Spiramycin Disarms Pseudomonas aeruginosa without Inhibiting Growth
Abstract
1. Introduction
2. Results
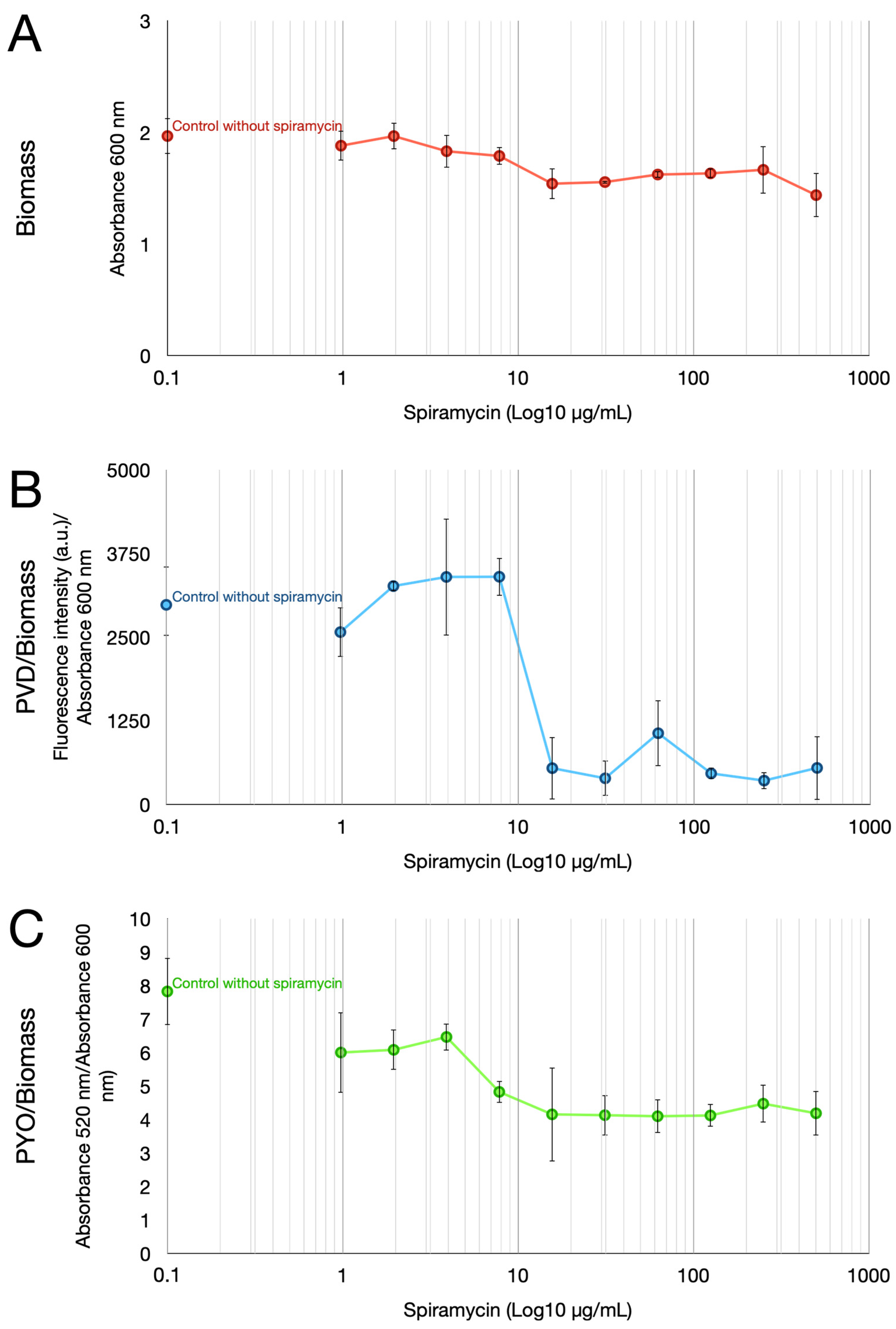
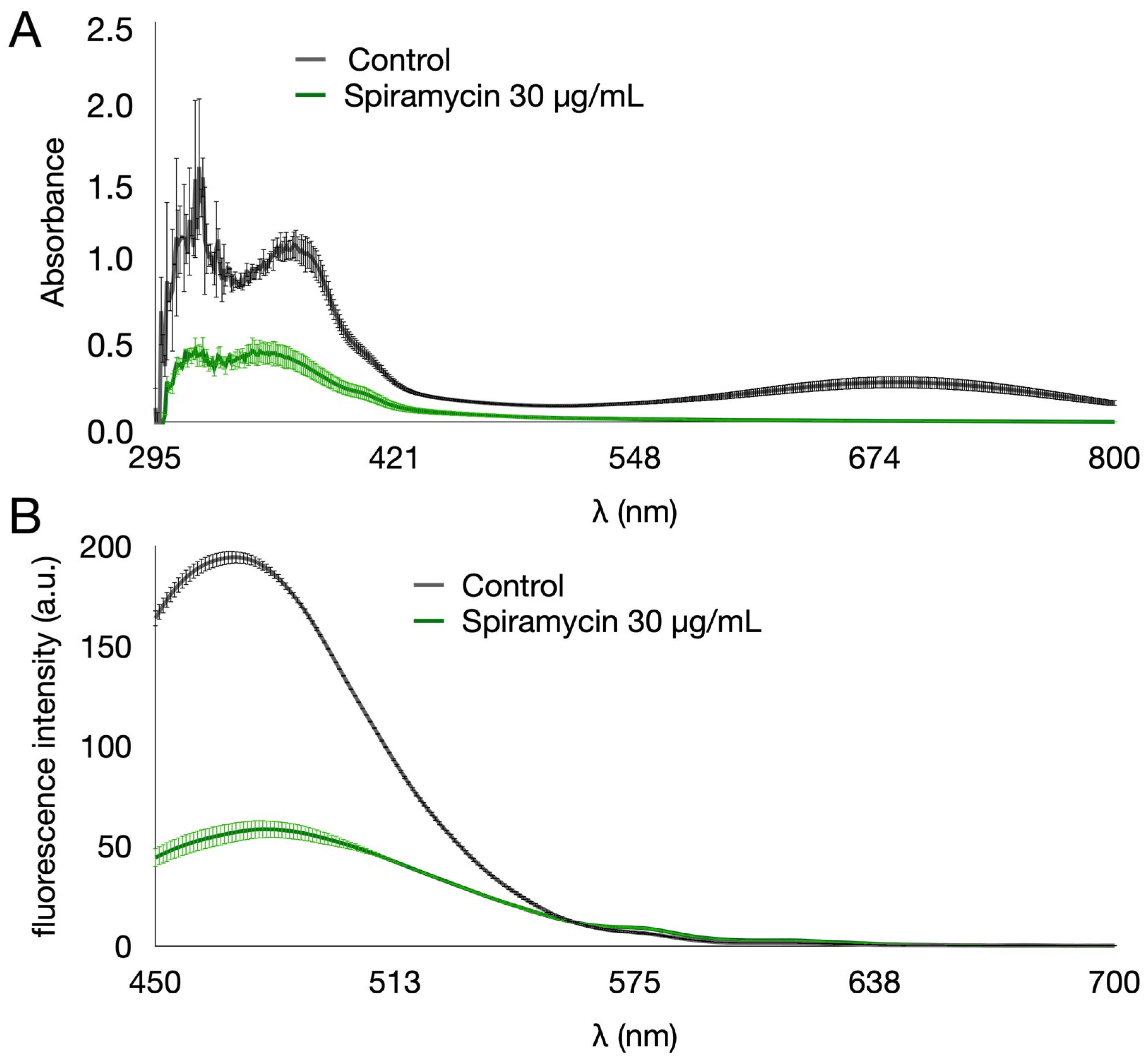
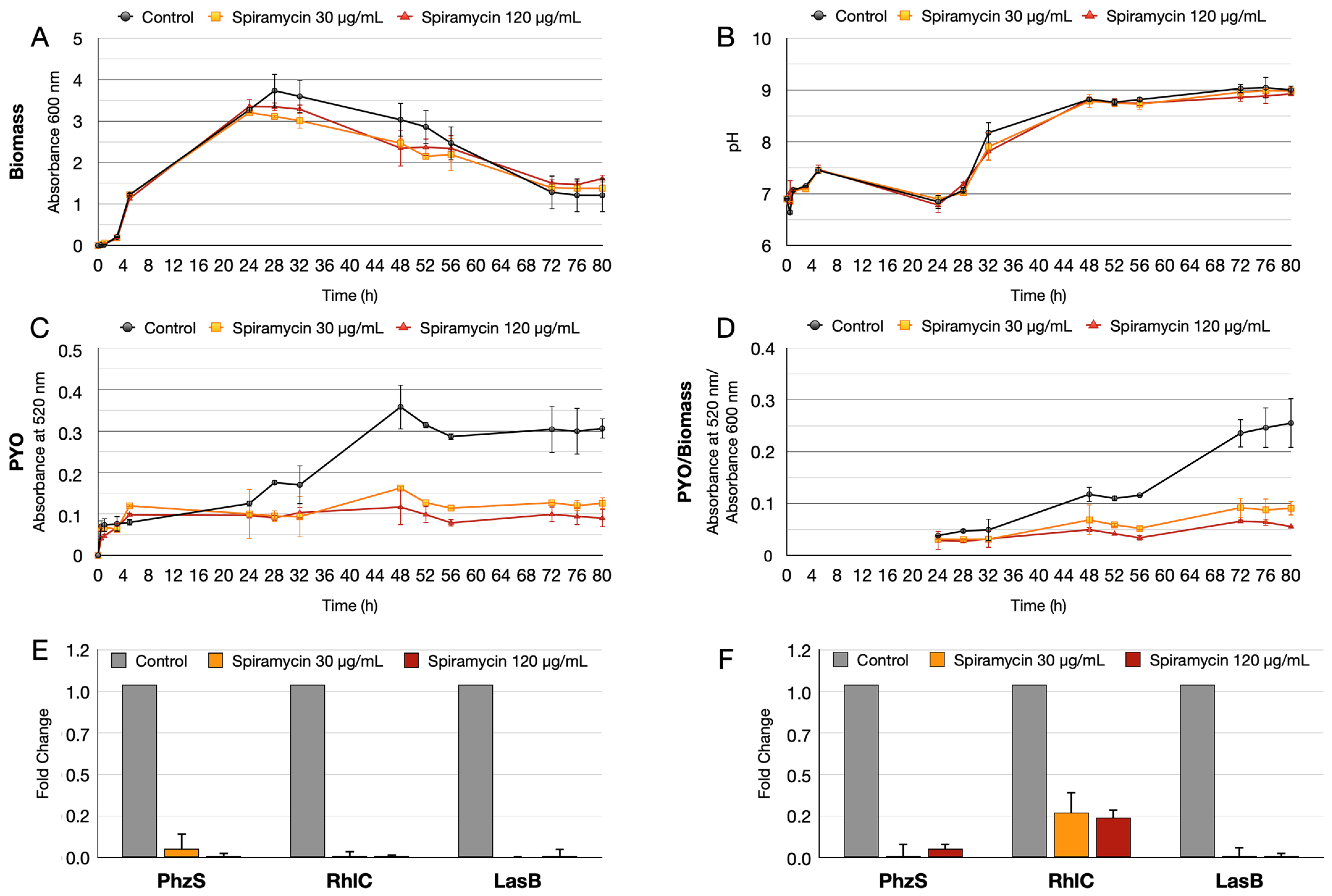
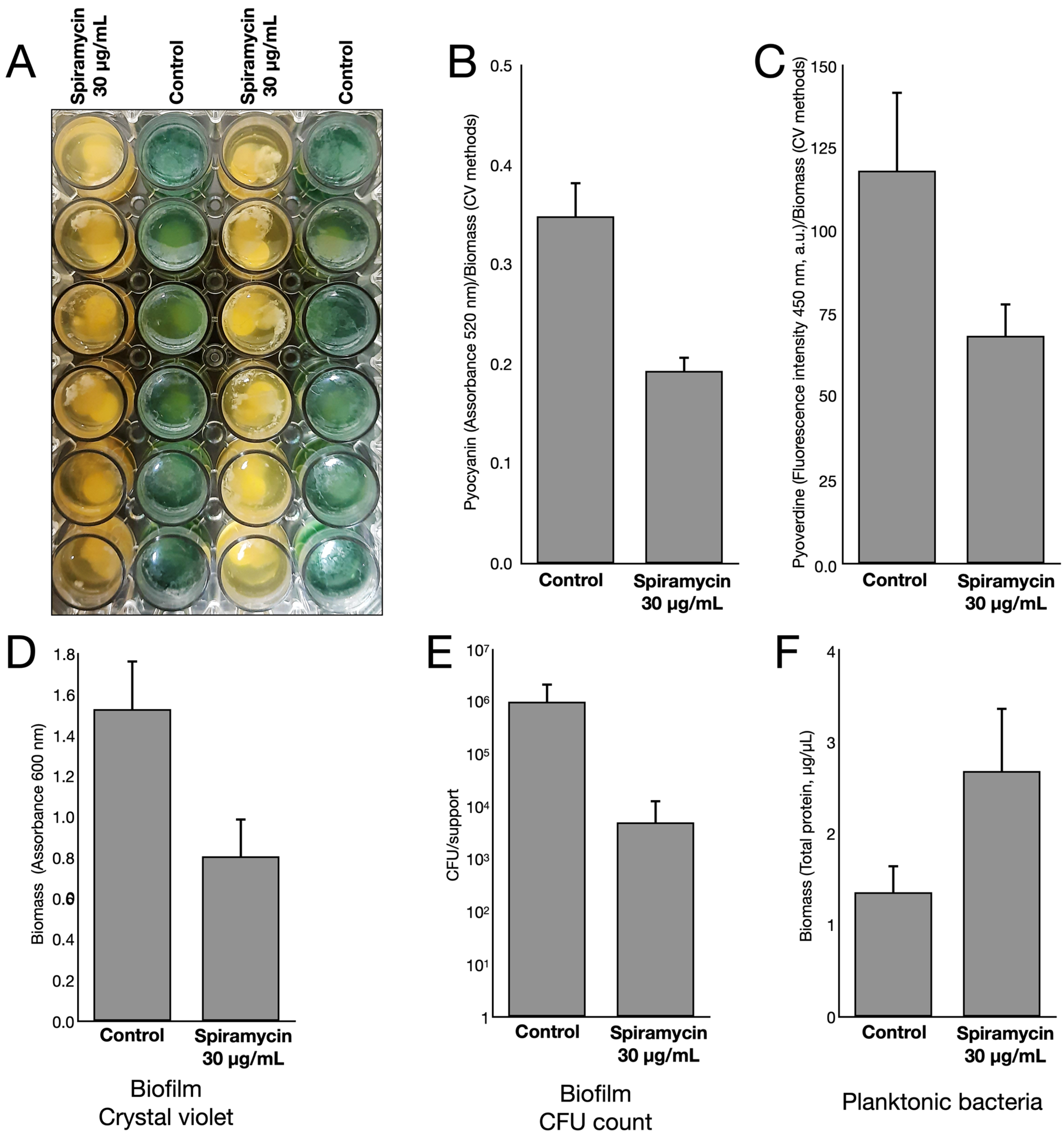
2.1. Spiramycin Inhibits the Production of Pyocyanin and Pyoverdine in P. aeruginosa
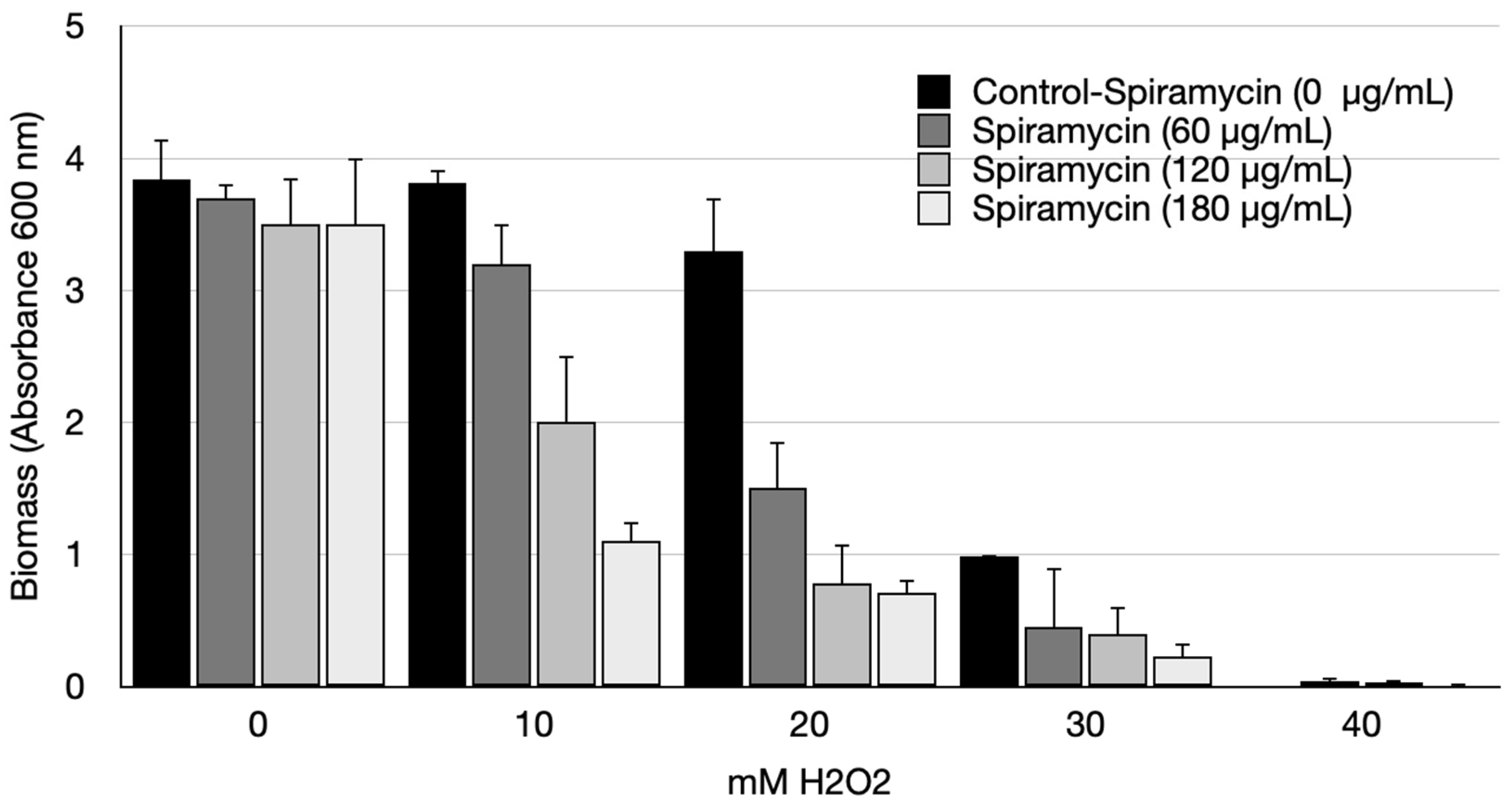
2.2. Spiramycin Sensitizes P. aeruginosa to Oxidative Damage
2.3. Spiramycin Inhibits the Formation of P. aeruginosa Biofilm on Hydroxyapatite
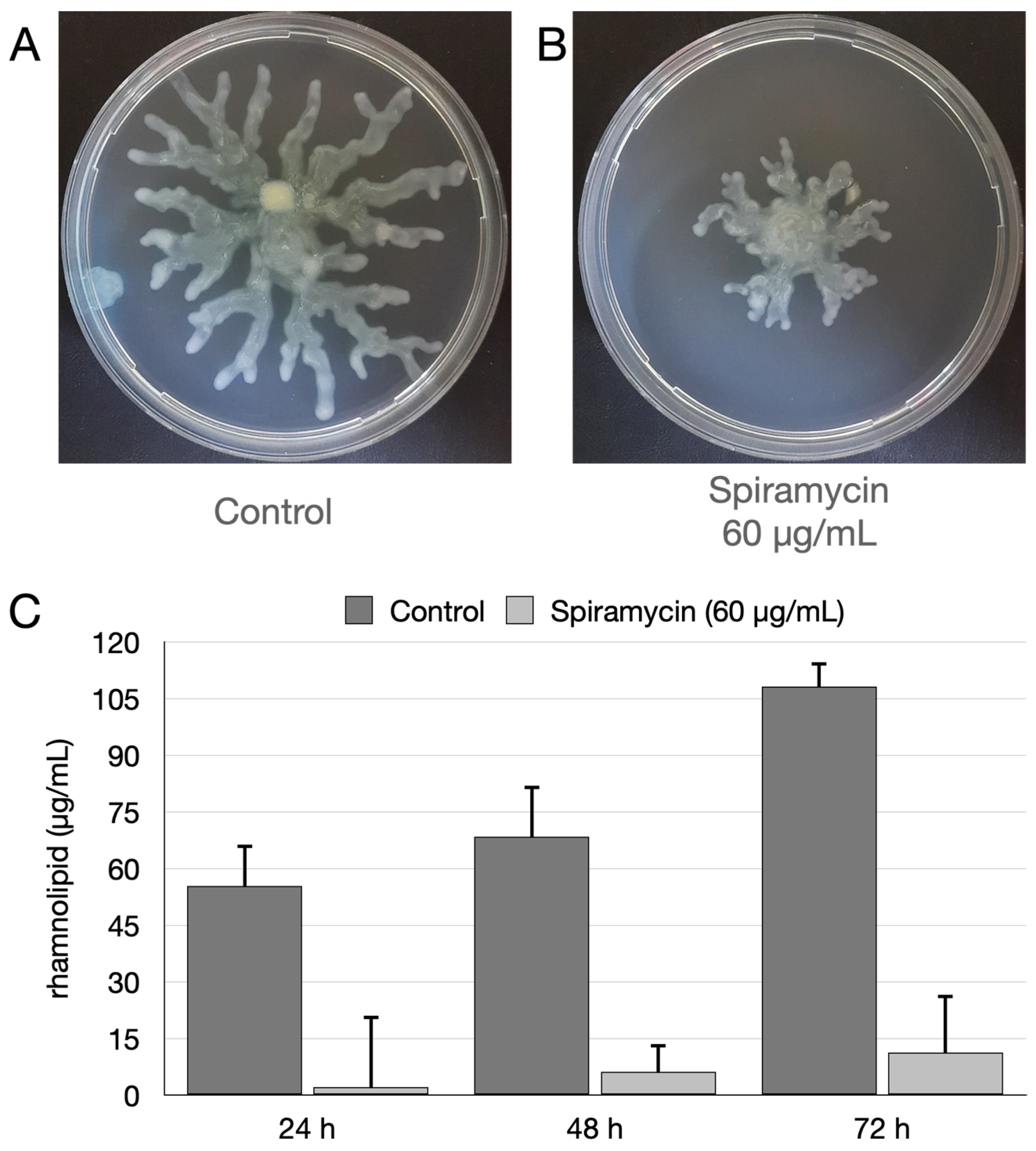
2.4. Spiramycin Inhibits the Swarming Motility and the Biosynthesis of Rhamnolipids of P. aeruginosa
2.5. Spiramycin Attenuates P. aeruginosa Virulence and Expression of Antimibrobial Peptides in Galleria mellonella Animal Model
3. Discussion and Conclusions
4. Materials and Methods
4.1. Strain, Media, Growth Condition and General Procedure
4.2. Determination of MIC, and Measurement of PVD and PYO Production in Multiwell Plates
4.3. Determination of Growth Curves and Measurement of PVD, PYO and Rhamnolipid Production in Shake Flask Experiments
4.4. Resistance to Oxidative Stress
4.5. Biofilm Experiment and Biomass Estimation
4.6. Swarming Motility Evaluation
4.7. Inoculum and Management of Galleria Mellonella Larvae
4.8. RNA Extraction from G. mellonella Larvae and RT-qPCR
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ashburn, T.T.; Thor, K.B. Drug repositioning: Identifying and developing new uses for existing drugs. Nat. Rev. Drug. Discov. 2004, 3, 673–683. [Google Scholar] [CrossRef]
- Cheng, Y.-S.; Williamson, P.R.; Zheng, W. Improving therapy of severe infections through drug repurposing of synergistic combinations. Curr. Opin. Pharmacol. 2019, 48, 92–98. [Google Scholar] [CrossRef]
- Liu, Y.; Tong, Z.; Shi, J.; Li, R.; Upton, M.; Wang, Z. Drug repurposing for next-generation combination therapies against multidrug-resistant bacteria. Theranostics 2021, 11, 4910–4928. [Google Scholar] [CrossRef]
- Serafin, M.B.; Hörner, R. Drug repositioning, a new alternative in infectious diseases. Braz. J. Infect. Dis. 2018, 22, 252–256. [Google Scholar] [CrossRef]
- Zheng, W.; Sun, W.; Simeonov, A. Drug repurposing screens and synergistic drug-combinations for infectious diseases. Br. J. Pharmacol. 2018, 175, 181–191. [Google Scholar] [CrossRef]
- Hijazi, S.; Visaggio, D.; Pirolo, M.; Frangipani, E.; Bernstein, L.; Visca, P. Antimicrobial Activity of Gallium Compounds on ESKAPE Pathogens. Front. Cell. Infect. Microbiol. 2018, 8, 316. [Google Scholar] [CrossRef]
- Chalmers, J.D. Macrolide resistance in Pseudomonas aeruginosa: Implications for practice. Eur. Respir. J. 2017, 49, 1700689. [Google Scholar] [CrossRef]
- Mustafa, M.-H.; Khandekar, S.; Tunney, M.M.; Elborn, J.S.; Kahl, B.C.; Denis, O.; Plésiat, P.; Traore, H.; Tulkens, P.M.; Vanderbist, F.; et al. Acquired resistance to macrolides in Pseudomonas aeruginosa from cystic fibrosis patients. Eur. Respir. J. 2017, 49, 1601847. [Google Scholar] [CrossRef]
- Kobayashi, H. Biofilm disease: Its clinical manifestation and therapeutic possibilities of macrolides. Am. J. Med. 1995, 99, 26S–30S. [Google Scholar] [CrossRef]
- Hirakata, Y.; Kaku, M.; Tomono, K.; Tateda, K.; Furuya, N.; Matsumoto, T.; Araki, R.; Yamaguchi, K. Efficacy of erythromycin lactobionate for treating Pseudomonas aeruginosa bacteremia in mice. Antimicrob. Agents Chemother. 1992, 36, 1198–1203. [Google Scholar] [CrossRef] [PubMed]
- Howe, R.A.; Spencer, R.C. Macrolides for the treatment of Pseudomonas aeruginosa infections? J. Antimicrob. Chemother. 1997, 40, 153–155. [Google Scholar] [CrossRef]
