Gut Microbiota Composition Can Predict Colonization by Multidrug-Resistant Bacteria in SARS-CoV-2 Patients in Intensive Care Unit: A Pilot Study
Abstract
1. Introduction
2. Results
2.1. Study Cohort Description
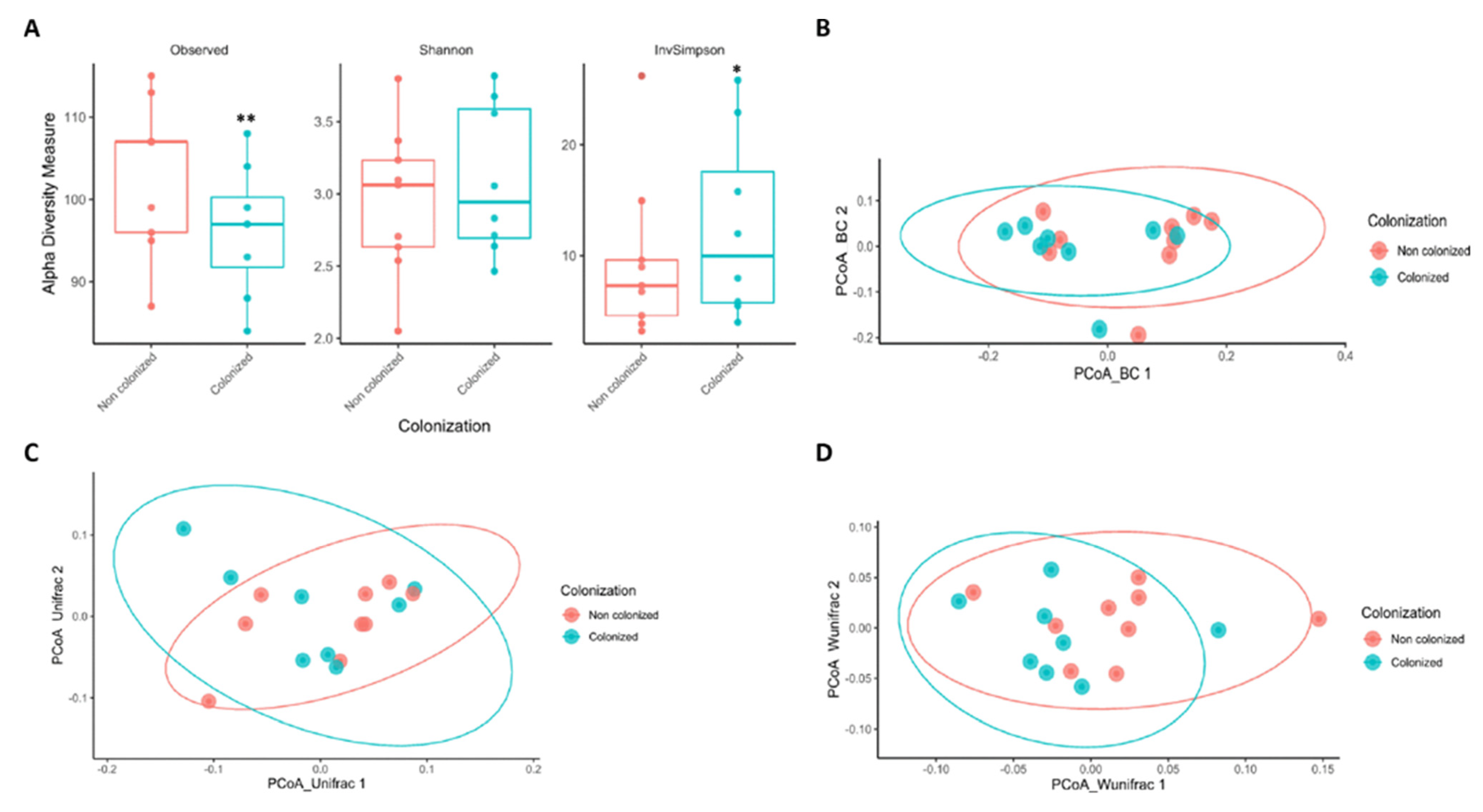
2.2. Lower Gut Microbiota Diversity Is Associated with a High Risk of Colonization by MDRB in Critical SARS-CoV-2 Patients
2.3. SARS-CoV-2 Associated Shifts in the Initial Gut Microbiota Are Related to MDRB Colonization in ICU Facilities
2.4. Identification of Specific Bacteria as Possible Markers for Predicting MDRB Colonization in SARS-CoV-2 Patients in ICUs
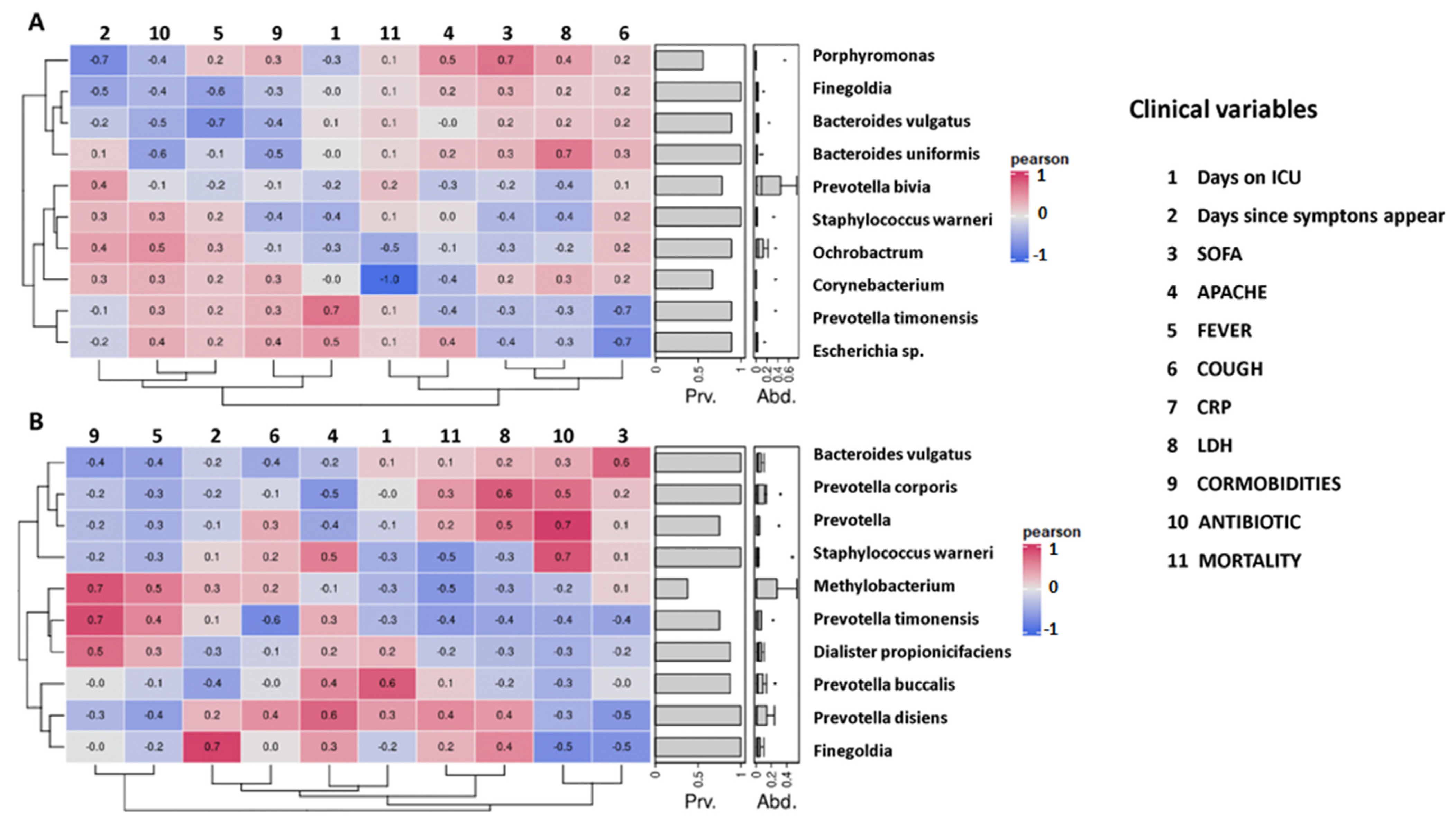
2.5. Correlation between Initial Gut Microbiota Composition and Clinical Variables of Non-Colonized and Colonized Patients
3. Discussion
4. Materials and Methods
4.1. Patient Population
4.2. Ethics Statement
4.3. Study Design
4.4. SARS-CoV-2 Diagnosis
4.5. Microbial DNA Extraction, Library Preparation, and Next Generation Sequencing
4.6. Bioinformatics and Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhao, W.; Zhang, J.; Meadows, M.E.; Liu, Y.; Hua, T.; Fu, B. A systematic approach is needed to contain COVID-19 globally. Sci. Bull. 2020, 65, 876–878. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. COVID-19 Weekly Epidemiological Update. Available online: https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19 (accessed on 23 November 2022).
