Worldwide Dissemination of blaKPC Gene by Novel Mobilization Platforms in Pseudomonas aeruginosa: A Systematic Review
Abstract
1. Introduction
2. Results
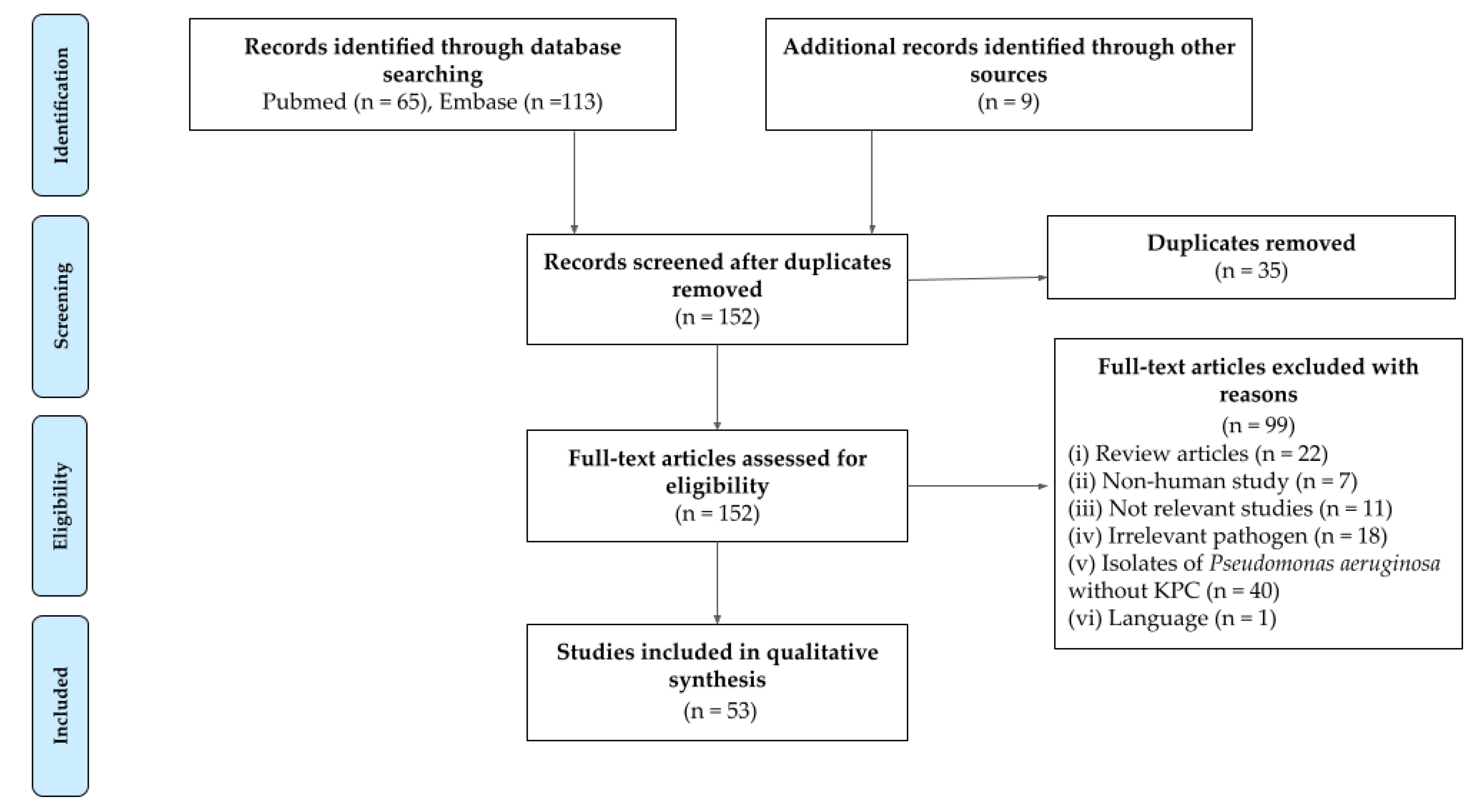
2.1. Search Results
2.2. Geographical Distribution and Genetic Relationship of blaKPC-Harboring P. aeruginosa Isolates
| First Author | Year | Continent | Country | Collection Date 1 | Isolates (n = 704) 2 | KPC Variant 3 | Sequence Types | Ref |
|---|---|---|---|---|---|---|---|---|
| Villegas | 2007 | South America | Colombia | 2006 | 3 (0.4) | KPC (2/3), KPC-2 (1/3) | NS (3/3) | [22] |
| Naas * | 2008 | NS | NS | NS | 1 (0.1) | KPC-2 (1/1) | NS (1/1) | [68] |
| Wolter | 2009 | Middle America | Puerto Rico | 2006–2007 | 25 (3.5) | KPC (18/25), KPC-2 (6/25), KPC-5 (1/25) | NS (25/25) | [55] |
| Wolter * | 2009 | Middle America | Puerto Rico | 2006 | 2 (0.2) | KPC-2 (1/2), KPC-5 (1/2) | NS (2/2) | [56] |
| Akpaka | 2009 | South America | Trinidad and Tobago | NS | 1 (0.1) | KPC-2 (1/1) | NS (1/1) | [64] |
| Poirel * | 2010 | North America | USA | 2009 | 1 (0.1) | KPC-2 (1/1) | NS (1/1) | [67] |
| Ge * | 2011 | Asia | China | 2009 | 3 (0.4) | KPC-2 (3/3) | ST463 (3/3) | [32] |
| Cuzon * | 2011 | South America | Colombia | 2006–2010 | 10 (1.4) | KPC-2 (10/10) | ST308 (6/10), ST235 (2/10), ST1006 (1/10), ST1060 (1/10) | [26] |
| Robledo | 2011 | Middle America | Puerto Rico | 2009 | 89 (12.6) | KPC (89/89) | NS (89/89) | [57] |
| Martínez * | 2012 | Middle America | Puerto Rico | 2009 | 1 (0.1) | KPC-2 (1/1) | NS (1/1) | [58] |
| Jácome | 2012 | South America | Brazil | 2010 | 2 (0.2) | KPC-2 (2/2) | NS (2/2) | [42] |
| Pasteran * | 2012 | South America | Argentina | 2006–2011 | 30 (4.2) | KPC-2 (30/30) | ST654 (29/30), ST162 (1/30) | [61] |
| Correa * | 2012 | South America | Colombia | 2010 | 1 (0.1) | KPC-2 (1/1) | ST111 (1/1) | [51] |
| Roth * | 2013 | NS | NS | NS | 1 (0.1) | KPC-2 (1/1) | NS (1/1) | [69] |
| Naas * | 2013 | South America | Colombia | NS | 2 (0.2) | KPC-2 (2/2) | ST308 (1/2), ST1006 (1/2) | [27] |
| Buelvas | 2013 | South America | Colombia | 2008 | 1 (0.1) | KPC-2 (1/1) | NS (1/1) | [52] |
| Vanegas | 2014 | South America | Colombia | 2012–2014 | 25 (3.5) | KPC-2 (25/25) | ST1801 (7/25), ST235 (5/25), ST362 (3/25), ST111 (1/25), ST1803 (1/25), NS (8/25) | [53] |
