Characteristics of tet(X4)−Producing Escherichia coli in Chicken and Pig Farms in Hunan Province, China
Abstract
1. Introduction
2. Results and Discussion
2.1. Bacteria Isolates
2.2. Antimicrobial Susceptibility Testing and Resistance Genes
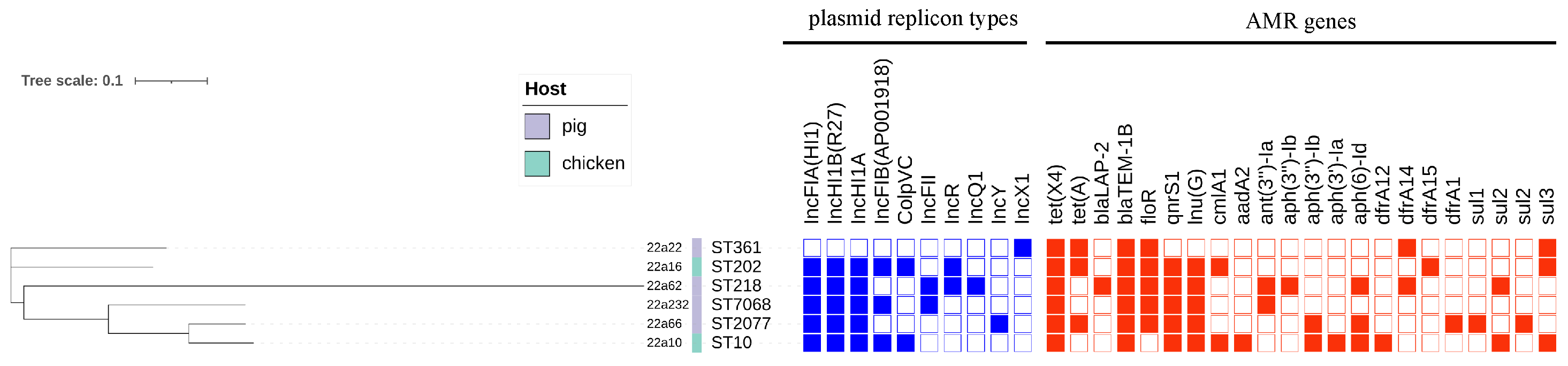
2.3. Multilocus Sequence Typing and Plasmid Replicons
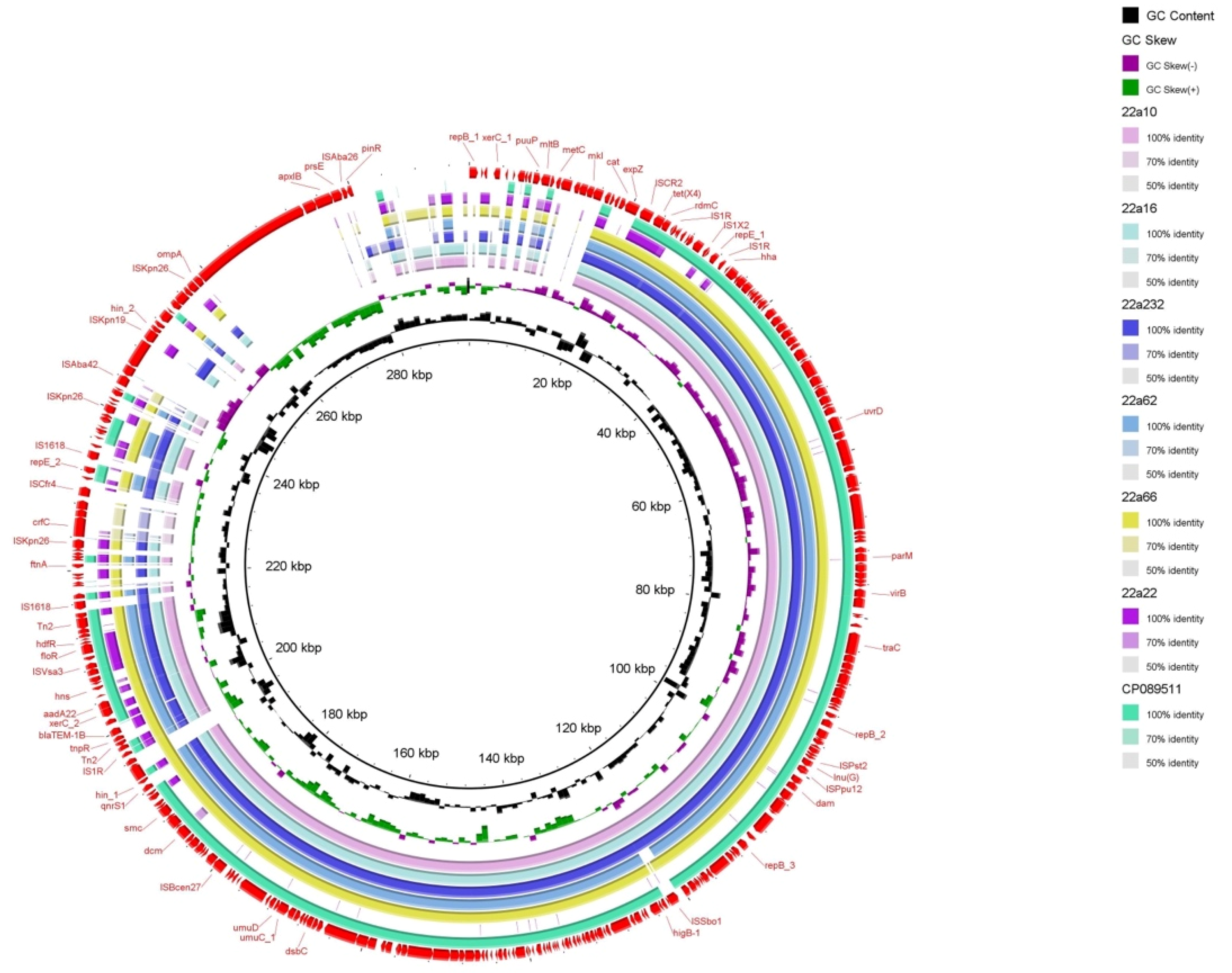
2.4. Genetic Contexts and Conjugation
3. Materials and Methods
3.1. Sample Collection and tet(X) Detection
3.2. Antimicrobial Susceptibility Testing
3.3. Whole−Genome Sequencing and Analysis
3.4. Conjugation Experiment
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, S.; Wen, J.; Wang, Y.; Wang, M.; Jia, R.; Chen, S.; Liu, M.; Zhu, D.; Zhao, X.; Wu, Y.; et al. Dissemination and prevalence of plasmid−mediated high−level tigecycline resistance gene tet (X4). Front. Microbiol. 2022, 13, 969769. [Google Scholar] [CrossRef] [PubMed]
- He, T.; Wang, R.; Liu, D.; Walsh, T.R.; Zhang, R.; Lv, Y.; Ke, Y.; Ji, Q.; Wei, R.; Liu, Z.; et al. Emergence of plasmid−mediated high−level tigecycline resistance genes in animals and humans. Nat. Microbiol. 2019, 4, 1450–1456. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Chen, C.; Cui, C.Y.; Zhang, Y.; Liu, X.; Cui, Z.H.; Ma, X.Y.; Feng, Y.; Fang, L.X.; Lian, X.L.; et al. Plasmid−encoded tet(X) genes that confer high−level tigecycline resistance in Escherichia coli. Nat. Microbiol. 2019, 4, 1457–1464. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, D.; Lv, Y.; Cui, L.; Li, Y.; Li, T.; Song, H.; Hao, Y.; Shen, J.; Wang, Y.; et al. Novel Plasmid−Mediated tet(X5) Gene Conferring Resistance to Tigecycline, Eravacycline, and Omadacycline in a Clinical Acinetobacter baumannii Isolate. Antimicrob. Agents Chemother. 2019, 64, e01326-19. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.Y.; Liu, Y.; Chen, Y.; Huang, F.M.; Chen, R.C.; Xiao, Y.H.; Zhou, K. Sporadic Dissemination of tet(X3) and tet(X6) Mediated by Highly Diverse Plasmidomes among Livestock−Associated Acinetobacter. Microbiol. Spectr. 2021, 9, e0114121. [Google Scholar] [CrossRef]