- NIH Clinical Spectrum of SARS-CoV-2 Infection. 2022. Available online: https://www.covid19treatmentguidelines.nih.gov/overview/clinical-spectrum/ (accessed on 1 July 2022).
- Wu, Z.; McGoogan, J.M. Characteristics of and Important Lessons from the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72 314 Cases from the Chinese Center for Disease Control and Prevention. JAMA 2020, 323, 1239–1242. [Google Scholar] [CrossRef] [PubMed]
- The World Bank. Drug Resistant Infections: A Threat to Our Economic Future. 2016. Available online: https://www.worldbank.org/en/topic/health/publication/drug-resistant-infections-a-threat-to-our-economic-future (accessed on 1 July 2022).
- Antimicrobial Resistance, C. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Monnet, D.L.; Harbarth, S. Will coronavirus disease (COVID-19) have an impact on antimicrobial resistance? Eurosurveillance 2020, 25, 2001886. [Google Scholar] [CrossRef] [PubMed]
- Rawson, T.M.; Ming, D.; Ahmad, R.; Moore, L.S.P.; Holmes, A.H. Antimicrobial use, drug-resistant infections and COVID-19. Nat. Rev. Microbiol. 2020, 18, 409–410. [Google Scholar] [CrossRef]
- Nori, P.; Cowman, K.; Chen, V.; Bartash, R.; Szymczak, W.; Madaline, T.; Punjabi Katiyar, C.; Jain, R.; Aldrich, M.; Weston, G.; et al. Bacterial and fungal coinfections in COVID-19 patients hospitalized during the New York City pandemic surge. Infect. Control. Hosp. Epidemiol. 2021, 42, 84–88. [Google Scholar] [CrossRef]
- Rawson, T.M.; Wilson, R.C.; Holmes, A. Understanding the role of bacterial and fungal infection in COVID-19. Clin. Microbiol. Infect. 2021, 27, 9–11. [Google Scholar] [CrossRef]
- McDonald, D.; Ackermann, G.; Khailova, L.; Baird, C.; Heyland, D.; Kozar, R.; Lemieux, M.; Derenski, K.; King, J.; Vis-Kampen, C.; et al. Extreme Dysbiosis of the Microbiome in Critical Illness. mSphere 2016, 1, e00199-16. [Google Scholar] [CrossRef]
- Dickson, R.P. The microbiome and critical illness. Lancet Respir. Med. 2016, 4, 59–72. [Google Scholar] [CrossRef]
- Fay, K.T.; Ford, M.L.; Coopersmith, C.M. The intestinal microenvironment in sepsis. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 2574–2583. [Google Scholar] [CrossRef]
- Bouza, E.; Munoz, P. Epidemiology of candidemia in intensive care units. Int. J. Antimicrob. Agents 2008, 32 (Suppl. 2), S87–S91. [Google Scholar] [CrossRef] [PubMed]
- Kondori, N.; Nowrouzian, F.; Ajdari, M.; Hesselmar, B.; Saalman, R.; Wold, A.E.; Adlerberth, I. Candida species as commensal gut colonizers: A study of 133 longitudinally followed Swedish infants. Med. Mycol. 2020, 58, 485–492. [Google Scholar] [CrossRef] [PubMed]
- Bassetti, M.; De Waele, J.J.; Eggimann, P.; Garnacho-Montero, J.; Kahlmeter, G.; Menichetti, F.; Nicolau, D.P.; Paiva, J.A.; Tumbarello, M.; Welte, T.; et al. Preventive and therapeutic strategies in critically ill patients with highly resistant bacteria. Intensive Care Med. 2015, 41, 776–795. [Google Scholar] [CrossRef] [PubMed]
- Davison, J.M.; Wischmeyer, P.E. Probiotic and synbiotic therapy in the critically ill: State of the art. Nutrition 2019, 59, 29–36. [Google Scholar] [CrossRef]