| Cavalcanti | 2015 | South America | Brazil | 2008–2010 | 3 (0.4) | KPC-2 (3/3) | ST235 (2/3), ST244 (1/3) | [43] |
| Hu * | 2015 | Asia | China | 2013 | 39 (5.5) | KPC-2 (39/39) | ST463 (31/39), ST1076 (2/39), ST1755 (1/39), ST850 (1/39), ST357 (1/39), ST836 (1/31), ST209 (1/39), ST244 (1/39) | [33] |
| Paul * | 2015 | Asia | India | 2012–2013 | 2 (0.2) | KPC-2 (2/2) | NS (2/2) | [66] |
| Dai * | 2016 | Asia | China | 2013 | 1 (0.1) | KPC-2 (1/1) | NS (1/1) | [17] |
| Kazmierczak | 2016 | America/Asia | Global data | 2012–2014 | 29 (4.1) | KPC-2 (29/29) | NS (29/29) | [71] |
| Galetti * | 2016 | South America | Brazil | 2011 | 1 (0.1) | KPC-2 (1/1) | ST244 (1/1) | [44] |
| Hagemann * | 2018 | Europe | Germany | NS | 1 (0.1) | KPC-2 (1/1) | ST235 (1/1) | [18] |
| de Oliveira Santos * | 2018 | South America | Brazil | 2014 | 1 (0.1) | KPC-2 (1/1) | ST2584 (1/1) | [21] |
| Shi * | 2018 | Asia | China | 2016 | 1 (0.1) | KPC-2 (1/1) | NS (1/1) | [34] |
| de Paula-Petroli | 2018 | South America | Brazil | 2008 | 1 (0.1) | KPC-2 (1/1) | ST235 (1/1) | [45] |
| Galetti * | 2019 | South America | Brazil | 2011 | 1 (0.1) | KPC-2 (1/1) | ST381 (1/1) | [46] |
| Hu * | 2019 | Asia | China | 2010 | 1 (0.1) | KPC-2 (1/1) | ST463 (1/1) | [35] |
| Pacheco | 2019 | South America | Colombia | 2017 | 5 (0.7) | KPC-2 (5/5) | NS (5/5) | [54] |
| Abril * | 2019 | South America | Colombia | 2014–2016 | 4 (0.5) | KPC-2 (4/4) | ST235 (4/4) | [29] |
| Li * | 2020 | Asia | China | 2018 | 21 (2.9) | KPC-2 (21/21) | ST664 (21/21) | [19] |
| Pérez-Vázquez | 2020 | Europe | Spain | 2016 | 2 (0.2) | KPC-2 (2/2) | ST244 (2/2) | [6] |
| Tartari * | 2021 | South America | Brazil | 2018 | 1 (0.1) | KPC-2 (1/1) | ST312 (1/1) | [49] |
| Cai * | 2021 | Asia | China | 2019 | 4 (0.5) | KPC-2 (4/4) | ST463 (4/4) | [14] |
| Wozniak * | 2021 | South America | Chile | 2015 | 2 (0.2) | KPC-2 (2/2) | ST654 (2/2) | [60] |
| Rada * | 2021 | South America | Colombia | 2013–2015 | 12 (1.7) | KPC-2 (12/12) | ST308 (2/12), ST699 (2/12), ST309 (1/12), ST313 (1/12), ST3512 (1/12), NS (5/12) | [28] |
| Hu * | 2021 | Asia | China | 2007–2018 | 105 (14.9) | KPC-2 (105/105) | ST463 (71/105), ST1212 (13/105), ST1076 (10/105), ST9 (1/105), ST209 (1/105), ST244 (1/1015), ST274 (1/105), ST277 (1/105), ST360 (1/105), ST377 (1/105), ST836 (1/105), ST1642 (1/105) ST2235 (1/105), NS (1/105) | [15] |