- Soliman, A.M.; Ramadan, H.; Zarad, H.; Sugawara, Y.; Yu, L.; Sugai, M.; Shimamoto, T.; Hiott, L.M.; Frye, J.G.; Jackson, C.R.; et al. Coproduction of Tet(X7) Conferring High−Level Tigecycline Resistance, Fosfomycin FosA4, and Colistin Mcr−1.1 in Escherichia coli Strains from Chickens in Egypt. Antimicrob. Agents Chemother. 2021, 65, e02084-20. [Google Scholar] [CrossRef]
- Gasparrini, A.J.; Markley, J.L.; Kumar, H.; Wang, B.; Fang, L.; Irum, S.; Symister, C.T.; Wallace, M.; Burnham, C.D.; Andleeb, S.; et al. Tetracycline−inactivating enzymes from environmental, human commensal, and pathogenic bacteria cause broad−spectrum tetracycline resistance. Commun. Biol. 2020, 3, 241. [Google Scholar] [CrossRef]
- Cheng, Y.; Chen, Y.; Liu, Y.; Guo, Y.; Zhou, Y.; Xiao, T.; Zhang, S.; Xu, H.; Chen, Y.; Shan, T.; et al. Identification of novel tetracycline resistance gene tet(X14) and its co−occurrence with tet(X2) in a tigecycline−resistant and colistin−resistant Empedobacter stercoris. Emerg. Microbes. Infect. 2020, 9, 1843–1852. [Google Scholar] [CrossRef]
- Li, R.; Peng, K.; Xiao, X.; Wang, Y.; Wang, Z. Characterization of novel ISAba1−bounded tet(X15)−bearing composite transposon Tn6866 in Acinetobacter variabilis. J. Antimicrob. Chemother. 2021, 76, 2481–2483. [Google Scholar] [CrossRef]
- Ma, J.; Zhou, W.; Wu, J.; Liu, X.; Lin, J.; Ji, X.; Lin, H.; Wang, J.; Jiang, H.; Zhou, Q.; et al. Large−Scale Studies on Antimicrobial Resistance and Molecular Characterization of Escherichia coli from Food Animals in Developed Areas of Eastern China. Microbiol. Spectr. 2022, 10, e0201522. [Google Scholar] [CrossRef]
- Li, W.; Yang, Z.; Hu, J.; Wang, B.; Rong, H.; Li, Z.; Sun, Y.; Wang, Y.; Zhang, X.; Wang, M.; et al. Evaluation of culturable 'last−resort' antibiotic resistant pathogens in hospital wastewater and implications on the risks of nosocomial antimicrobial resistance prevalence. J. Hazard. Mater. 2022, 438, 129477. [Google Scholar] [CrossRef]