- McClave, S.A.; Patel, J.; Bhutiani, N. Should fecal microbial transplantation be used in the ICU? Curr. Opin. Crit. Care 2018, 24, 105–111. [Google Scholar] [CrossRef]
- Shimizu, K.; Yamada, T.; Ogura, H.; Mohri, T.; Kiguchi, T.; Fujimi, S.; Asahara, T.; Yamada, T.; Ojima, M.; Ikeda, M.; et al. Synbiotics modulate gut microbiota and reduce enteritis and ventilator-associated pneumonia in patients with sepsis: A randomized controlled trial. Crit. Care 2018, 22, 239. [Google Scholar] [CrossRef]
- Giannos, P.; Prokopidis, K. Gut dysbiosis and long COVID-19: Feeling gutted. J. Med. Virol. 2022, 94, 2917–2918. [Google Scholar] [CrossRef]
- Kerneis, S.; Lucet, J.C. Controlling the Diffusion of Multidrug-Resistant Organisms in Intensive Care Units. Semin. Respir. Crit. Care Med. 2019, 40, 558–568. [Google Scholar] [CrossRef]
- Russo, A.; Bassetti, M.; Bellelli, V.; Bianchi, L.; Marincola Cattaneo, F.; Mazzocchetti, S.; Paciacconi, E.; Cottini, F.; Schiattarella, A.; Tufaro, G.; et al. Efficacy of a Fosfomycin-Containing Regimen for Treatment of Severe Pneumonia Caused by Multidrug-Resistant Acinetobacter baumannii: A Prospective, Observational Study. Infect. Dis. Ther. 2021, 10, 187–200. [Google Scholar] [CrossRef]
- Russo, A.; Gavaruzzi, F.; Ceccarelli, G.; Borrazzo, C.; Oliva, A.; Alessandri, F.; Magnanimi, E.; Pugliese, F.; Venditti, M. Multidrug-resistant Acinetobacter baumannii infections in COVID-19 patients hospitalized in intensive care unit. Infection 2022, 50, 83–92. [Google Scholar] [CrossRef]
- Langford, B.J.; So, M.; Raybardhan, S.; Leung, V.; Soucy, J.R.; Westwood, D.; Daneman, N.; MacFadden, D.R. Antibiotic prescribing in patients with COVID-19: Rapid review and meta-analysis. Clin. Microbiol. Infect. 2021, 27, 520–531. [Google Scholar] [CrossRef] [PubMed]
- Pettit, N.N.; Nguyen, C.T.; Mutlu, G.M.; Wu, D.; Kimmig, L.; Pitrak, D.; Pursell, K. Late onset infectious complications and safety of tocilizumab in the management of COVID-19. J. Med. Virol. 2021, 93, 1459–1464. [Google Scholar] [CrossRef] [PubMed]
- Contou, D.; Claudinon, A.; Pajot, O.; Micaelo, M.; Longuet Flandre, P.; Dubert, M.; Cally, R.; Logre, E.; Fraisse, M.; Mentec, H.; et al. Bacterial and viral co-infections in patients with severe SARS-CoV-2 pneumonia admitted to a French ICU. Ann. Intensive Care 2020, 10, 119. [Google Scholar] [CrossRef] [PubMed]
- Ceccarelli, G.; Alessandri, F.; Moretti, S.; Borsetti, A.; Maggiorella, M.T.; Fabris, S.; Russo, A.; Ruberto, F.; De Meo, D.; Ciccozzi, M.; et al. Clinical Impact of Colonization with Carbapenem-Resistant Gram-Negative Bacteria in Critically Ill Patients Admitted for Severe Trauma. Pathogens 2022, 11, 1295. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Villodres, A.; Martin-Gandul, C.; Penalva, G.; Guisado-Gil, A.B.; Crespo-Rivas, J.C.; Pachon-Ibanez, M.E.; Lepe, J.A.; Cisneros, J.M. Prevalence and Risk Factors for Multidrug-Resistant Organisms Colonization in Long-Term Care Facilities Around the World: A Review. Antibiotics 2021, 10, 680. [Google Scholar] [CrossRef]