| Tran | 2021 | Asia | Vietnam | 2011–2013 | 7 (0.9) | KPC-2 (7/7) | ST3151 (7/7) | [63] |
| Souza | 2021 | South America | Brazil | 2015–2016 | 3 (0.4) | KPC-2 (3/3) | NS (3/3) | [48] |
| Costa-Júnior | 2021 | South America | Brazil | 2018–2019 | 11 (1.5) | KPC (11/11) | NS (11/11) | [47] |
| Hu * | 2021 | Asia | China | 2014–2019 | 16 (2.2) | KPC-2 (16/16) | ST463 (7/16), ST1076 (3/16), ST1212 (3/16), ST633 (2/16), NS (1/16) | [37] |
| Yuan * | 2021 | Asia | China | 2015 | 1 (0.1) | KPC-2 (1/1) | NS (1/1) | [39] |
| Zhu * | 2021 | Asia | China | 2017–2018 | 151 (21.4) | KPC-2 (151/151) | ST463 (107/151), ST485 (14/151), ST1212 (12/151), ST244 (7/151), ST234 (2/151), ST1076 (2/151), ST606 (1/151), ST1631 (1/151), ST3217 (1/151), NS (4/151) | [40] |
| Hu * | 2021 | Asia | China | 2019–2020 | 24 (3.4) | KPC-2 (23/24), KPC-33 (1/24) | ST463 (23/24), ST1076 (1/24) | [36] |
| Costa | 2021 | South America | Chile | 2015–2018 | 19 (2.7) | KPC-2 (19/19) | NS (19/19) | [59] |
| Wang * | 2021 | Asia | China | 2017 | 1 (0.1) | KPC-2 (1/1) | NS (1/1) | [38] |
| Cardinal | 2021 | South America | Global data | 2017–2019 | 24 (3.4) | KPC-2 (24/24) | NS (24/24) | [72] |
| Takahashi * | 2021 | Asia | Nepal | 2018–2020 | 4 (0.5) | KPC-2 (4/4) | ST235 (4/4) | [65] |
| Cejas * | 2022 | South America | Argentina | 2008 and 2018 | 2 (0.2) | KPC-2 (2/2) | ST654 (1/2), ST235 (1/2) | [62] |
| Tu * | 2022 | Asia | China | 2021 | 1 (0.1) | KPC-90 (1/1) | ST463 (1/1) | [41] |
| Li * | 2022 | NS | NS | NS | 2 (0.2) | KPC-2 (2/2) | ST463 (2/2) | [70] |
| Silveira * | 2022 | South America | Brazil | 2020 | 3 (0.4) | KPC-2 (3/3) | ST277 (3/3) | [50] |
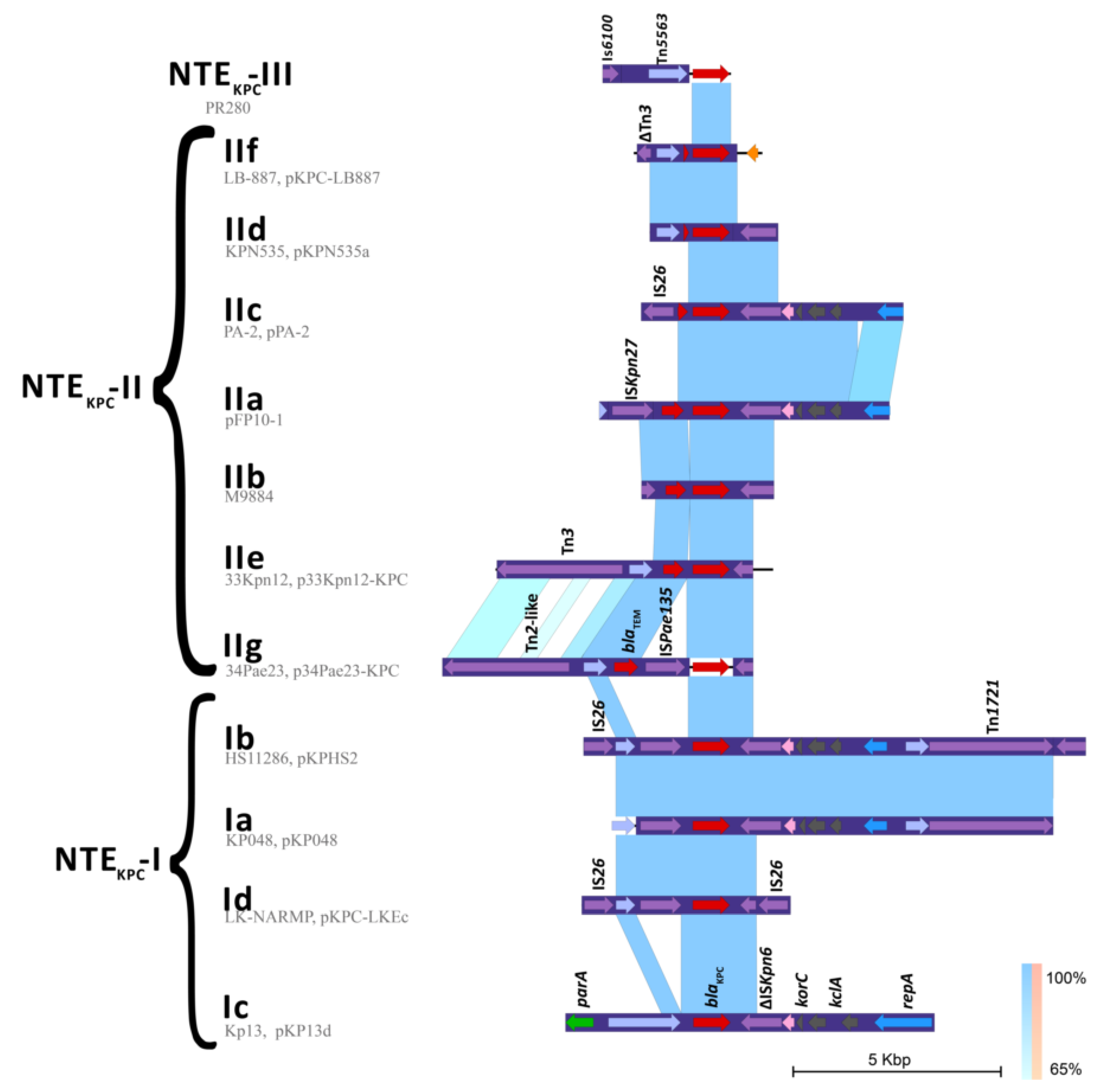
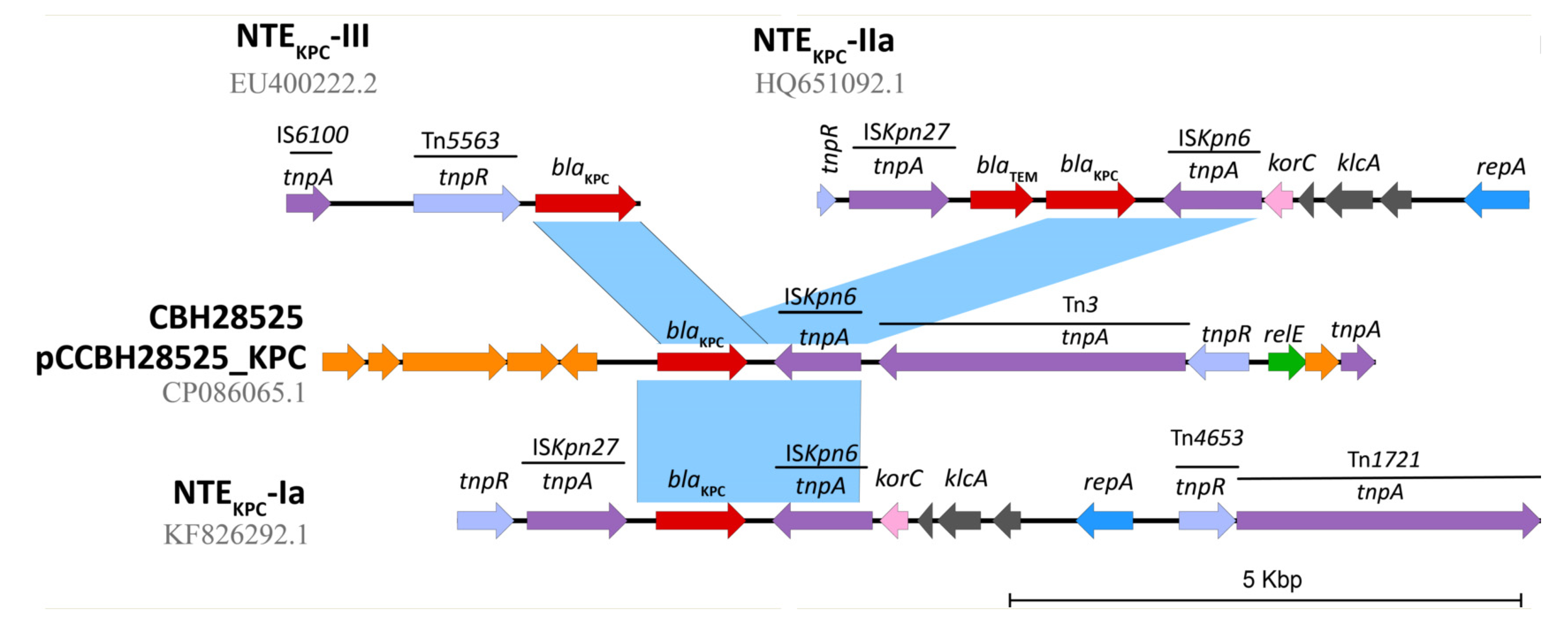
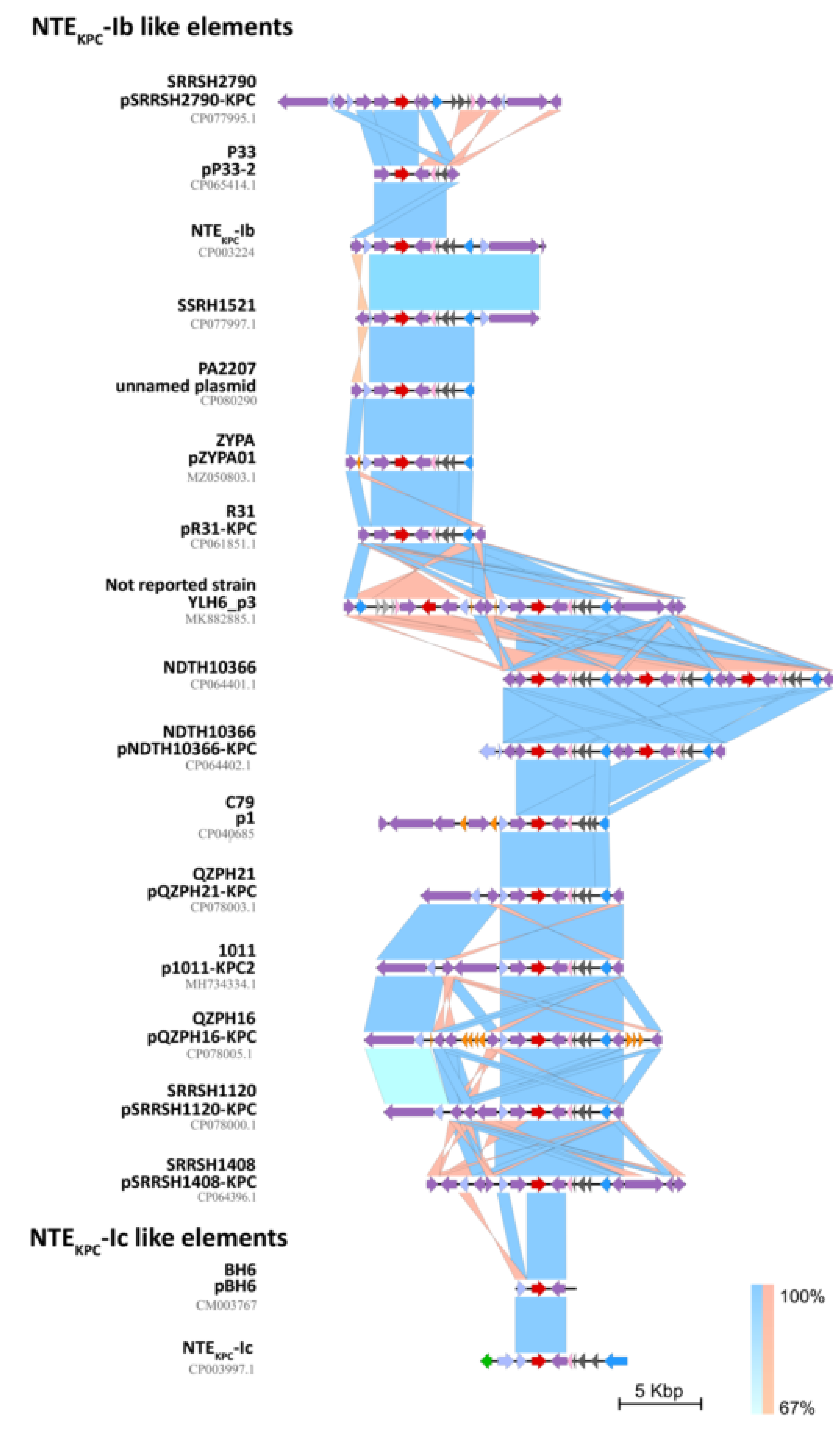
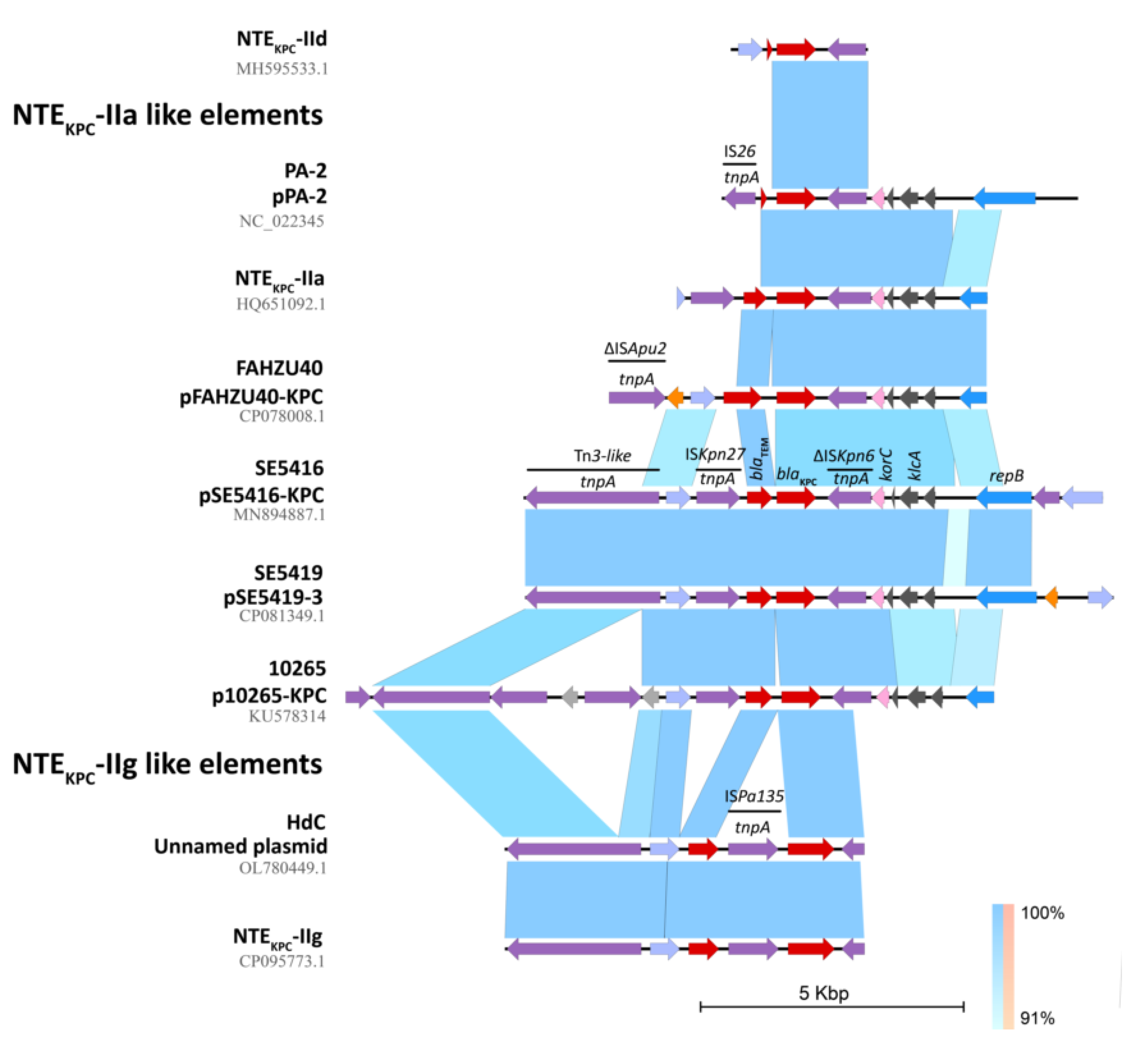
2.3. Genetic Platforms Mobilizing blaKPC Gene in P. aeruginosa
2.4. In Silico Assessment of the blaKPC Genetic Environment on P. aeruginosa Isolates
2.5. Interactive Online Map Construction
3. Discussion
4. Materials and Methods
4.1. Search Strategy
4.2. Inclusion and Exclusion Criteria
4.3. Data Extraction and Analysis
4.4. Exploration of the blaKPC Genetic Environment for P. aeruginosa in GenBank
4.5. Participation of Patients in the Study
