Recent Trends of Antibiotic Resistance in Staphylococcus aureus Causing Clinical Mastitis in Dairy Herds in Abruzzo and Molise Regions, Italy
Abstract
1. Introduction
2. Results
2.1. Rate of Mastitis Caused by S. aureus in 2021 and 2022
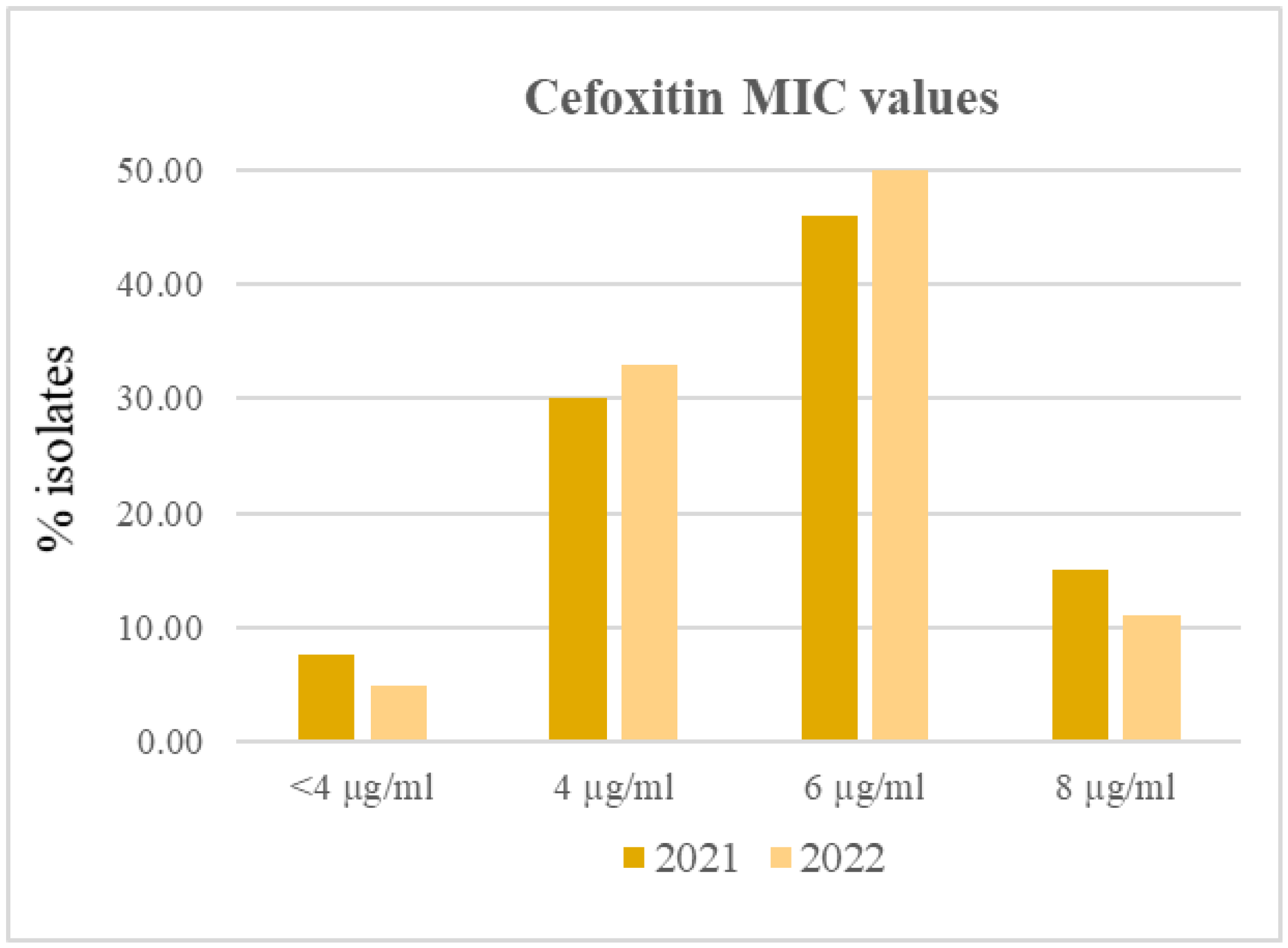
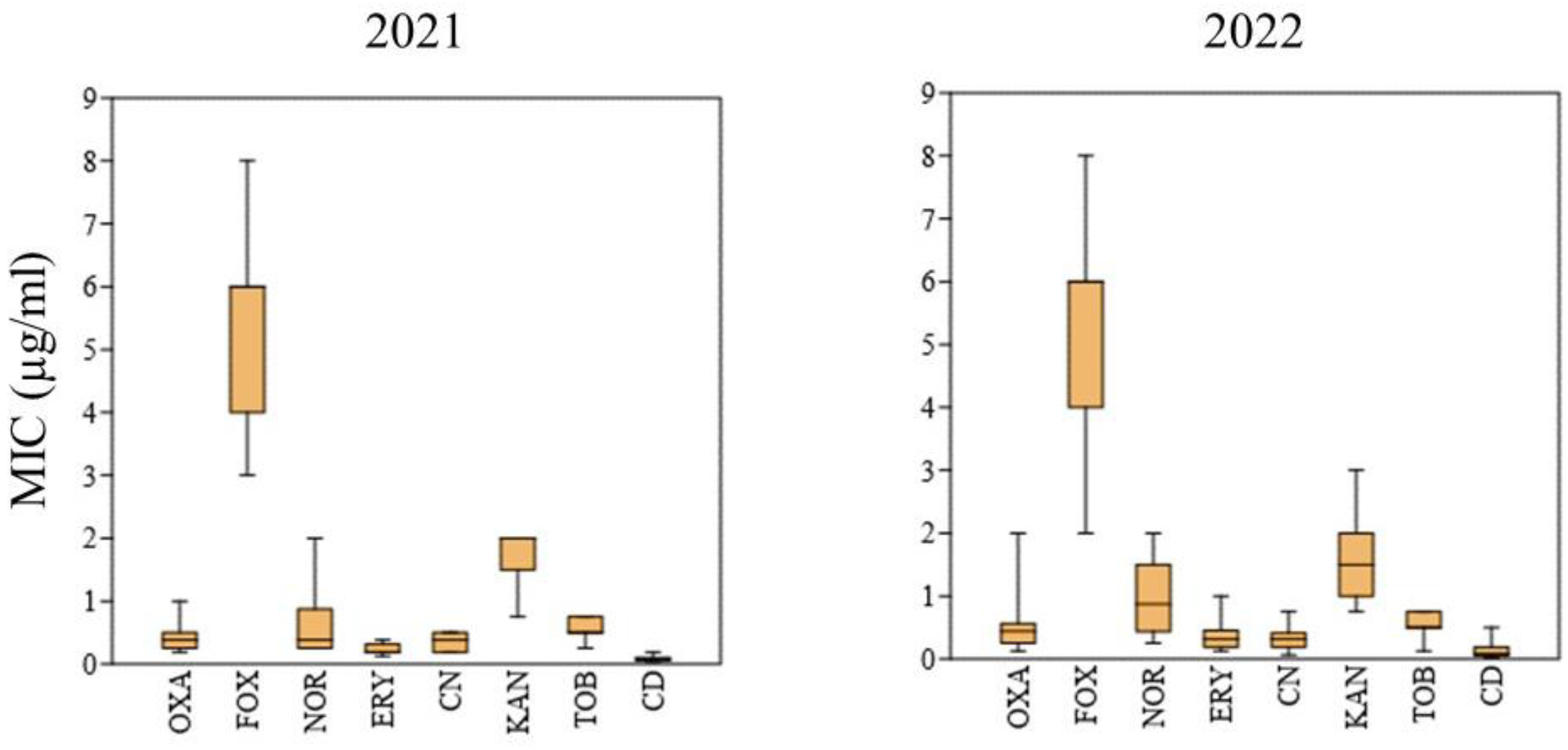
2.2. Phenotypic AR of S. aureus Isolates
2.3. Occurrence of AR Genes in the S. aureus Isolates
2.4. Evaluation of Antibiotic Management via Veterinarian Interview
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Culture Conditions
4.2. Phenotypic AR Testing
4.3. Quantitative PCR Primer Design
4.4. DNA Extraction
4.5. Quantitative PCR Conditions
4.6. Veterinarian Questionnaire
4.7. Statistical Analyses
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zaatout, N.; Ayachi, A.; Kecha, M. Staphylococcus aureus persistence properties associated with bovine mastitis and alternative therapeutic modalities. J. Appl. Microbiol. 2020, 129, 1102–1119. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.Y.C.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef] [PubMed]