- Giamarellos-Bourboulis, E.J. Macrolides beyond the conventional antimicrobials: A class of potent immunomodulators. Int. J. Antimicrob. Agents 2008, 31, 12–20. [Google Scholar] [CrossRef]
- Ishiguro, M.; Koga, H.; Kohno, S.; Hayashi, T.; Yamaguchi, K.; Hirota, M. Penetration of macrolides into human poly- morphonuclear leucocytes. J. Antimicrob. Chemother. 1989, 24, 719–729. [Google Scholar] [CrossRef]
- Pocidalo, J.-J.; Albert, F.; Desnottes, J.F.; Kernbaum, S. Intraphagocytic penetration of macrolides: In-vivo comparison of erythromycin and spiramycin. J. Antimicrob. Chemother. 1985, 16, 167–173. [Google Scholar] [CrossRef]
- Hirakata, Y.; Kaku, M.; Mizukane, R.; Ishida, K.; Furuya, N.; Matsumoto, T.; Tateda, K.; Yamaguchi, K. Potential effects of erythromycin on host defense systems and virulence of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1992, 36, 1922–1927. [Google Scholar] [CrossRef]
- Mizukane, R.; Hirakata, Y.; Kaku, M.; Ishii, Y.; Furuya, N.; Ishida, K.; Koga, H.; Kohno, S.; Yamaguchi, K. Comparative in vitro exoenzyme-suppressing activities of azithromycin and other macrolide antibiotics against Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1994, 38, 528–533. [Google Scholar] [CrossRef]
- Molinari, G.; Guzmán, C.A.; Pesce, A.; Schito, G.C. Inhibition of Pseudomonas aeruginosa virulence factors by subinhibitory concentrations of azithromycin and other macrolide antibiotics. J. Antimicrob. Chemother. 1993, 31, 681–688. [Google Scholar] [CrossRef]
- Schneierson, S.S.; Amsterdam, D.; Perlman, E. Inhibition of Pseudomonas aeruginosa pigment formation by chloramphenicol and erythromycin. Antibiot. Chemother. 1960, 10, 30–33. [Google Scholar]
- Tanaka, E.; Kanthakumar, K.; Cundell, D.R.; Tsang, K.W.; Taylor, G.W.; Kuze, F.; Cole, P.J.; Wilson, R. The effect of erythromycin on Pseudomonas aeruginosa and neutrophil mediated epithelial damage. J. Antimicrob. Chemother. 1994, 33, 765–775. [Google Scholar] [CrossRef]
- Kita, E.; Sawaki, M.; Oku, D.; Hamuro, A.; Mikasa, K.; Konishi, M.; Emoto, M.; Takeuchi, S.; Narita, N.; Kashiba, S. Suppression of virulence factors of Pseudomonas aeruginosa by erythromycin. J. Antimicrob. Chemother. 1991, 27, 273–284. [Google Scholar] [CrossRef]
- Menninger, J.R.; Coleman, R.A.; Tsai, L.-N. Erythromycin, lincosamides, peptidyl-tRNA dissociation, and ribosome editing. Mol. Gen. Genet. 1994, 243, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Tateda, K.; Comte, R.; Pechere, J.-C.; Köhler, T.; Yamaguchi, K.; Van Delden, C. Azithromycin inhibits quorum sensing in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2001, 45, 1930–1933. [Google Scholar] [CrossRef] [PubMed]
- Pechère, J.C. Azithromycin reduces the production of virulence factors in Pseudomonas aeruginosa by inhibiting quorum sensing. Jpn. J. Antibiot. 2001, 54, 87–89. [Google Scholar] [PubMed]
- Imamura, Y.; Yanagihara, K.; Mizuta, Y.; Seki, M.; Ohno, H.; Higashiyama, Y.; Miyazaki, Y.; Tsukamoto, K.; Hirakata, Y.; Tomono, K.; et al. Azithromycin inhibits MUC5AC production induced by the Pseudomonas aeruginosa autoinducer N-(3-Oxododecanoyl) homoserine lactone in NCI-H292 Cells. Antimicrob. Agents Chemother. 2004, 48, 3457–3461. [Google Scholar] [CrossRef]
- Leroy, A.-G.; Caillon, J.; Caroff, N.; Broquet, A.; Corvec, S.; Asehnoune, K.; Roquilly, A.; Crémet, L. Could azithromycin be part of Pseudomonas aeruginosa acute pneumonia treatment? Front. Microbiol. 2021, 12, 642541. [Google Scholar] [CrossRef]