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Peleg, A.Y.; Hooper, D.C. Hospital-acquired infections due to gram-negative bacteria. N. Engl. J. Med. 2010, 362, 1804–1813. [Google Scholar] [CrossRef] [PubMed]
- Friedrich, A.W. Control of hospital acquired infections and antimicrobial resistance in Europe: The way to go. Wien. Med. Wochenschr. 2019, 169, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Voidazan, S.; Albu, S.; Toth, R.; Grigorescu, B.; Rachita, A.; Moldovan, I. Healthcare associated infections—A new pathology in medical practice? Int. J. Environ. Res. Public Health 2020, 17, 760. [Google Scholar] [CrossRef]
- Szabó, S.; Feier, B.; Capatina, D.; Tertis, M.; Cristea, C.; Popa, A. An overview of healthcare associated infections and their detection methods caused by pathogen bacteria in Romania and Europe. J. Clin. Med. 2022, 11, 3204. [Google Scholar] [CrossRef] [PubMed]
- HAI and Antibiotic Use Prevalence Survey. Available online: https://www.cdc.gov/hai/eip/antibiotic-use.html (accessed on 15 December 2022).
- Perez-Vazquez, M.; Sola-Campoy, P.J.; Zurita, Á.M.; Avila, A.; Gomez-Bertomeu, F.; Solis, S.; Lopez-Urrutia, L.; GÓnzalez-BarberÁ, E.M.; Cercenado, E.; Bautista, V. Carbapenemase-producing Pseudomonas aeruginosa in Spain: Interregional dissemination of the high-risk clones ST175 and ST244 carrying blaVIM-2, blaVIM-1, blaIMP-8, blaVIM-20 and blaKPC-2. Int. J. Antimicrob. Agents 2020, 56, 106026. [Google Scholar] [CrossRef]
- Antibiotic Resistance Threats in the United States. 2019. Available online: https://stacks.cdc.gov/view/cdc/82532 (accessed on 10 December 2022).
- Talebi Bezmin Abadi, A.; Rizvanov, A.A.; Haertlé, T.; Blatt, N.L. World Health Organization report: Current crisis of antibiotic resistance. BioNanoScience 2019, 9, 778–788. [Google Scholar] [CrossRef]
- WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed. Available online: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 12 January 2023).
- COVID-19: U.S. Impact on Antimicrobial Resistance, Special Report 2022. Available online: https://www.cdc.gov/drugresistance/pdf/covid19-impact-report-508.pdf (accessed on 10 January 2023).
- Vigilancia por WHONET de Resistencia Antimicrobiana en el Ámbito Hospitalario, Colombia 2021. Available online: https://www.ins.gov.co/buscador-eventos/Informacin%20de%20laboratorio/Forms/AllItems.aspx (accessed on 27 December 2022).