- Zeng, Y.; Deng, L.; Zhou, X.; Zhang, C.; Hu, Z.; Chen, Y.; Zheng, W. Prevalence and risk factors of tet(X4)−positive Enterobacteriaceae in human gut microbiota. J. Glob. Antimicrob. Resist. 2022, 31, 15–21. [Google Scholar] [CrossRef]
- Dong, N.; Zeng, Y.; Cai, C.; Sun, C.; Lu, J.; Liu, C.; Zhou, H.; Sun, Q.; Shu, L.; Wang, H.; et al. Prevalence, transmission, and molecular epidemiology of tet(X)−positive bacteria among humans, animals, and environmental niches in China: An epidemiological, and genomic−based study. Sci. Total Environ. 2022, 818, 151767. [Google Scholar] [CrossRef]
- Dao, T.D.; Kasuga, I.; Hirabayashi, A.; Nguyen, D.T.; Tran, H.T.; Vu, H.; Pham, L.T.N.; Vu, T.M.H.; Hasebe, F.; Nguyen, H.T.; et al. Emergence of mobile tigecycline resistance gene tet(X4)−harbouring Shewanella xiamenensis in a water environment. J. Glob. Antimicrob. Resist. 2022, 28, 140–142. [Google Scholar] [CrossRef]
- Li, R.; Li, Y.; Peng, K.; Yin, Y.; Liu, Y.; He, T.; Bai, L.; Wang, Z. Comprehensive Genomic Investigation of Tigecycline Resistance Gene tet(X4)−Bearing Strains Expanding among Different Settings. Microbiol. Spectr. 2021, 9, e01633-21. [Google Scholar] [CrossRef]
- Liu, D.; Wang, T.; Shao, D.; Song, H.; Zhai, W.; Sun, C.; Zhang, Y.; Zhang, M.; Fu, Y.; Zhang, R.; et al. Structural diversity of the ISCR2−mediated rolling−cycle transferable unit carrying tet(X4). Sci. Total Environ. 2022, 826, 154010. [Google Scholar] [CrossRef]
- Li, Y.; Wang, Q.; Peng, K.; Liu, Y.; Xiao, X.; Mohsin, M.; Li, R.; Wang, Z. Distribution and genomic characterization of tigecycline−resistant tet(X4)−positive Escherichia coli of swine farm origin. Microb. Genom. 2021, 7, 000667. [Google Scholar] [CrossRef]
- Wang, J.; Lu, M.J.; Wang, Z.Y.; Jiang, Y.; Wu, H.; Pan, Z.M.; Jiao, X. Tigecycline−resistant Escherichia coli ST761 carrying tet(X4) in a pig farm, China. Front. Microbiol. 2022, 13, 967313. [Google Scholar] [CrossRef]