- Hoffmann, N.; Lee, B.; Hentzer, M.; Rasmussen, T.B.; Song, Z.; Johansen, H.K.; Givskov, M.; Høiby, N. Azithromycin blocks quorum sensing and alginate polymer formation and increases the sensitivity to serum and stationary-growth-phase killing of Pseudomonas aeruginosa and attenuates chronic P. aeruginosa lung infection in Cftr−/− mice. Antimicrob. Agents Chemother. 2007, 51, 3677–3687. [Google Scholar] [CrossRef]
- Bala, A.; Kumar, R.; Harjai, K. Inhibition of quorum sensing in Pseudomonas aeruginosa by azithromycin and its effectiveness in urinary tract infections. J. Med. Microbiol. 2011, 60, 300–306. [Google Scholar] [CrossRef]
- Burr, L.D.; Rogers, G.B.; Chen, A.C.-H.; Hamilton, B.R.; Pool, G.F.; Taylor, S.L.; Venter, D.; Bowler, S.D.; Biga, S.; McGuckin, M.A. Macrolide Treatment Inhibits Pseudomonas aeruginosa Quorum Sensing in Non-Cystic Fibrosis Bronchiectasis. An Analysis from the Bronchiectasis and Low-Dose Erythromycin Study Trial. Ann. Am. Thorac. Soc. 2016, 13, 1697–1703. [Google Scholar] [CrossRef]
- Nalca, Y.; Jänsch, L.; Bredenbruch, F.; Geffers, R.; Buer, J.; Häussler, S. Quorum-sensing antagonistic activities of azithromycin in Pseudomonas aeruginosa PAO1: A global approach. Antimicrob. Agents Chemother. 2006, 50, 1680–1688. [Google Scholar] [CrossRef]
- Chua, S.L.; Yam, J.K.H.; Hao, P.; Adav, S.S.; Salido, M.M.; Liu, Y.; Givskov, M.; Sze, S.K.; Tolker-Nielsen, T.; Yang, L. Selective labelling and eradication of antibiotic-tolerant bacterial populations in Pseudomonas aeruginosa biofilms. Nat. Commun. 2016, 7, 10750. [Google Scholar] [CrossRef]
- Li, Y.; Huang, J.; Li, L.; Liu, L. Synergistic Activity of Berberine with Azithromycin against Pseudomonas Aeruginosa Isolated from Patients with Cystic Fibrosis of Lung In Vitro and In Vivo. Cell. Physiol. Biochem. 2017, 42, 1657–1669. [Google Scholar] [CrossRef]
- van Delden, C.; Köhler, T.; Brunner-Ferber, F.; François, B.; Carlet, J.; Pechère, J.-C. Azithromycin to prevent Pseudomonas aeruginosa ventilator-associated pneumonia by inhibition of quorum sensing: A randomized controlled trial. Intensive Care Med. 2012, 38, 1118–1125. [Google Scholar] [CrossRef] [PubMed]
- Saini, H.; Vadekeetil, A.; Chhibber, S.; Harjai, K. Azithromycin-Ciprofloxacin-Impregnated Urinary Catheters Avert Bacterial Colonization, Biofilm Formation, and Inflammation in a Murine Model of Foreign-Body-Associated Urinary Tract Infections Caused by Pseudomonas aeruginosa. Antimicrob. Agents. Chemother. 2017, 61, e01906-16. [Google Scholar] [CrossRef] [PubMed]
- Köhler, T.; Perron, G.G.; Buckling, A.; van Delden, C. Quorum sensing inhibition selects for virulence and cooperation in Pseudomonas aeruginosa. PLoS Pathog. 2010, 6, e1000883. [Google Scholar] [CrossRef] [PubMed]
- Pinnert-Sindico, S.; Ninet, L.; Preud’Homme, J.; Cosar, C. A new antibiotic-spiramycin. In Antibiotics annual; Welch, H., Marti-Ibanez, F., Eds.; Medical Encyclopedia: New York, USA, 1995; pp. 827–830. [Google Scholar]
- Poulet, P.-P.; Duffaut, D.; Barthet, P.; Brumpt, I. Concentrations and in vivo antibacterial activity of spiramycin and metronidazole in patients with periodontitis treated with high-dose metronidazole and the spiramycin/metronidazole combination. J. Antimicrob. Chemother. 2005, 55, 347–351. [Google Scholar] [CrossRef] [PubMed]
- Chew, W.K.; Segarra, I.; Ambu, S.; Mak, J.W. Significant reduction of brain cysts caused by Toxoplasma gondii after treatment with spiramycin coadministered with metronidazole in a mouse model of chronic toxoplasmosis. Antimicrob. Agents Chemother. 2012, 56, 1762–1768. [Google Scholar] [CrossRef] [PubMed]