- Hofer, U. Insights into the mechanism of superantigen. Nat. Rev. Microbiol. 2022, 20, 253. [Google Scholar] [CrossRef] [PubMed]
- Tuffs, S.W.; Goncheva, M.I.; Xu, S.X.; Craig, H.C.; Kasper, K.J.; Choi, J.; Flannagan, R.S.; Kerfoot, S.M.; Heinrichs, D.E.; McCormick, J.K. Superantigens promote Staphylococcus aureus bloodstream infection by eliciting pathogenic interferon-gamma production. Proc. Natl. Acad. Sci. USA 2022, 119, e2115987119. [Google Scholar] [CrossRef] [PubMed]
- Loffler, B.; Hussain, M.; Grundmeier, M.; Brück, M.; Holzinger, D.; Varga, G.; Roth, J.; Kahl, B.C.; Proctor, R.A.; Peters, G. Staphylococcus aureus Panton–Valentine leukocidin is a very potent cytotoxic factor for human neutrophils. PLoS Pathog. 2010, 6, e1000715. [Google Scholar] [CrossRef]
- Cheung, G.Y.C.; Bae, J.S.; Otto, M. Pathogenicity and virulence of Staphylococcus aureus. Virulence 2021, 12, 547–569. [Google Scholar] [CrossRef]
- World Health Organization. WHO Publishes List of Bacteria for Which New Antibiotics are Urgently Needed. 27 February 2017 News Release, Geneva. Available online: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 27 December 2022).