- Grasselli, G.; Scaravilli, V.; Mangioni, D.; Scudeller, L.; Alagna, L.; Bartoletti, M.; Bellani, G.; Biagioni, E.; Bonfanti, P.; Bottino, N.; et al. Hospital-Acquired Infections in Critically Ill Patients With COVID-19. Chest 2021, 160, 454–465. [Google Scholar] [CrossRef]
- Yeoh, Y.K.; Zuo, T.; Lui, G.C.; Zhang, F.; Liu, Q.; Li, A.Y.; Chung, A.C.; Cheung, C.P.; Tso, E.Y.; Fung, K.S.; et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut 2021, 70, 698–706. [Google Scholar] [CrossRef]
- Fernandez, P.; Moreno, L.; Yague, G.; Andreu, E.; Jara, R.; Segovia, M. Colonization by multidrug-resistant microorganisms in ICU patients during the COVID-19 pandemic. Med. Intensiv. Engl. Ed. 2021, 45, 313–315. [Google Scholar] [CrossRef]
- Venzon, M.; Bernard-Raichon, L.; Klein, J.; Axelrad, J.E.; Hussey, G.A.; Sullivan, A.P.; Casanovas-Massana, A.; Noval, M.G.; Valero-Jimenez, A.M.; Gago, J.; et al. Gut microbiome dysbiosis during COVID-19 is associated with increased risk for bacteremia and microbial translocation. Res. Sq. 2021, 13, 5926. [Google Scholar] [CrossRef]
- Liu, T.F.D.; Philippou, E.; Kolokotroni, O.; Siakallis, G.; Rahima, K.; Constantinou, C. Gut and airway microbiota and their role in COVID-19 infection and pathogenesis: A scoping review. Infection 2022, 50, 815–847. [Google Scholar] [CrossRef]
- Zuo, T.; Zhang, F.; Lui, G.C.Y.; Yeoh, Y.K.; Li, A.Y.L.; Zhan, H.; Wan, Y.; Chung, A.C.K.; Cheung, C.P.; Chen, N.; et al. Alterations in Gut Microbiota of Patients With COVID-19 During Time of Hospitalization. Gastroenterology 2020, 159, 944–955.e8. [Google Scholar] [CrossRef] [PubMed]
- Scaldaferri, F.; Ianiro, G.; Privitera, G.; Lopetuso, L.R.; Vetrone, L.M.; Petito, V.; Pugliese, D.; Neri, M.; Cammarota, G.; Ringel, Y.; et al. The Thrilling Journey of SARS-CoV-2 into the Intestine: From Pathogenesis to Future Clinical Implications. Inflamm. Bowel Dis. 2020, 26, 1306–1314. [Google Scholar] [CrossRef] [PubMed]
- Zuo, T.; Liu, Q.; Zhang, F.; Lui, G.C.; Tso, E.Y.; Yeoh, Y.K.; Chen, Z.; Boon, S.S.; Chan, F.K.; Chan, P.K.; et al. Depicting SARS-CoV-2 faecal viral activity in association with gut microbiota composition in patients with COVID-19. Gut 2021, 70, 276–284. [Google Scholar] [CrossRef] [PubMed]
- Murphy, E.C.; Frick, I.M. Gram-positive anaerobic cocci--commensals and opportunistic pathogens. FEMS Microbiol. Rev. 2013, 37, 520–553. [Google Scholar] [CrossRef] [PubMed]
- Bosak, J.; Micenkova, L.; Hrala, M.; Pomorska, K.; Kunova Bosakova, M.; Krejci, P.; Gopfert, E.; Faldyna, M.; Smajs, D. Colicin F(Y) inhibits pathogenic Yersinia enterocolitica in mice. Sci. Rep. 2018, 8, 12242. [Google Scholar] [CrossRef] [PubMed]
- Hanchi, H.; Mottawea, W.; Sebei, K.; Hammami, R. The Genus Enterococcus: Between Probiotic Potential and Safety Concerns-An Update. Front. Microbiol. 2018, 9, 1791. [Google Scholar] [CrossRef]
- Ryan, M.P.; Pembroke, J.T. The Genus Ochrobactrum as Major Opportunistic Pathogens. Microorganisms 2020, 8, 1797. [Google Scholar] [CrossRef]