- Tenson, T.; Lovmar, M.; Ehrenberg, M. The mechanism of action of macrolides, lincosamides and streptogramin B reveals the nascent peptide exit path in the ribosome. J. Mol. Biol. 2003, 330, 1005–1014. [Google Scholar] [CrossRef]
- Chabbert, Y.A. Early studies on in-vitro and experimental activity of spiramycin: A review. J. Antimicrob. Chemother. 1988, 22, 1–11. [Google Scholar] [CrossRef]
- Calcagnile, M.; Alifano, P. Off-Target Activity of Spiramycin Disarms Pseudomonas aeruginosa by Inhibition of Biofilm Formation, Pigment Production and Phenotypic Differentiation. Med. Sci. Forum 2022, 12, 42. [Google Scholar]
- Wadday, A.K.; Saleh, Z.A.; Al-Marjani, M.F. Spectroscopic characteristics and energy transfer of bacterial pigment:(pyocyanin/curcumin). In AIP Conference Proceedings; AIP Publishing: Melville, NY, USA, 2019; p. 020014. [Google Scholar]
- Gonçalves, T.; Vasconcelos, U. Colour Me Blue: The History and the Biotechnological Potential of Pyocyanin. Molecules 2021, 26, 927. [Google Scholar] [CrossRef]
- Eslami, P.; Hajfarajollah, H.; Bazsefidpar, S. Recent advancements in the production of rhamnolipid biosurfactants by Pseudomonas aeruginosa. RSC Adv. 2020, 10, 34014–34032. [Google Scholar] [CrossRef]
- Sood, U.; Singh, D.N.; Hira, P.; Lee, J.-K.; Kalia, V.C.; Lal, R.; Shakarad, M. Rapid and solitary production of mono-rhamnolipid biosurfactant and biofilm inhibiting pyocyanin by a taxonomic outlier Pseudomonas aeruginosa strain CR1. J. Biotechnol. 2020, 307, 98–106. [Google Scholar] [CrossRef]
- Yu, H.; He, X.; Xie, W.; Xiong, J.; Sheng, H.; Guo, S.; Huang, C.; Zhang, D.; Zhang, K. Elastase LasB of Pseudomonas aeruginosa promotes biofilm formation partly through rhamnolipid-mediated regulation. Can. J. Microbiol. 2014, 60, 227–235. [Google Scholar] [CrossRef]
- Sun, J.; LaRock, D.L.; Skowronski, E.A.; Kimmey, J.M.; Olson, J.; Jiang, Z.; O’Donoghue, A.J.; Nizet, V.; LaRock, C.N. The Pseudomonas aeruginosa protease LasB directly activates IL-1β. EBioMedicine 2020, 60, 102984. [Google Scholar] [CrossRef]
- Mateu-Borrás, M.; Zamorano, L.; González-Alsina, A.; Sánchez-Diener, I.; Doménech-Sánchez, A.; Oliver, A.; Albertí, S. Molecular Analysis of the Contribution of Alkaline Protease A and Elastase B to the Virulence of Pseudomonas aeruginosa Bloodstream Infections. Front. Cell. Infect. Microbiol. 2022, 11, 816356. [Google Scholar] [CrossRef]
- Galdino, A.C.M.; Viganor, L.; De Castro, A.A.; da Cunha, E.F.F.; Mello, T.P.; Mattos, L.M.; Pereira, M.D.; Hunt, M.C.; O’Shaughnessy, M.; Howe, O.; et al. Disarming Pseudomonas aeruginosa virulence by the inhibitory action of 1, 10-phenanthroline-5, 6-dione-based compounds: Elastase B (lasB) as a chemotherapeutic target. Front. Microbiol. 2019, 10, 1701. [Google Scholar] [CrossRef]
- Everett, M.J.; Davies, D.T. Pseudomonas aeruginosa elastase (LasB) as a therapeutic target. Drug Discov. Today 2021, 26, 2108–2123. [Google Scholar] [CrossRef]
- Orlandi, V.T.; Bolognese, F.; Chiodaroli, L.; Tolker-Nielsen, T.; Barbieri, P. Pigments influence the tolerance of Pseudomonas aeruginosa PAO1 to photodynamically induced oxidative stress. Microbiology 2015, 161, 2298–2309. [Google Scholar] [CrossRef]
- Ichimiya, T.; Takeoka, K.; Hiramatsu, K.; Hirai, K.; Yamasaki, T.; Nasu, M. The influence of azithromycin on the biofilm formation of Pseudomonas aeruginosa in vitro. Chemotherapy 1996, 42, 186–191. [Google Scholar] [CrossRef]
- Gillis, R.J.; Iglewski, B.H. Azithromycin retards Pseudomonas aeruginosa biofilm formation. J. Clin. Microbiol. 2004, 42, 5842–5845. [Google Scholar] [CrossRef]