- Zhai, W.; Wang, T.; Yang, D.; Zhang, Q.; Liang, X.; Liu, Z.; Sun, C.; Wu, C.; Liu, D.; Wang, Y. Clonal relationship of tet(X4)−positive Escherichia coli ST761 isolates between animals and humans. J. Antimicrob. Chemother. 2022, 77, 2153–2157. [Google Scholar] [CrossRef]
- Zhai, W.; Tian, Y.; Shao, D.; Zhang, M.; Li, J.; Song, H.; Sun, C.; Wang, Y.; Liu, D.; Zhang, Y. Fecal Carriage of Escherichia coli Harboring the tet(X4)−IncX1 Plasmid from a Tertiary Class−A Hospital in Beijing, China. Antibiotics 2022, 11, 1068. [Google Scholar] [CrossRef]
- Du, P.; Liu, D.; Song, H.; Zhang, P.; Li, R.; Fu, Y.; Liu, X.; Jia, J.; Li, X.; Fanning, S.; et al. Novel IS26−mediated hybrid plasmid harbouring tet(X4) in Escherichia coli. J. Glob. Antimicrob. Resist. 2020, 21, 162–168. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhan, Z.; Shi, C. International Spread of Tet(X4)−Producing Escherichia coli Isolates. Foods 2022, 11, 2010. [Google Scholar] [CrossRef] [PubMed]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin. Microbiol. Rev. 2018, 31, e00088-17. [Google Scholar] [CrossRef] [PubMed]
- Bailey, J.K.; Pinyon, J.L.; Anantham, S.; Hall, R.M. Distribution of the blaTEM gene and blaTEM−containing transposons in commensal Escherichia coli. J. Antimicrob. Chemother. 2011, 66, 745–751. [Google Scholar] [CrossRef]
- Hu, J.; Yang, J.; Chen, W.; Liu, Z.; Zhao, Q.; Yang, H.; Sun, Z.; Chen, X.; Li, J. Prevalence and Characteristics of mcr−1−Producing Escherichia coli in Three Kinds of Poultry in Changsha, China. Front. Microbiol. 2022, 13, 840520. [Google Scholar] [CrossRef]
- Sun, C.; Cui, M.; Zhang, S.; Liu, D.; Fu, B.; Li, Z.; Bai, R.; Wang, Y.; Wang, H.; Song, L.; et al. Genomic epidemiology of animal−derived tigecycline−resistant Escherichia coli across China reveals recent endemic plasmid−encoded tet(X4) gene. Commun. Biol. 2020, 3, 412. [Google Scholar] [CrossRef]
- Srivastava, S.; Singh, V.; Kumar, V.; Verma, P.C.; Srivastava, R.; Basu, V.; Gupta, V.; Rawat, A.K. Identification of regulatory elements in 16S rRNA gene of Acinetobacter species isolated from water sample. Bioinformation 2008, 3, 173–176. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; CLSI Supplement M100; CLSI: Wayne, PA, USA, 2020. [Google Scholar]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single−cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]