- Yagel, Y.; Sestito, S.; Motro, Y.; Shnaiderman-Torban, A.; Khalfin, B.; Sagi, O.; Navon-Venezia, S.; Steinman, A.; Moran-Gilad, J. Genomic Characterization of Antimicrobial Resistance, Virulence, and Phylogeny of the Genus Ochrobactrum. Antibiotics 2020, 9, 177. [Google Scholar] [CrossRef]
- Alves, P.M.; Al-Badi, E.; Withycombe, C.; Jones, P.M.; Purdy, K.J.; Maddocks, S.E. Interaction between Staphylococcus aureus and Pseudomonas aeruginosa is beneficial for colonisation and pathogenicity in a mixed biofilm. Pathog. Dis. 2018, 76, fty003. [Google Scholar] [CrossRef]
- Guo, P.; Zhang, K.; Ma, X.; He, P. Clostridium species as probiotics: Potentials and challenges. J. Anim. Sci. Biotechnol. 2020, 11, 24. [Google Scholar] [CrossRef]
- Djukovic, A.; Garzon, M.J.; Canlet, C.; Cabral, V.; Lalaoui, R.; Garcia-Garcera, M.; Rechenberger, J.; Tremblay-Franco, M.; Penaranda, I.; Puchades-Carrasco, L.; et al. Lactobacillus supports Clostridiales to restrict gut colonization by multidrug-resistant Enterobacteriaceae. Nat. Commun. 2022, 13, 5617. [Google Scholar] [CrossRef] [PubMed]
- Bomar, L.; Brugger, S.D.; Yost, B.H.; Davies, S.S.; Lemon, K.P. Corynebacterium accolens Releases Antipneumococcal Free Fatty Acids from Human Nostril and Skin Surface Triacylglycerols. mBio 2016, 7, e01725-15. [Google Scholar] [CrossRef] [PubMed]
- Grice, E.A.; Kong, H.H.; Conlan, S.; Deming, C.B.; Davis, J.; Young, A.C.; Program, N.C.S.; Bouffard, G.G.; Blakesley, R.W.; Murray, P.R.; et al. Topographical and temporal diversity of the human skin microbiome. Science 2009, 324, 1190–1192. [Google Scholar] [CrossRef]
- Jumas-Bilak, E.; Carlier, J.P.; Jean-Pierre, H.; Citron, D.; Bernard, K.; Damay, A.; Gay, B.; Teyssier, C.; Campos, J.; Marchandin, H. Jonquetella anthropi gen. nov., sp. nov., the first member of the candidate phylum ‘Synergistetes’ isolated from man. Int. J. Syst. Evol. Microbiol. 2007, 57, 2743–2748. [Google Scholar] [CrossRef] [PubMed]
- Marchandin, H.; Damay, A.; Roudiere, L.; Teyssier, C.; Zorgniotti, I.; Dechaud, H.; Jean-Pierre, H.; Jumas-Bilak, E. Phylogeny, diversity and host specialization in the phylum Synergistetes with emphasis on strains and clones of human origin. Res. Microbiol. 2010, 161, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Badri, M.; Nilson, B.; Ragnarsson, S.; Senneby, E.; Rasmussen, M. Clinical and microbiological features of bacteraemia with Gram-positive anaerobic cocci: A population-based retrospective study. Clin. Microbiol. Infect. 2019, 25, 760.e1–760.e6. [Google Scholar] [CrossRef] [PubMed]
- Morio, F.; Jean-Pierre, H.; Dubreuil, L.; Jumas-Bilak, E.; Calvet, L.; Mercier, G.; Devine, R.; Marchandin, H. Antimicrobial susceptibilities and clinical sources of Dialister species. Antimicrob. Agents Chemother. 2007, 51, 4498–4501. [Google Scholar] [CrossRef]
- Larsen, J.M. The immune response to Prevotella bacteria in chronic inflammatory disease. Immunology 2017, 151, 363–374. [Google Scholar] [CrossRef]