- lchimiya, T.; Yamasaki, T.; Nasu, M. In-vitro effects of antimicrobial agents on Pseudomonas aeruginosa biofilm formation. J. Antimicrob. Chemother. 1994, 34, 331–341. [Google Scholar] [CrossRef]
- Gödeke, J.; Pustelny, C.; Häussler, S. Recycling of peptidyl-tRNAs by peptidyl-tRNA hydrolase counteracts azithromycin-mediated effects on Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2013, 57, 1617–1624. [Google Scholar] [CrossRef] [PubMed]
- Glick, R.; Gilmour, C.; Tremblay, J.; Satanower, S.; Avidan, O.; Déziel, E.; Greenberg, E.P.; Poole, K.; Banin, E. Increase in rhamnolipid synthesis under iron-limiting conditions influences surface motility and biofilm formation in Pseudomonas aeruginosa. J. Bacteriol. 2010, 192, 2973–2980. [Google Scholar] [CrossRef] [PubMed]
- Davey, M.E.; Caiazza, N.C.; O’Toole, G.A. Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J. Bacteriol. 2003, 185, 1027–1036. [Google Scholar] [CrossRef]
- Boles, B.R.; Thoendel, M.; Singh, P.K. Rhamnolipids mediate detachment of Pseudomonas aeruginosa from biofilms. Mol. Microbiol. 2005, 57, 1210–1223. [Google Scholar] [CrossRef]
- Caiazza, N.C.; Shanks, R.M.Q.; O’Toole, G.A. Rhamnolipids modulate swarming motility patterns of Pseudomonas aeruginosa. J. Bacteriol. 2005, 187, 7351–7361. [Google Scholar] [CrossRef]
- Liao, C.; Huang, X.; Wang, Q.; Yao, D.; Lu, W. Virulence Factors of Pseudomonas Aeruginosa and Antivirulence Strategies to Combat Its Drug Resistance. Front. Cell. Infect. Microbiol. 2022, 12, 926758. [Google Scholar] [CrossRef]
- Sheehan, G.; Garvey, A.; Croke, M.; Kavanagh, K. Innate humoral immune defences in mammals and insects: The same, with differences? Virulence 2018, 9, 1625–1639. [Google Scholar] [CrossRef]
- Hancock, R.E.W.; Haney, E.F.; Gill, E.E. The immunology of host defence peptides: Beyond antimicrobial activity. Nat. Rev. Immunol. 2016, 16, 321–334. [Google Scholar] [CrossRef]
- Reuter, K.; Steinbach, A.; Helms, V. Interfering with bacterial quorum sensing. Perspect. Med. Chem. 2016, 8, 1–15. [Google Scholar] [CrossRef]
- Wang, L.; Hu, C.; Shao, L. The antimicrobial activity of nanoparticles: Present situation and prospects for the future. Int. J. Nanomed. 2017, 12, 1227–1249. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Raudonis, R.; Glick, B.R.; Lin, T.J.; Cheng, Z. Antibiotic resistance in Pseudomonas aeruginosa: Mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 2019, 37, 177–192. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.; Xiao, W.; Zhou, C.; Pu, Q.; Deng, X.; Lan, L.; Liang, H.; Song, X.; Wu, M. Pseudomonas aeruginosa: Pathogenesis, virulence factors, antibiotic resistance, interaction with host, technology advances and emerging therapeutics. Signal Transduct. Target. Ther. 2022, 7, 199. [Google Scholar] [CrossRef] [PubMed]
- Kohanski, M.A.; Dwyer, D.J.; Hayete, B.; Lawrence, C.A.; Collins, J.J. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 2007, 130, 797–810. [Google Scholar] [CrossRef]
- Kint, C.I.; Verstraeten, N.; Fauvart, M.; Michiels, J. New-found fundamentals of bacterial persistence. Trends Microbiol. 2012, 20, 577–585. [Google Scholar] [CrossRef] [PubMed]
- Wagner, T.; Soong, G.; Sokol, S.; Saiman, L.; Prince, A. Effects of azithromycin on clinical isolates of Pseudomonas aeruginosa from cystic fibrosis patients. Chest 2005, 128, 912–919. [Google Scholar] [CrossRef]
- Kai, T.; Tateda, K.; Kimura, S.; Ishii, Y.; Ito, H.; Yoshida, H.; Kimura, T.; Yamaguchi, K. A low concentration of azithromycin inhibits the mRNA expression of N-acyl homoserine lactone synthesis enzymes, upstream of lasI or rhlI, in Pseudomonas aeruginosa. Pulm. Pharmacol. Ther. 2009, 22, 483–486. [Google Scholar] [CrossRef]