- Lynch, J.P.; Zhanel, G.G.; Clark, N.M. Emergence of antimicrobial resistance among Pseudomonas aeruginosa: Implications for therapy. Semin. Respir. Crit. Care Med. 2017, 38, 326–345. [Google Scholar]
- Yoon, E.J.; Jeong, S.H. Mobile Carbapenemase Genes in Pseudomonas aeruginosa. Front. Microbiol. 2021, 12, 614058. [Google Scholar] [CrossRef]
- Cai, H.; Zhu, Y.; Hu, D.; Li, Y.; Leptihn, S.; Loh, B.; Hua, X.; Yu, Y. Co-harboring of Novel blaKPC–2 Plasmid and Integrative and Conjugative Element Carrying Tn6203 in Multidrug-Resistant Pseudomonas aeruginosa. Front. Microbiol. 2021, 12, 674974. [Google Scholar] [CrossRef]
- Hu, Y.; Liu, C.; Wang, Q.; Zeng, Y.; Sun, Q.; Shu, L.; Lu, J.; Cai, J.; Wang, S.; Zhang, R.; et al. Emergence and expansion of a carbapenem-resistant Pseudomonas aeruginosa clone are associated with plasmid-borne bla KPC-2 and virulence-related genes. mSystems 2021, 6, e00154-21. [Google Scholar] [CrossRef]
- Boucher, H.W.; Talbot, G.H.; Bradley, J.S.; Edwards, J.E.; Gilbert, D.; Rice, L.B.; Scheld, M.; Spellberg, B.; Bartlett, J. Bad bugs, no drugs: No ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009, 48, 1–12. [Google Scholar] [CrossRef]
- Dai, X.; Zhou, D.; Xiong, W.; Feng, J.; Luo, W.; Luo, G.; Wang, H.; Sun, F.; Zhou, X. The IncP-6 plasmid p10265-KPC from Pseudomonas aeruginosa carries a novel ΔISEc33-associated blaKPC-2 gene cluster. Front. Microbiol. 2016, 7, 310. [Google Scholar] [CrossRef] [PubMed]
- Hagemann, J.B.; Pfennigwerth, N.; Gatermann, S.G.; von Baum, H.; Essig, A. KPC-2 carbapenemase-producing Pseudomonas aeruginosa reaching Germany. J. Antimicrob. Chemother. 2018, 73, 1812–1814. [Google Scholar] [CrossRef]
- Li, Z.; Cai, Z.; Cai, Z.; Zhang, Y.; Fu, T.; Jin, Y.; Cheng, Z.; Jin, S.; Wu, W.; Yang, L. Molecular genetic analysis of an XDR Pseudomonas aeruginosa ST664 clone carrying multiple conjugal plasmids. J. Antimicrob. Chemother. 2020, 75, 1443–1452. [Google Scholar] [CrossRef]
- Yigit, H.; Queenan, A.M.; Anderson, G.J.; Domenech-Sanchez, A.; Biddle, J.W.; Steward, C.D.; Alberti, S.; Bush, K.; Tenover, F.C. Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2001, 45, 1151–1161. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira Santos, I.C.; Albano, R.M.; Asensi, M.D.; Carvalho-Assef, A.P.D.A. Draft genome sequence of KPC-2-producing Pseudomonas aeruginosa recovered from a bloodstream infection sample in Brazil. J. Glob. Antimicrob. Resist. 2018, 15, 99–100. [Google Scholar] [CrossRef] [PubMed]
- Villegas, M.V.; Lolans, K.; Correa, A.; Kattan, J.N.; Lopez, J.A.; Quinn, J.P. First identification of Pseudomonas aeruginosa isolates producing a KPC-type carbapenem-hydrolyzing β-lactamase. Antimicrob. Agents Chemother. 2007, 51, 1553–1555. [Google Scholar] [CrossRef] [PubMed]
- Cuzon, G.; Naas, T.; Nordmann, P. Functional characterization of Tn 4401, a Tn 3-based transposon involved in bla KPC gene mobilization. Antimicrob. Agents Chemother. 2011, 55, 5370–5373. [Google Scholar] [CrossRef]
- Chen, L.; Mathema, B.; Chavda, K.D.; DeLeo, F.R.; Bonomo, R.A.; Kreiswirth, B.N. Carbapenemase-producing Klebsiella pneumoniae: Molecular and genetic decoding. Trends Microbiol. 2014, 22, 686–696. [Google Scholar] [CrossRef]
- Tang, Y.; Li, G.; Shen, P.; Zhang, Y.; Jiang, X. Replicative transposition contributes to the evolution and dissemination of KPC-2-producing plasmid in Enterobacterales. Emerg. Microbes Infect. 2022, 11, 113–122. [Google Scholar] [CrossRef]
- Cuzon, G.; Naas, T.; Villegas, M.-V.; Correa, A.; Quinn, J.P.; Nordmann, P. Wide dissemination of Pseudomonas aeruginosa producing β-lactamase bla KPC-2 gene in Colombia. Antimicrob. Agents Chemother. 2011, 55, 5350–5353. [Google Scholar] [CrossRef]
- Naas, T.; Bonnin, R.A.; Cuzon, G.; Villegas, M.-V.; Nordmann, P. Complete sequence of two KPC-harbouring plasmids from Pseudomonas aeruginosa. J. Antimicrob. Chemother. 2013, 68, 1757–1762. [Google Scholar] [CrossRef]
- Rada, A.M.; De La Cadena, E.; Agudelo, C.A.; Pallares, C.; Restrepo, E.; Correa, A.; Villegas, M.V.; Capataz, C. Genetic Diversity of Multidrug-Resistant Pseudomonas aeruginosa Isolates Carrying blaVIM–2 and blaKPC–2 Genes That Spread on Different Genetic Environment in Colombia. Front. Microbiol. 2021, 12, 663020. [Google Scholar] [CrossRef] [PubMed]
- Abril, D.; Marquez-Ortiz, R.A.; Castro-Cardozo, B.; Moncayo-Ortiz, J.I.; Olarte Escobar, N.M.; Corredor Rozo, Z.L.; Reyes, N.; Tovar, C.; Sánchez, H.F.; Castellanos, J. Genome plasticity favours double chromosomal Tn4401b-blaKPC-2 transposon insertion in the Pseudomonas aeruginosa ST235 clone. BMC Microbiol. 2019, 19, 45. [Google Scholar] [CrossRef]
- Chen, S.; Larsson, M.; Robinson, R.C.; Chen, S.L. Direct and convenient measurement of plasmid stability in lab and clinical isolates of E. coli. Sci. Rep. 2017, 7, 4788. [Google Scholar] [CrossRef]
- de Lima, G.J.; Scavuzzi, A.M.L.; Beltrão, E.M.B.; Firmo, E.F.; de Oliveira, É.M.; de Oliveira, S.R.; de Rezende, A.M.; Lopes, A.C.S. Identification of plasmid IncQ1 and NTE KPC-IId harboring bla KPC-2 in isolates from Klebsiella pneumoniae infections in patients from Recife-PE, Brazil. Rev. Soc. Bras. Med. Trop. 2020, 53, e20190526. [Google Scholar] [CrossRef]
- Ge, C.; Wei, Z.; Jiang, Y.; Shen, P.; Yu, Y.; Li, L. Identification of KPC-2-producing Pseudomonas aeruginosa isolates in China. J. Antimicrob. Chemother. 2011, 66, 1184–1186. [Google Scholar] [CrossRef]
- Hu, Y.-Y.; Gu, D.-X.; Cai, J.-C.; Zhou, H.-W.; Zhang, R. Emergence of KPC-2-producing Pseudomonas aeruginosa sequence type 463 isolates in Hangzhou, China. Antimicrob. Agents Chemother. 2015, 59, 2914–2917. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Liang, Q.; Feng, J.; Zhan, Z.; Zhao, Y.; Yang, W.; Yang, H.; Chen, Y.; Huang, M.; Tong, Y. Coexistence of two novel resistance plasmids, bla KPC-2-carrying p14057A and tetA (A)-carrying p14057B, in Pseudomonas aeruginosa. Virulence 2018, 9, 306–311. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.-Y.; Wang, Q.; Sun, Q.-L.; Chen, G.-X.; Zhang, R. A novel plasmid carrying carbapenem-resistant gene blaKPC-2 in Pseudomonas aeruginosa. Infect. Drug Resist. 2019, 12, 1285. [Google Scholar] [CrossRef]