- Khan, A.A.; Khan, Z. COVID-2019-associated overexpressed Prevotella proteins mediated host-pathogen interactions and their role in coronavirus outbreak. Bioinformatics 2020, 36, 4065–4069. [Google Scholar] [CrossRef]
- Todorov, S.D.; de Melo Franco, B.D.G.; Tagg, J.R. Bacteriocins of Gram-positive bacteria having activity spectra extending beyond closely-related species. Benef. Microbes 2019, 10, 315–328. [Google Scholar] [CrossRef]
- Romani, L.; Del Chierico, F.; Macari, G.; Pane, S.; Ristori, M.V.; Guarrasi, V.; Gardini, S.; Pascucci, G.R.; Cotugno, N.; Perno, C.F.; et al. The Relationship Between Pediatric Gut Microbiota and SARS-CoV-2 Infection. Front. Cell. Infect. Microbiol. 2022, 12, 908492. [Google Scholar] [CrossRef]
- Ferreira-Junior, A.S.; Borgonovi, T.F.; De Salis, L.V.V.; Leite, A.Z.; Dantas, A.S.; De Salis, G.V.V.; Cruz, G.N.F.; De Oliveira, L.F.V.; Gomes, E.; Penna, A.L.B.; et al. Detection of Intestinal Dysbiosis in Post-COVID-19 Patients One to Eight Months after Acute Disease Resolution. Int. J. Environ. Res. Public Health 2022, 19, 10189. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Forst, C.V.; Gordon, A.; Gussin, G.; Geber, A.B.; Fernandez, P.J.; Ding, T.; Lashua, L.; Wang, M.; Balmaseda, A.; et al. Characterization of antibiotic resistance and host-microbiome interactions in the human upper respiratory tract during influenza infection. Microbiome 2020, 8, 39. [Google Scholar] [CrossRef]
- Jing, R.; Yan, Y. Metagenomic analysis reveals antibiotic resistance genes in the bovine rumen. Microb. Pathog. 2020, 149, 104350. [Google Scholar] [CrossRef] [PubMed]
- Papadimitriou-Olivgeris, M.; Aretha, D.; Zotou, A.; Koutsileou, K.; Zbouki, A.; Lefkaditi, A.; Sklavou, C.; Marangos, M.; Fligou, F. The Role of Obesity in Sepsis Outcome among Critically Ill Patients: A Retrospective Cohort Analysis. Biomed. Res. Int. 2016, 2016, 5941279. [Google Scholar] [CrossRef] [PubMed]
- Bartoletti, M.; Giannella, M.; Scudeller, L.; Tedeschi, S.; Rinaldi, M.; Bussini, L.; Fornaro, G.; Pascale, R.; Pancaldi, L.; Pasquini, Z.; et al. Development and validation of a prediction model for severe respiratory failure in hospitalized patients with SARS-CoV-2 infection: A multicentre cohort study (PREDI-CO study). Clin. Microbiol. Infect. 2020, 26, 1545–1553. [Google Scholar] [CrossRef]
- Rodriguez-Nogales, A.; Algieri, F.; Garrido-Mesa, J.; Vezza, T.; Utrilla, M.P.; Chueca, N.; Garcia, F.; Olivares, M.; Rodriguez-Cabezas, M.E.; Galvez, J. Differential intestinal anti-inflammatory effects of Lactobacillus fermentum and Lactobacillus salivarius in DSS mouse colitis: Impact on microRNAs expression and microbiota composition. Mol. Nutr. Food Res. 2017, 61, 1700144. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef] [PubMed]
- Glockner, F.O.; Yilmaz, P.; Quast, C.; Gerken, J.; Beccati, A.; Ciuprina, A.; Bruns, G.; Yarza, P.; Peplies, J.; Westram, R.; et al. 25 years of serving the community with ribosomal RNA gene reference databases and tools. J. Biotechnol. 2017, 261, 169–176. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022; Available online: https://www.R-project.org/ (accessed on 1 July 2022).