- Bahari, S.; Zeighami, H.; Mirshahabi, H.; Roudashti, S.; Haghi, F. Inhibition of Pseudomonas aeruginosa quorum sensing by subinhibitory concentrations of curcumin with gentamicin and azithromycin. J. Glob. Antimicrob. Resist. 2017, 10, 21–28. [Google Scholar] [CrossRef]
- Zeng, J.; Zhang, N.; Huang, B.; Cai, R.; Wu, B.; Shunmei, E.; Fang, C.; Chen, C. Mechanism of azithromycin inhibition of HSL synthesis in Pseudomonas aeruginosa. Sci. Rep. 2016, 6, 24299. [Google Scholar] [CrossRef]
- Skindersoe, M.E.; Alhede, M.; Phipps, R.; Yang, L.; Jensen, P.O.; Rasmussen, T.B.; Bjarnsholt, T.; Tolker-Nielsen, T.; Høiby, N.; Givskov, M. Effects of antibiotics on quorum sensing in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2008, 52, 3648–3663. [Google Scholar] [CrossRef]
- Pérez-Martínez, I.; Haas, D. Azithromycin inhibits expression of the GacA-dependent small RNAs RsmY and RsmZ in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2011, 55, 3399–3405. [Google Scholar] [CrossRef] [PubMed]
- Köhler, T.; Dumas, J.-L.; Van Delden, C. Ribosome protection prevents azithromycin-mediated quorum-sensing modulation and stationary-phase killing of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2007, 51, 4243–4248. [Google Scholar] [CrossRef] [PubMed]
- Imperi, F.; Leoni, L.; Visca, P. Antivirulence activity of azithromycin in Pseudomonas aeruginosa. Front. Microbiol. 2014, 5, 178. [Google Scholar] [CrossRef] [PubMed]
- Baldelli, V.; D’Angelo, F.; Pavoncello, V.; Fiscarelli, E.V.; Visca, P.; Rampioni, G.; Leoni, L. Identification of FDA-approved antivirulence drugs targeting the Pseudomonas aeruginosa quorum sensing effector protein PqsE. Virulence 2020, 11, 652–668. [Google Scholar] [CrossRef] [PubMed]
- Rampioni, G.; Falcone, M.; Heeb, S.; Frangipani, E.; Fletcher, M.P.; Dubern, J.-F.; Visca, P.; Leoni, L.; Cámara, M.; Williams, P. Unravelling the Genome-Wide Contributions of Specific 2-Alkyl-4-Quinolones and PqsE to Quorum Sensing in Pseudomonas aeruginosa. PLoS Pathog. 2016, 12, e1006029. [Google Scholar] [CrossRef] [PubMed]
- Jander, G.; Rahme, L.G.; Ausubel, F.M. Positive correlation between virulence of Pseudomonas aeruginosa mutants in mice and insects. J. Bacteriol. 2000, 182, 3843–3845. [Google Scholar] [CrossRef]
- Andrejko, M.; Mak, P.; Siemińska-Kuczer, A.; Iwański, B.; Wojda, I.; Suder, P.; Kuleta, P.; Regucka, K.; Cytryńska, M. A comparison of the production of antimicrobial peptides and proteins by Galleria mellonella larvae in response to infection with two Pseudomonas aeruginosa strains differing in the profile of secreted proteases. J. Insect Physiol. 2021, 131, 104239. [Google Scholar] [CrossRef]
- Freitak, D.; Schmidtberg, H.; Dickel, F.; Lochnit, G.; Vogel, H.; Vilcinskas, A. The maternal transfer of bacteria can mediate trans-generational immune priming in insects. Virulence 2014, 5, 547–554. [Google Scholar] [CrossRef]
- Lange, A.; Schäfer, A.; Bender, A.; Steimle, A.; Beier, S.; Parusel, R.; Frick, J.-S. Galleria mellonella: A Novel Invertebrate Model to Distinguish Intestinal Symbionts From Pathobionts. Front. Immunol. 2018, 9, 2114. [Google Scholar] [CrossRef]
- Pollock, J.; Chalmers, J.D. The immunomodulatory effects of macrolide antibiotics in respiratory disease. Pulm. Pharmacol. Ther. 2021, 71, 102095. [Google Scholar] [CrossRef]