| Strain | Antimicrobial Agents (mg/L) a | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| TGC | AMP | AMK | CPL | CTX | NAL | FFC | COL | MEM | GEN | STX | |
| 22a10 | 4 | >256 | 0.5 | 256 | 0.06 | >256 | 128 | 0.25 | 0.06 | >128 | >16 |
| 22a16 | 8 | >256 | 0.5 | 128 | 0.06 | 32 | 128 | 0.125 | 0.5 | >128 | >16 |
| 22a22 | 8 | >256 | 0.5 | 256 | 0.06 | >256 | 128 | 0.125 | 0.06 | 64 | >16 |
| 22a62 | 4 | >256 | 0.5 | 128 | 0.125 | >256 | 64 | 0.125 | 0.03 | >128 | >16 |
| 22a66 | 8 | 256 | 0.5 | 128 | 0.06 | 64 | 128 | 0.125 | 0.06 | >128 | >16 |
| 22a232 | 4 | >256 | 0.5 | 256 | 0.125 | 8 | 128 | 0.125 | 0.06 | >128 | >16 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, J.; Xiao, G.; Xiao, N.; Jiang, Z.; Jiang, C.; Li, Y.; Chen, W.; Lin, H.; Sun, Z.; Li, J. Characteristics of tet(X4)−Producing Escherichia coli in Chicken and Pig Farms in Hunan Province, China. Antibiotics 2023, 12, 147. https://doi.org/10.3390/antibiotics12010147
Yang J, Xiao G, Xiao N, Jiang Z, Jiang C, Li Y, Chen W, Lin H, Sun Z, Li J. Characteristics of tet(X4)−Producing Escherichia coli in Chicken and Pig Farms in Hunan Province, China. Antibiotics. 2023; 12(1):147. https://doi.org/10.3390/antibiotics12010147
Chicago/Turabian StyleYang, Jie, Gang Xiao, Ning Xiao, Zonghan Jiang, Chao Jiang, Yujuan Li, Wenxin Chen, Hongguang Lin, Zhiliang Sun, and Jiyun Li. 2023. "Characteristics of tet(X4)−Producing Escherichia coli in Chicken and Pig Farms in Hunan Province, China" Antibiotics 12, no. 1: 147. https://doi.org/10.3390/antibiotics12010147
APA StyleYang, J., Xiao, G., Xiao, N., Jiang, Z., Jiang, C., Li, Y., Chen, W., Lin, H., Sun, Z., & Li, J. (2023). Characteristics of tet(X4)−Producing Escherichia coli in Chicken and Pig Farms in Hunan Province, China. Antibiotics, 12(1), 147. https://doi.org/10.3390/antibiotics12010147






