A.L.; Petrich, A.; Hübner, J.; Berning, M.; Friedmann, A.; Göbel, U.B.; Moter, A. Filifactor alocis—involvement in periodontal biofilms. BMC Microbiol. 2010, 10, 66. [Google Scholar] [CrossRef]
- Gray, R.; Vidwans, M. Mixed anaerobic thoracic empyema: The first report of Filifactor alocis causing extra-oral disease. New Microbes New Infect. 2019, 29, 100528. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhu, Y.; Wan, H. Case report:Multiple abscesses caused by Porphyromonas gingivalis diagnosed by metagenomic next-generation sequencing. Front. Med. 2023, 9, 1089863. [Google Scholar] [CrossRef]
- Yusuf, K.; Sampath, V.; Umar, S. Bacterial Infections and Cancer: Exploring This Association And Its Implications for Cancer Patients. Int. J. Mol. Sci. 2023, 24, 3110. [Google Scholar] [CrossRef]
- Mesturino, M.A.; Bitetti, C.; Clemente, A.; Krzysztofiak, A.; Lancella, L.; Lombardi, R.; Cursi, L.; Boccuzzi, E.; Musolino, A.M.; Villani, A. Aggregatibacter actinomycetemcomitans infection in a 15-year-old boy with pulmonary empyema: A case report and review of literature. Ital. J. Pediatr. 2023, 49, 42. [Google Scholar] [CrossRef] [PubMed]
- Inubushi, J.; Liang, K. Update on minocycline in vitro activity against odontogenic bacteria. J. Infect. Chemother. 2020, 26, 1334–1337. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, M.I.; Römling, U.; Nadeem, F.; Bilal, H.M.; Zafar, M.; Jahan, H.; Ur-Rahman, A. Innovative Strategies to Overcome Antimicrobial Resistance and Tolerance. Microorganisms 2022, 11, 16. [Google Scholar] [CrossRef]
- European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint Tables for Interpretation of MICs and Zone Diameters, Version 13.0. EFSA Journal. Available online: http://www.eucast.org (accessed on 20 February 2023).
- Àlvarez, G.; Arredondo, A.; Isabal, S.; Teughels, W.; Laleman, I.; Contreras, M.J.; Isbej, L.; Huapaya, E.; Mendoza, G.; Mor, C.; et al. Association of nine pathobionts with periodontitis in four South American and European countries. J. Oral Microbiol. 2023, 15, 2188630. [Google Scholar] [CrossRef] [PubMed]
- Lafaurie, G.I.; Neuta, Y.; Ríos, R.; Pacheco-Montealegre, M.; Pianeta, R.; Castillo, D.M.; Herrera, D.; Reyes, J.; Diaz, L.; Castillo, Y.; et al. Differences in the subgingival microbiome according to stage of periodontitis: A comparison of two geographic regions. PLoS ONE 2022, 17, e0273523. [Google Scholar] [CrossRef]
- Thaker, M.; Spanogiannopoulos, P.; Wright, G.D. The tetracycline resistome. Cell. Mol. Life Sci. 2010, 67, 419–431. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, X.; Zong, Z. Genome sequence and virulence factors of a group G Streptococcus dysgalactiae subsp. equisimilis strain with a new element carrying erm(B). Sci. Rep. 2016, 6, 20389. [Google Scholar] [CrossRef]
- Arredondo, A.; Blanc, V.; Mor, C.; Nart, J.; León, R. Tetracycline and multidrug resistance in the oral microbiota: Differences between healthy subjects and patients with periodontitis in Spain. J. Oral Microbiol. 2021, 13, 1847431. [Google Scholar] [CrossRef]
- Warburton, P.; Roberts, A.P.; Allan, E.; Seville, L.; Lancaster, H.; Mullany, P. Characterization of tet (32) Genes from the Oral Metagenome. Antimicrob. Agents Chemother. 2009, 53, 273–276. [Google Scholar] [CrossRef]
- Speer, B.S.; Shoemaker, N.B.; Salyers, A.A. Bacterial Resistance to Tetracycline: Mechanisms, Transfer, and Clinical Significance. Clin. Microbiol. Rev. 1992, 5, 387–399. [Google Scholar] [CrossRef]
- Roberts, M.C. Resistance to Tetracycline, Macrolide-Lincosamide-Streptogramin, Trimethoprim, and Sulfonamide Drug Classes. Appl. Biochem. Biotechnol. Part B Mol. Biotechnol. 2002, 20, 261–284. [Google Scholar] [CrossRef] [PubMed]
- Weisblum, B. Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 1995, 39, 577–585. [Google Scholar] [CrossRef] [PubMed]
- Ciric, L.; Ellatif, M.; Sharma, P.; Patel, R.; Song, X.; Mullany, P.; Roberts, A.P. Tn916-like elements from human, oral, commensal streptococci possess a variety of antibiotic and antiseptic resistance genes. Int. J. Antimicrob. Agents 2012, 39, 360–361. [Google Scholar] [CrossRef] [PubMed]
- Arredondo, A.; Blanc, V.; Mor, C.; Nart, J.; León, R. Azithromycin and erythromycin susceptibility and macrolide resistance genes in Prevotella from patients with periodontal disease. Oral Dis. 2019, 25, 860–867. [Google Scholar] [CrossRef] [PubMed]
- Roberts, M.C. Location of the Various Genes Table. Available online: http://faculty.washington.edu/marilynr (accessed on 20 February 2023).
- Tansirichaiya, S.; Rahman, A.; Roberts, A.P. The Transposon Registry. Mob. DNA 2019, 10, 40. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control (ECDC); European Food Safety Authority (EFSA); European Medicines Agency (EMA). ECDC/EFSA/EMA Second Joint Report on the Integrated Analysis of the Consumption of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Bacteria from Humans and Food-Producing Animals. EFSA J. 2017, 15, e04872. [Google Scholar] [CrossRef]
- Oscarsson, J.; Claesson, R.; Bao, K.; Brundin, M.; Belibasakis, G. Phylogenetic Analysis of Filifactor alocis Strains Isolated from Several Oral Infections Identified a Novel RTX Toxin, FtxA. Toxins 2020, 12, 687. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Feldgarden, M.; Brover, V.; Gonzalez-Escalona, N.; Frye, J.G.; Haendiges, J.; Haft, D.H.; Hoffmann, M.; Pettengill, J.B.; Prasad, A.B.; Tillman, G.E.; et al. AMRFinderPlus and the Reference Gene Catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci. Rep. 2021, 11, 12728. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).