- Hu, H.; Zhang, Y.; Zhang, P.; Wang, J.; Yuan, Q.; Shi, W.; Zhang, S.; Feng, H.; Chen, Y.; Yu, M. Bloodstream Infections Caused by Klebsiella pneumoniae Carbapenemase–Producing P. aeruginosa Sequence Type 463, Associated with High Mortality Rates in China: A Retrospective Cohort Study. Front. Cell. Infect. Microbiol. 2021, 11, 756782. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Qing, Y.; Chen, J.; Liu, C.; Lu, J.; Wang, Q.; Zhen, S.; Zhou, H.; Huang, L.; Zhang, R. Prevalence, Risk Factors, and Molecular Epidemiology of Intestinal Carbapenem-Resistant Pseudomonas aeruginosa. Microbiol. Spectr. 2021, 9, e01344-21. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.-J.; Chen, E.-Z.; Yang, L.; Feng, D.-H.; Xu, Z.; Chen, D.-Q. Emergence of clinical Pseudomonas aeruginosa isolate Guangzhou-PaeC79 carrying crpP, bla GES-5, and bla KPC-2 in Guangzhou of China. Microb. Drug Resist. 2021, 27, 965–970. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Guan, H.; Sha, D.; Cao, W.; Song, X.; Che, J.; Kan, B.; Li, J. Characterization of bla KPC-2-Carrying Plasmid pR31-KPC from a Pseudomonas aeruginosa Strain Isolated in China. Antibiotics 2021, 10, 1234. [Google Scholar] [CrossRef]
- Zhu, Y.; Chen, J.; Shen, H.; Chen, Z.; Yang, Q.-w.; Zhu, J.; Li, X.; Yang, Q.; Zhao, F.; Ji, J. Emergence of ceftazidime-and avibactam-resistant Klebsiella pneumoniae carbapenemase-producing Pseudomonas aeruginosa in China. mSystems 2021, 6, e00787-21. [Google Scholar] [CrossRef]
- Tu, Y.; Wang, D.; Zhu, Y.; Li, J.; Jiang, Y.; Wu, W.; Li, X.; Zhou, H. Emergence of a KPC-90 Variant that Confers Resistance to Ceftazidime-Avibactam in an ST463 Carbapenem-Resistant Pseudomonas aeruginosa Strain. Microbiol. Spectr. 2022, 10, e01869-21. [Google Scholar] [CrossRef]
- Jácome, P.R.L.d.A.; Alves, L.R.; Cabral, A.B.; Lopes, A.C.S.; Maciel, M.A.V. First report of KPC-producing Pseudomonas aeruginosa in Brazil. Antimicrob. Agents Chemother. 2012, 56, 4990. [Google Scholar] [CrossRef]
- Cavalcanti, F.L.d.S.; Mirones, C.R.; Paucar, E.R.; Montes, L.Á.; Leal-Balbino, T.C.; Morais, M.M.C.d.; Martínez-Martínez, L.; Ocampo-Sosa, A.A. Mutational and acquired carbapenem resistance mechanisms in multidrug resistant Pseudomonas aeruginosa clinical isolates from Recife, Brazil. Memórias Inst. Oswaldo Cruz 2015, 110, 1003–1009. [Google Scholar] [CrossRef]
- Galetti, R.; Andrade, L.N.; Chandler, M.; Varani, A.d.M.; Darini, A.L.C. New small plasmid harboring bla KPC-2 in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2016, 60, 3211–3214. [Google Scholar] [CrossRef]
- de Paula-Petroli, S.B.; Campana, E.H.; Bocchi, M.; Bordinhão, T.; Picão, R.C.; Yamada-Ogatta, S.F.; Carrara-Marroni, F.E. Early detection of a hypervirulent KPC-2-producing Pseudomonas aeruginosa ST235 in Brazil. J. Glob. Antimicrob. Resist. 2018, 12, 153–154. [Google Scholar] [CrossRef]
- Galetti, R.; Andrade, L.N.; Varani, A.M.; Darini, A.L.C. A phage-like plasmid carrying blaKPC-2 gene in carbapenem-resistant Pseudomonas aeruginosa. Front. Microbiol. 2019, 10, 572. [Google Scholar] [CrossRef]
- Costa-Júnior, S.D.; da Silva, A.M.C.M.; Niedja da Paz Pereira, J.; da Costa Lima, J.L.; Cavalcanti, I.M.F.; Maciel, M.A.V. Emergence of rmtD1 gene in clinical isolates of Pseudomonas aeruginosa carrying blaKPC and/or blaVIM-2 genes in Brazil. Braz. J. Microbiol. 2021, 52, 1959–1965. [Google Scholar] [CrossRef] [PubMed]
- Souza, G.H.A.; Rossato, L.; Brito, G.T.; Bet, G.; Simionatto, S. Carbapenem-resistant Pseudomonas aeruginosa strains: A worrying health problem in intensive care units. Rev. Inst. Med. Trop. São Paulo 2021, 63, e71. [Google Scholar] [CrossRef]
- Tartari, D.C.; Zamparette, C.P.; Martini, G.; Christakis, S.; Costa, L.H.; de Oliveira Silveira, A.C.; Sincero, T.C.M. Genomic analysis of an extensively drug-resistant Pseudomonas aeruginosa ST312 harbouring IncU plasmid-mediated blaKPC-2 isolated from ascitic fluid. J. Glob. Antimicrob. Resist. 2021, 25, 151–153. [Google Scholar] [CrossRef]
- Silveira, M.C.; Albano, R.M.; Rocha-de-Souza, C.M.; Leão, R.S.; Marques, E.A.; Picão, R.C.; Kraychete, G.B.; de Oliveira Santos, I.C.; e Oliveira, T.R.T.; Tavares-Teixeira, C.B. Description of a novel IncP plasmid harboring blaKPC-2 recovered from a SPM-1-producing Pseudomonas aeruginosa from ST277. Infect. Genet. Evol. 2022, 102, 105302. [Google Scholar] [CrossRef] [PubMed]
- Correa, A.; Montealegre, M.C.; Mojica, M.F.; Maya, J.J.; Rojas, L.J.; De La Cadena, E.P.; Ruiz, S.J.; Recalde, M.; Rosso, F.; Quinn, J.P. First report of a Pseudomonas aeruginosa isolate coharboring KPC and VIM carbapenemases. Antimicrob. Agents Chemother. 2012, 56, 5422–5423. [Google Scholar] [CrossRef] [PubMed]
- Buelvas Doria, F.A.; Díaz Osorio, M.Á.; Muñoz Delgado, Á.B.; Tovar Acero, C. Clinical Isolation of KPC-2-Producing Pseudomonas aeruginosa in the City of Montería, Córdoba, Colombia. Infectio 2013, 17, 35–38. [Google Scholar] [CrossRef]
- Vanegas, J.M.; Cienfuegos, A.V.; Ocampo, A.M.; López, L.; del Corral, H.; Roncancio, G.; Sierra, P.; Echeverri-Toro, L.; Ospina, S.; Maldonado, N. Similar frequencies of Pseudomonas aeruginosa isolates producing KPC and VIM carbapenemases in diverse genetic clones at tertiary-care hospitals in Medellín, Colombia. J. Clin. Microbiol. 2014, 52, 3978–3986. [Google Scholar] [CrossRef]
- Pacheco, T.; Bustos-Cruz, R.H.; Abril, D.; Arias, S.; Uribe, L.; Rincón, J.; García, J.-C.; Escobar-Perez, J. Pseudomonas aeruginosa coharboring blaKPC-2 and blaVIM-2 carbapenemase genes. Antibiotics 2019, 8, 98. [Google Scholar] [CrossRef]