| N = 17 | ||
|---|---|---|
| Demographics | Non-Colonized (n = 9) | Colonized (n = 8) |
| Age (median, IQR) | 64 (58–67) | 67 (65–69) |
| Gender (male) | 5 (55%) | 6 (75%) |
| Comorbidities | ||
| Obesity (Yes) | 2 (22%) | 3 (38%) |
| Pulmonar disease (Yes) | 1 (11%) | 3 (38%) |
| Diabetes (Yes) | 1 (11%) | 3 (38%) |
| Hypertension (Yes) | 3 (33%) | 4 (50%) |
| Days since symptoms appear (mean ± SD) | 7 ± 1 | 10 ± 4 |
| Days in ICU (mean ± SD) | 36 ± 23 | 31 ± 22 |
| Symptoms at hospitalization | ||
| Fever (Yes) | 2 (22%) | 5 (63%) |
| Cough (Yes) | 2 (22%) | 2 (25%) |
| Biochemistry parameters | ||
| LDH (U/L) (mean ± SD) | 389 ± 87 | 559 ± 152 |
| Lymphocytes (%) (median, IQR) | 7.8 (5.7–14.2) | 7.5 (6.6–8) |
| CRP (mg/dL) (median, IQR) | 87 (26.4–147.8) | 93.8 (76.2–152.3) |
| Severity index at hospitalization | ||
| SOFA (median, IQR) | 7 (6–8) | 7 (6–9) |
| APACHE (mean ± SD) | 15 ± 3 | 17 ± 4 |
| Care measures | ||
| Mechanical ventilation (Yes) | 9 (100%) | 8 (100%) |
| Ionotropic support (Yes) | 8 (90%) | 6 (75%) |
| Renal replacement therapy (TRR) (Yes) | 1 (11%) | 2 (25%) |
| Antibiotic administration (Yes) | 4 (44%) | 6 (75%) |
| Outcome | ||
| Mortality (Yes) | 1 (11%) | 3 (38%) |
| MDRB Colonizing Intestinal Tract | Colonized Patients (n = 8) |
|---|---|
| ESBL | |
| Escherichia coli | 1 (12 %) |
| Carbapenemase producer | |
| Enterobacter cloacae (OXA 48) | 2 (25 %) |
| Klebsiella pneumoniae (OXA 48) | 1 (12 %) |
| Serratia marcescens (OXA 48) | 1 (12 %) |
| Enterobacter cloacae (OXA 48/VIM) | 2 (25 %) |
| Non-fermenter GNB | |
| Pseudomonas aeruginosa | 1 (12 %) |
| Stenotrophomonas maltophila | 1 (12 %) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-García, J.; Diez-Echave, P.; Yuste, M.E.; Chueca, N.; García, F.; Cabeza-Barrera, J.; Fernández-Varón, E.; Gálvez, J.; Colmenero, M.; Rodríguez-Cabezas, M.E.; et al. Gut Microbiota Composition Can Predict Colonization by Multidrug-Resistant Bacteria in SARS-CoV-2 Patients in Intensive Care Unit: A Pilot Study. Antibiotics 2023, 12, 498. https://doi.org/10.3390/antibiotics12030498
García-García J, Diez-Echave P, Yuste ME, Chueca N, García F, Cabeza-Barrera J, Fernández-Varón E, Gálvez J, Colmenero M, Rodríguez-Cabezas ME, et al. Gut Microbiota Composition Can Predict Colonization by Multidrug-Resistant Bacteria in SARS-CoV-2 Patients in Intensive Care Unit: A Pilot Study. Antibiotics. 2023; 12(3):498. https://doi.org/10.3390/antibiotics12030498
Chicago/Turabian StyleGarcía-García, Jorge, Patricia Diez-Echave, María Eugenia Yuste, Natalia Chueca, Federico García, Jose Cabeza-Barrera, Emilio Fernández-Varón, Julio Gálvez, Manuel Colmenero, Maria Elena Rodríguez-Cabezas, and et al. 2023. "Gut Microbiota Composition Can Predict Colonization by Multidrug-Resistant Bacteria in SARS-CoV-2 Patients in Intensive Care Unit: A Pilot Study" Antibiotics 12, no. 3: 498. https://doi.org/10.3390/antibiotics12030498
APA StyleGarcía-García, J., Diez-Echave, P., Yuste, M. E., Chueca, N., García, F., Cabeza-Barrera, J., Fernández-Varón, E., Gálvez, J., Colmenero, M., Rodríguez-Cabezas, M. E., Rodríguez-Nogales, A., & Morón, R. (2023). Gut Microbiota Composition Can Predict Colonization by Multidrug-Resistant Bacteria in SARS-CoV-2 Patients in Intensive Care Unit: A Pilot Study. Antibiotics, 12(3), 498. https://doi.org/10.3390/antibiotics12030498