- Hubble, V.B.; Hubbard, B.A.; Minrovic, B.M.; Melander, R.J.; Melander, C. Using Small-Molecule Adjuvants to Repurpose Azithromycin for Use against Pseudomonas aeruginosa. ACS Infect. Dis. 2019, 5, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Belanger, C.R.; Lee, A.H.-Y.; Pletzer, D.; Dhillon, B.K.; Falsafi, R.; Hancock, R.E.W. Identification of novel targets of azithromycin activity against Pseudomonas aeruginosa grown in physiologically relevant media. Proc. Natl. Acad. Sci. USA 2020, 117, 33519–33529. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Miyoshi-Akiyama, T.; Ogura, K.; Murata, S.; Ishige, S.; Kai, K.; Mitsutsuka, K.; Tomita, H.; Tanimoto, K.; Matsumoto, A. Effect of Sub-MICs of Macrolides on the Sensitivity of Pseudomonas aeruginosa to Nitrosative Stress: Effectiveness against P. aeruginosa with and without Multidrug Resistance. Antimicrob. Agents Chemother. 2020, 64, e01180-20. [Google Scholar] [CrossRef] [PubMed]
- Yazdanian, M.; Barzegar, P.E.F.; Ranjbar, R.; Tahmasebi, E.; Tebyaniyan, H.; Barzegar, K.E.F.; Hayati, F.Z.; Farjanikish, G. The comparative effects of erythromycin and amikacin on acute respiratory Pseudomonas aeruginosa infection. Vet. Med. Sci. 2022, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Rao, M.; Mathur, T.; Barman, T.K.; Joshi, V.; Chaira, T.; Singhal, S.; Pandya, M.; Al Khodor, S.; Upadhyay, D.J.; et al. Azithromycin Exhibits Activity Against Pseudomonas aeruginosa in Chronic Rat Lung Infection Model. Front. Microbiol. 2021, 12, 603151. [Google Scholar] [CrossRef] [PubMed]
- Sörensen, M.; Khakimov, B.; Nurjadi, D.; Boutin, S.; Yi, B.; Dalpke, A.H.; Eigenbrod, T. Comparative evaluation of the effect of different growth media on in vitro sensitivity to azithromycin in multi-drug resistant Pseudomonas aeruginosa isolated from cystic fibrosis patients. Antimicrob. Resist. Infect. Control. 2020, 9, 1–7. [Google Scholar] [CrossRef]
- Boczar, D.; Michalska, K. Cyclodextrin Inclusion Complexes with Antibiotics and Antibacterial Agents as Drug-Delivery Systems-A Pharmaceutical Perspective. Pharmaceutics 2022, 14, 1389. [Google Scholar] [CrossRef]
- Calcagnile, M.; Bettini, S.; Damiano, F.; Talà, A.; Tredici, S.M.; Pagano, R.; Di Salvo, M.; Siculella, L.; Fico, D.; De Benedetto, G.E.; et al. Stimulatory Effects of Methyl-β-cyclodextrin on Spiramycin Production and Physical-Chemical Characterization of Nonhost@Guest Complexes. ACS Omega 2018, 3, 2470–2478. [Google Scholar] [CrossRef]
- Song, W.; Yu, X.; Wang, S.; Blasier, R.; Markel, D.C.; Mao, G.; Shi, T.; Ren, W. Cyclodextrin-erythromycin complexes as a drug delivery device for orthopedic application. Int. J. Nanomed. 2011, 6, 3173–3186. [Google Scholar] [CrossRef]
- Tredici, S.M.; Buccolieri, A.; Tanini, L.; Calcagnile, M.; Manno, D.; Alifano, P. Calcite-forming Bacillus licheniformis thriving on underwater speleothems of a hydrothermal cave. Geomicrobiol. J. 2018, 35, 804–817. [Google Scholar] [CrossRef]
- Dao, K.-H.T.; Hamer, K.E.; Clark, C.L.; Harshman, L.G. Pyoverdine production by Pseudomonas aeruginosa exposed to metals or an oxidative stress agent. Ecol. Appl. 1999, 9, 441–448. [Google Scholar] [CrossRef]
- Cheng, X.; Liu, J.; Li, J.; Zhou, X.; Wang, L.; Liu, J.; Xu, X. Comparative effect of a stannous fluoride toothpaste and a sodium fluoride toothpaste on a multispecies biofilm. Arch. Oral Biol. 2017, 74, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Cotter, G.; Doyle, S.; Kavanagh, K. Development of an insect model for the in vivo pathogenicity testing of yeasts. FEMS Immunol. Med. Microbiol. 2000, 27, 163–169. [Google Scholar] [CrossRef]