- Wolter, D.J.; Khalaf, N.; Robledo, I.E.; Vázquez, G.J.; Santé, M.I.; Aquino, E.E.; Goering, R.V.; Hanson, N.D. Surveillance of carbapenem-resistant Pseudomonas aeruginosa isolates from Puerto Rican Medical Center Hospitals: Dissemination of KPC and IMP-18 β-lactamases. Antimicrob. Agents Chemother. 2009, 53, 1660–1664. [Google Scholar] [CrossRef] [PubMed]
- Wolter, D.J.; Kurpiel, P.M.; Woodford, N.; Palepou, M.-F.I.; Goering, R.V.; Hanson, N.D. Phenotypic and enzymatic comparative analysis of the novel KPC variant KPC-5 and its evolutionary variants, KPC-2 and KPC-4. Antimicrob. Agents Chemother. 2009, 53, 557–562. [Google Scholar] [CrossRef] [PubMed]
- Robledo, I.E.; Aquino, E.E.; Vázquez, G.J. Detection of the KPC gene in Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter baumannii during a PCR-based nosocomial surveillance study in Puerto Rico. Antimicrob. Agents Chemother. 2011, 55, 2968–2970. [Google Scholar] [CrossRef]
- Martínez, T.; Vázquez, G.J.; Aquino, E.E.; Ramírez-Ronda, R.; Robledo, I.E. First report of a Pseudomonas aeruginosa clinical isolate co-harbouring KPC-2 and IMP-18 carbapenemases. Int. J. Antimicrob. Agents 2012, 39, 542–543. [Google Scholar] [CrossRef] [PubMed]
- Costa, J.; Lima, C.A.; Vera-Leiva, A.; San Martin Magdalena, I.; Bello-Toledo, H.; Opazo-Capurro, A.; Quezada-Aguiluz, M.; González-Rocha, G. Carbapenemases produced by Carbapenem-resistant Pseudomonas aeruginosa isolated from hospitals in Chile. Rev. Chil. Infectol. Organo Soc. Chil. nfectol. 2021, 38, 81–87. [Google Scholar] [CrossRef]
- Wozniak, A.; Figueroa, C.; Moya-Flores, F.; Guggiana, P.; Castillo, C.; Rivas, L.; Munita, J.M.; Garcia, P.C. A multispecies outbreak of carbapenem-resistant bacteria harboring the blaKPC gene in a non-classical transposon element. BMC Microbiol. 2021, 21, 107. [Google Scholar] [CrossRef]
- Pasteran, F.; Faccone, D.; Gomez, S.; De Bunder, S.; Spinelli, F.; Rapoport, M.; Petroni, A.; Galas, M.; Corso, A. Detection of an international multiresistant clone belonging to sequence type 654 involved in the dissemination of KPC-producing Pseudomonas aeruginosa in Argentina. J. Antimicrob. Chemother. 2012, 67, 1291–1293. [Google Scholar] [CrossRef]
- Cejas, D.; Elena, A.; González-Espinosa, F.E.; Pallecchi, L.; Vay, C.; Rossolini, G.M.; Gutkind, G.; Di Pilato, V.; Radice, M. Characterisation of blaKPC-2–harbouring plasmids recovered from Pseudomonas aeruginosa ST654 and ST235 high-risk clones. J. Glob. Antimicrob. Resist. 2022, 29, 310–312. [Google Scholar] [CrossRef] [PubMed]
- Tran, H.A.; Vu, T.N.B.; Trinh, S.T.; Tran, D.L.; Pham, H.M.; Ngo, T.H.H.; Nguyen, M.T.; Tran, N.D.; Pham, D.T.; Dang, D.A. Resistance mechanisms and genetic relatedness among carbapenem-resistant Pseudomonas aeruginosa isolates from three major hospitals in Hanoi, Vietnam (2011–15). JAC-Antimicrob. Resist. 2021, 3, dlab103. [Google Scholar] [CrossRef]
- Akpaka, P.E.; Swanston, W.H.; Ihemere, H.N.; Correa, A.; Torres, J.A.; Tafur, J.D.; Montealegre, M.C.; Quinn, J.P.; Villegas, M.V. Emergence of KPC-producing Pseudomonas aeruginosa in Trinidad and Tobago. J. Clin. Microbiol. 2009, 47, 2670–2671. [Google Scholar] [CrossRef]
- Takahashi, T.; Tada, T.; Shrestha, S.; Hishinuma, T.; Sherchan, J.B.; Tohya, M.; Kirikae, T.; Sherchand, J.B. Molecular characterisation of carbapenem-resistant Pseudomonas aeruginosa clinical isolates in Nepal. J. Glob. Antimicrob. Resist. 2021, 26, 279–284. [Google Scholar] [CrossRef]
- Paul, D.; Dhar Chanda, D.; Maurya, A.P.; Mishra, S.; Chakravarty, A.; Sharma, G.D.; Bhattacharjee, A. Co-Carriage of bla KPC-2 and bla NDM-1 in Clinical Isolates of Pseudomonas aeruginosa Associated with Hospital Infections from India. PLoS ONE 2015, 10, e0145823. [Google Scholar] [CrossRef]
- Poirel, L.; Nordmann, P.; Lagrutta, E.; Cleary, T.; Munoz-Price, L.S. Emergence of KPC-producing Pseudomonas aeruginosa in the United States. Antimicrob. Agents Chemother. 2010, 54, 3072. [Google Scholar] [CrossRef]
- Naas, T.; Cuzon, G.; Villegas, M.-V.; Lartigue, M.-F.; Quinn, J.P.; Nordmann, P. Genetic structures at the origin of acquisition of the β-lactamase bla KPC gene. Antimicrob. Agents Chemother. 2008, 52, 1257–1263. [Google Scholar] [CrossRef] [PubMed]
- Roth, A.L.; Lister, P.D.; Hanson, N.D. Effect of drug treatment options on the mobility and expression of bla KPC. J. Antimicrob. Chemother. 2013, 68, 2779–2785. [Google Scholar] [CrossRef]
- Li, Y.; Zhu, Y.; Zhou, W.; Chen, Z.; Moran, R.A.; Ke, H.; Feng, Y.; van Schaik, W.; Shen, H.; Ji, J. Alcaligenes faecalis metallo-β-lactamase in extensively drug-resistant Pseudomonas aeruginosa isolates. Clin. Microbiol. Infect. 2022, 28, 880.e1–880.e8. [Google Scholar] [CrossRef]
- Kazmierczak, K.M.; Biedenbach, D.J.; Hackel, M.; Rabine, S.; de Jonge, B.L.M.; Bouchillon, S.K.; Sahm, D.F.; Bradford, P.A. Global dissemination of bla KPC into bacterial species beyond Klebsiella pneumoniae and in vitro susceptibility to ceftazidime-avibactam and aztreonam-avibactam. Antimicrob. Agents Chemother. 2016, 60, 4490–4500. [Google Scholar] [CrossRef] [PubMed]
- Cardinal, L.; Marcelino, C.P.; Okuma, A.; Mizuno, G.; Tuon, F.; Gales, A.C.; Gales, A.C.; Negra, M.D.; Polis, T.; Beirao, E. 1217. Molecular Epidemiology of Pseudomonas aeruginosa in Latin America: Clinical Isolates From Respiratory Tract Infection. Open Forum Infect. Dis. 2021, 8 (Suppl. 1), 697–698. [Google Scholar] [CrossRef]
- del Barrio-Tofiño, E.; López-Causapé, C.; Oliver, A. Pseudomonas aeruginosa epidemic high-risk clones and their association with horizontally-acquired β-lactamases: 2020 update. Int. J. Antimicrob. Agents 2020, 56, 106196. [Google Scholar] [CrossRef]
- Campana, E.H.; Kraychete, G.B.; Montezzi, L.F.; Xavier, D.E.; Picão, R.C. Description of a new non-Tn4401 element (NTEKPC-IIe) harboured on IncQ plasmid in Citrobacter werkmanii from recreational coastal water. J. Glob. Antimicrob. Resist. 2022, 29, 207–211. [Google Scholar] [CrossRef]
- Antimicrobial Testing Leadership and Surveillance. Available online: https://atlas-surveillance.com/ (accessed on 22 December 2022).
- Álvarez-Otero, J.; Lamas-Ferreiro, J.; González-González, L.; Rodríguez-Code, I.; Fernández-Soneira, M.; Arca-Blanco, A.; Bermúdez-Sanjuro, J.; de la Fuente-Aguado, J. Resistencia a carbapenemas en Pseudomonas aeruginosa aisladas en urocultivos: Prevalencia y factores de riesgo. Rev. Esp. Quim. 2017, 30, 195–200. [Google Scholar]
- Venter, H. Reversing resistance to counter antimicrobial resistance in the World Health Organisation’s critical priority of most dangerous pathogens. Biosci. Rep. 2019, 39, BSR20180474. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, C.; Bavlovič, J.; Machado, E.; Amorim, J.; Peixe, L.; Novais, Â. KPC-3-producing Klebsiella pneumoniae in Portugal linked to previously circulating non-CG258 lineages and uncommon genetic platforms (Tn 4401d-IncFIA and Tn 4401d-IncN). Front. Microbiol. 2016, 7, 1000. [Google Scholar] [CrossRef]
- Faccone, D.; de Mendieta, J.M.; Albornoz, E.; Chavez, M.; Genero, F.; Echegorry, M.; Ceriana, P.; Mora, A.; Seah, C.; Corso, A. Emergence of KPC-31, a KPC-3 Variant Associated with Ceftazidime-Avibactam Resistance, in an Extensively Drug-Resistant ST235 Pseudomonas aeruginosa Clinical Isolate. Antimicrob. Agents Chemother. 2022, 66, e00648-22. [Google Scholar] [CrossRef]
- Feil, E.J.; Li, B.C.; Aanensen, D.M.; Hanage, W.P.; Spratt, B.G. eBURST: Inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 2004, 186, 1518–1530. [Google Scholar] [CrossRef] [PubMed]
- Roy Chowdhury, P.; Scott, M.; Worden, P.; Huntington, P.; Hudson, B.; Karagiannis, T.; Charles, I.G.; Djordjevic, S.P. Genomic islands 1 and 2 play key roles in the evolution of extensively drug-resistant ST235 isolates of Pseudomonas aeruginosa. Open Biol. 2016, 6, 150175. [Google Scholar] [CrossRef] [PubMed]
- Rada, A.M.; De La Cadena, E.; Agudelo, C.; Capataz, C.; Orozco, N.; Pallares, C.; Dinh, A.Q.; Panesso, D.; Ríos, R.; Diaz, L. Dynamics of bla KPC-2 dissemination from non-CG258 Klebsiella pneumoniae to other Enterobacterales via IncN plasmids in an area of high endemicity. Antimicrob. Agents Chemother. 2020, 64, e01743-20. [Google Scholar] [CrossRef]
- Sheppard, A.E.; Stoesser, N.; Wilson, D.J.; Sebra, R.; Kasarskis, A.; Anson, L.W.; Giess, A.; Pankhurst, L.J.; Vaughan, A.; Grim, C.J. Nested Russian doll-like genetic mobility drives rapid dissemination of the carbapenem resistance gene bla KPC. Antimicrob. Agents Chemother. 2016, 60, 3767–3778. [Google Scholar] [CrossRef]
- He, S.; Hickman, A.B.; Varani, A.M.; Siguier, P.; Chandler, M.; Dekker, J.P.; Dyda, F. Insertion sequence IS 26 reorganizes plasmids in clinically isolated multidrug-resistant bacteria by replicative transposition. mBio 2015, 6, e00762. [Google Scholar] [CrossRef]
- Abril, D.; Vergara, E.; Palacios, D.; Leal, A.L.; Marquez-Ortiz, R.A.; Madroñero, J.; Corredor Rozo, Z.L.; De La Rosa, Z.; Nieto, C.A.; Vanegas, N. Within patient genetic diversity of bla KPC harboring Klebsiella pneumoniae in a Colombian hospital and identification of a new NTEKPC platform. Sci. Rep. 2021, 11, 21409. [Google Scholar] [CrossRef]
- Grant, M.J.; Booth, A. A typology of reviews: An analysis of 14 review types and associated methodologies. Health Inf. Libr. J. 2009, 26, 91–108. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST. org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef] [PubMed]
- Carver, T.J.; Rutherford, K.M.; Berriman, M.; Rajandream, M.A.; Barrell, B.G.; Parkhill, J. ACT: The Artemis Comparison Tool. Bioinformatics 2005, 21, 3422–3423. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef]
- Tansirichaiya, S.; Rahman, M.A.; Roberts, A.P. The Transposon Registry. Mob. DNA 2019, 10, 40. [Google Scholar] [CrossRef]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef]


| First Author | Year | Country | Strain | KPC Variant | ST | Plasmid Name | Length (bp) | Inc Group | Access Number | Ref |
|---|---|---|---|---|---|---|---|---|---|---|
| Naas | 2013 | Colombia | COL-1 | KPC-2 | ST308 | pCOL-1 | 31,529 | IncP-6 | KC609323 | [27] |
| Naas | 2013 | Colombia | PA-2 | KPC-2 | ST1006 | pPA-2 | 7995 | IncU | KC609322 | [27] |
| Dai | 2016 | China | 10265 | KPC-2 | NS | p10265-KPC | 38,939 | IncP-6 | KU578314 | [17] |
| Galetti | 2016 | Brazil | BH6 | KPC-2 | ST244 | pBH6 | 3652 | UI | LGVH01000782.1 | [44] |
| Shi | 2018 | China | 14057 | KPC-2 | NS | p14057A | 51,663 | UI | KY296095 | [34] |
| Galetti | 2019 | Brazil | BH9 | KPC-2 | ST381 | pBH6::Phage BH9 | 41,024 | UI | CP029714 | [46] |
| Hu | 2019 | China | PA1011 | KPC-2 | ST463 | pPA1011 | 62.793 | UI | MH734334 | [35] |
| Li | 2020 | China | NK546 | KPC-2 | ST664 | pNK546a | 475,027 | IncP-3-like (IncA/C) | MN433457 | [19] |
| Wang | 2021 | China | Guangzhou-PaeC79 | KPC-2 | NS | pPAEC79 | 40,180 | IncP-6 | CP040685.1 | [38] |
| Tartari | 2021 | Brazil | MIMA_PA2.1 | KPC-2 | ST312 | pMIMA_PA2.1 | 7975 | IncU | MT683857 | [49] |
| Cai | 2021 | China | P23 | KPC-2 | ST463 | pP23-KPC | 40,937 | UI | CP065418 | [14] |
| Cai | 2021 | China | P33 | KPC-2 | ST463 | pP33-2 | 48,306 | UI | CP065414.1 | [14] |
| Wozniak | 2021 | Chile | Pae-13 | KPC-2 | ST654 | pPae-13 | 35,034 | UI | MT949191 | [60] |
| Yuan | 2021 | China | R31 | KPC-2 | NS | pR31-KPC | 29,402 | UI | CP061851 | [39] |
| Zhu | 2021 | China | FAHZU31 | KPC-2 | ST244 | pFAHZU31-KPC | 24,350 | UI | CP078010 | [40] |
| Zhu | 2021 | China | FAHZU40 | KPC-2 | ST234 | pFAHZU40-KPC | 28,700 | UI | CP078008 | [40] |
| Zhu | 2021 | China | QZPH41 | KPC-2 | NS | pQZPH41-KPC | 88,210 | UI | CP064400 | [40] |
| Zhu | 2021 | China | WTJH12 | KPC-2 | ST485 | pWTJH12-KPC | 396,963 | UI | CP064404 | [40] |
| Zhu | 2021 | China | ZPPH1 | KPC-2 | ST1212 | pZPPH1-KPC | 52,415 | UI | CP077990 | [40] |
| Cejas | 2022 | Argentina | PA_2047 | KPC-2 | ST654 | pPA_2047 | 43,660 | UI | MN082782 | [62] |
| Cejas | 2022 | Argentina | PA_HdC | KPC-2 | ST235 | pPA_HdC | 42,750 | UI | OL780449 | [62] |
| Tu | 2022 | China | PA2207 | KPC-90 | ST463 | pPA2207_2 | 41,938 | UI | CP080290 | [41] |
| Li | 2022 | NS | NDTH10366 | KPC-2 | ST463 | pNDTH10366-KPC | 392,244 | UI | CP064402 | [70] |
| Silveira | 2022 | Brazil | CCBH28525 | KPC-2 | ST277 | pCCBH28525 | 60,312 | IncP | CP086065 | [50] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Forero-Hurtado, D.; Corredor-Rozo, Z.L.; Ruiz-Castellanos, J.S.; Márquez-Ortiz, R.A.; Abril, D.; Vanegas, N.; Lafaurie, G.I.; Chambrone, L.; Escobar-Pérez, J. Worldwide Dissemination of blaKPC Gene by Novel Mobilization Platforms in Pseudomonas aeruginosa: A Systematic Review. Antibiotics 2023, 12, 658. https://doi.org/10.3390/antibiotics12040658
Forero-Hurtado D, Corredor-Rozo ZL, Ruiz-Castellanos JS, Márquez-Ortiz RA, Abril D, Vanegas N, Lafaurie GI, Chambrone L, Escobar-Pérez J. Worldwide Dissemination of blaKPC Gene by Novel Mobilization Platforms in Pseudomonas aeruginosa: A Systematic Review. Antibiotics. 2023; 12(4):658. https://doi.org/10.3390/antibiotics12040658
Chicago/Turabian StyleForero-Hurtado, Daniela, Zayda Lorena Corredor-Rozo, Julián Santiago Ruiz-Castellanos, Ricaurte Alejandro Márquez-Ortiz, Deisy Abril, Natasha Vanegas, Gloria Inés Lafaurie, Leandro Chambrone, and Javier Escobar-Pérez. 2023. "Worldwide Dissemination of blaKPC Gene by Novel Mobilization Platforms in Pseudomonas aeruginosa: A Systematic Review" Antibiotics 12, no. 4: 658. https://doi.org/10.3390/antibiotics12040658
APA StyleForero-Hurtado, D., Corredor-Rozo, Z. L., Ruiz-Castellanos, J. S., Márquez-Ortiz, R. A., Abril, D., Vanegas, N., Lafaurie, G. I., Chambrone, L., & Escobar-Pérez, J. (2023). Worldwide Dissemination of blaKPC Gene by Novel Mobilization Platforms in Pseudomonas aeruginosa: A Systematic Review. Antibiotics, 12(4), 658. https://doi.org/10.3390/antibiotics12040658