- Mowlds, P.; Kavanagh, K. Effect of pre-incubation temperature on susceptibility of Galleria mellonella to infection by Candida albicans. Mycopathologia 2008, 165, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Kavanagh, K.; Fallon, J.P. Galleria mellonella larvae as models for studying fungal virulence. Fungal. Biol. Rev. 2010, 24, 79–83. [Google Scholar] [CrossRef]
- Siculella, L.; Giannotti, L.; Di Chiara Stanca, B.; Calcagnile, M.; Rochira, A.; Stanca, E.; Alifano, P.; Damiano, F. Evidence for a Negative Correlation between Human Reactive Enamine-Imine Intermediate Deaminase A (RIDA) Activity and Cell Proliferation Rate: Role of Lysine Succinylation of RIDA. Int. J. Mol. Sci. 2021, 22, 3804. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Yang, D.; Zhu, L.; Shi, F.; Ye, G.; Guo, H.; Deng, H.; Zhao, L.; Xu, Z.; Li, Y. Paeonol Interferes with Quorum-Sensing in Pseudomonas aeruginosa and Modulates Inflammatory Responses In Vitro and In Vivo. Front. Immunol. 2022, 13, 896874. [Google Scholar] [CrossRef]
- Lane, D.J.; Pace, B.; Olson, G.J.; Stahl, D.A.; Sagin, M.L.; Pace, N.R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analysis. Proc. Natl. Acad. Sci. USA 1985, 82, 6955–6959. [Google Scholar] [CrossRef]


| μg/mL | AMP | STR | RMP | ERY | SPM |
|---|---|---|---|---|---|
| 500 | - | - | - | - | + |
| 250 | + | - | - | - | + |
| 125 | + | - | - | + | + |
| 62.5 | + | - | - | + | + |
| 31.3 | + | + | - | + | + |
| 15.6 | + | + | + | + | + |
| 7.8 | + | + | + | + | + |
| 3.9 | + | + | + | + | + |
| 2.0 | + | + | + | + | + |
| 1.0 | + | + | + | + | + |
| 0.5 | + | + | + | + | + |
| 0.2 | + | + | + | + | + |
| Control | + | + | + | + | + |
| Sample | Primer Name | Sequence 5′-3′ | Reference |
|---|---|---|---|
| G. mellonella | Gallerimycin f | GAAGTCTACAGAATCACACGA | [79] |
| G. mellonella | Gallerimycin r | ATCGAAGACATTGACATCCA | |
| G. mellonella | ubiquitin f 1 | TCAATGCAAGTAGTCCGGTTC | [80] |
| G. mellonella | ubiquitin r 1 | CCAGTCTGCTGCTGATAAACC | |
| G. mellonella | Gloverin f | GTGTTGAGCCCGTATGGGAA | [79] |
| G. mellonella | Gloverin r | CCGTGCATCTGCTTGCTAAC | |
| G. mellonella | Lysozyme f | GGACTGGTCCGAGCACTTAG | [79] |
| G. mellonella | Lysozyme r | CGCATTTAGAGGCAACCGTG | |
| G. mellonella | Moricin f | GCTGTACTCGCTGCACTGAT | [79] |
| G. mellonella | Moricin r | TGGCGATCATTGCCCTCTTT | |
| P. aeruginosa | lasB r | AACCGTGCGTTCTACCTGTT | [99] |
| P. aeruginosa | lasB f | CGGTCCAGTAGTAGCGGTTG | |
| P. aeruginosa | rhlC f | GCCATCCATCTCGACGGAC | [99] |
| P. aeruginosa | rhlC r | CGCAGGCTGTATTCGGTG | |
| P. aeruginosa | phzS f | CCGAAGGCAAGTCGCTGGTGA | [99] |
| P. aeruginosa | phzS r | GGTCCCAGTCGGCGAAGAACG | |
| P. aeruginosa | COM1 1 | CAGCAGCCGCGGTAATAC | [100] |
| P. aeruginosa | COM2 1 | CCGTCAATTCCTTTGAGTTT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Calcagnile, M.; Jeguirim, I.; Tredici, S.M.; Damiano, F.; Alifano, P. Spiramycin Disarms Pseudomonas aeruginosa without Inhibiting Growth. Antibiotics 2023, 12, 499. https://doi.org/10.3390/antibiotics12030499
Calcagnile M, Jeguirim I, Tredici SM, Damiano F, Alifano P. Spiramycin Disarms Pseudomonas aeruginosa without Inhibiting Growth. Antibiotics. 2023; 12(3):499. https://doi.org/10.3390/antibiotics12030499
Chicago/Turabian StyleCalcagnile, Matteo, Inès Jeguirim, Salvatore Maurizio Tredici, Fabrizio Damiano, and Pietro Alifano. 2023. "Spiramycin Disarms Pseudomonas aeruginosa without Inhibiting Growth" Antibiotics 12, no. 3: 499. https://doi.org/10.3390/antibiotics12030499
APA StyleCalcagnile, M., Jeguirim, I., Tredici, S. M., Damiano, F., & Alifano, P. (2023). Spiramycin Disarms Pseudomonas aeruginosa without Inhibiting Growth. Antibiotics, 12(3), 499. https://doi.org/10.3390/antibiotics12030499