- Liu, J.; Chen, D.; Peters, B.M.; Li, L.; Li, B.; Xu, Z.; Shirliff, M.E. Staphylococcal chromosomal cassettes mec (SCCmec): A mobile genetic element in methicillin-resistant Staphylococcus aureus. Microb. Pathog. 2016, 101, 56–67. [Google Scholar] [CrossRef]
- Becker, K.; van Alen, S.; Idelevich, E.A.; Schleimer, N.; Seggewiß, J.; Mellmann, A.; Kaspar, U.; Peters, G. Plasmid-Encoded Transferable mecB-Mediated Methicillin Resistance in Staphylococcus aureus. Emerg. Infect Dis. 2018, 24, 242–248. [Google Scholar] [CrossRef]
- Schnitt, A.; Tenhagen, B.A. Risk Factors for the Occurrence of Methicillin-Resistant Staphylococcus aureus in Dairy Herds: An Update. Foodborne Pathog. Dis. 2020, 17, 585–596. [Google Scholar] [CrossRef]
- Lima, M.C.; de Barros, M.; Scatamburlo, T.M.; Polveiro, R.C.; de Castro, L.K.; Guimarães, S.H.S.; da Costa, S.L.; da Costa, M.M.; Moreira, M.A.S. Profiles of Staphyloccocus aureus isolated from goat persistent mastitis before and after treatment with enrofloxacin. BMC Microbiol. 2020, 20, 127. [Google Scholar] [CrossRef]
- Shepheard, M.A.; Fleming, V.M.; Connor, T.R.; Corander, J.; Feil, E.J.; Fraser, C.; Hanage, W.P. Historical zoonoses and other changes in host tropism of Staphylococcus aureus, identified by phylogenetic analysis of a population dataset. PLoS ONE 2013, 8, e62369. [Google Scholar] [CrossRef]
- The European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 13.0. 2023. Available online: https://www.eucast.org/clinical_breakpoints (accessed on 11 January 2023).
- Wang, Y.; Zhang, W.; Wang, J.; Wu, C.; Shen, Z.; Fu, X.; Yan, Y.; Zhang, Q.; Schwarz, S.; Shen, J. Distribution of the multidrug resistance gene cfr in Staphylococcus species isolates from swine farms in China. Antimicrob Agents Chemother 2012, 56, 1485–1490. [Google Scholar] [CrossRef]
- European Community. Regulation (EC) No 853/2004 of the European Parliament and of the Council of 29 April 2004 Laying Down Specific Hygiene Rules for Food of Animal Origin. Available online: https://www.legislation.gov.uk/eur/2004/853/contents# (accessed on 14 February 2023).
- Pennone, V.; Prieto, M.; Álvarez-Ordóñez, A.; Cobo-Diaz, J.F. Antimicrobial Resistance Genes Analysis of Publicly Available Staphylococcus aureus Genomes. Antibiotics 2022, 11, 1632. [Google Scholar] [CrossRef]
- Pereira, L.A.; Harnett, G.B.; Hodge, M.M.; Cattell, J.A.; Speers, D.J. Real-time PCR assay for detection of blaZ genes in Staphylococcus aureus clinical isolates. J. Clin. Microbiol. 2014, 52, 1259–1261. [Google Scholar] [CrossRef]
- Velasco, V.; Sherwood, J.S.; Rojas-García, P.P.; Logue, C.M. Multiplex real-time PCR for detection of Staphylococcus aureus, mecA and Panton-Valentine Leukocidin (PVL) genes from selective enrichments from animals and retail meat. PLoS ONE 2014, 9, e97617. [Google Scholar] [CrossRef]
- Yang, D.; Heederik, D.J.J.; Scherpenisse, P.; Van Gompel, L.; Luiken, R.E.C.; Wadepohl, K.; Skarżyńska, M.; Van Heijnsbergen, E.; Wouters, I.M.; Greve, G.D.; et al. Antimicrobial resistance genes aph(3′)-III, erm(B), sul2 and tet(W) abundance in animal faeces, meat, production environments and human faeces in Europe. J. Antimicrob Chemother 2022, 77, 1883–1893. [Google Scholar] [CrossRef]
- Gajewska, J.; Chajęcka-Wierzchowska, W.; Zadernowska, A. Occurrence and Characteristics of Staphylococcus aureus Strains along the Production Chain of Raw Milk Cheeses in Poland. Molecules 2022, 27, 6569. [Google Scholar] [CrossRef]
- Santos Costa, S.; Sobkowiak, B.; Parreira, R.; Edgeworth, J.D.; Viveiros, M.; Clark, T.G.; Couto, I. Genetic diversity of norA, coding for a main efflux pump of Staphylococcus aureus. Front. Genet. 2019, 9, 710. [Google Scholar] [CrossRef]
- Santos Costa, S.; Viveiros, M.; Rosato, A.E.; Melo-Cristino, J.; Couto, I. Impact of efflux in the development of multidrug resistance phenotypes in Staphylococcus aureus. BMC Microbiol. 2015, 15, 232. [Google Scholar]
- Titouche, Y.; Hakem, A.; Houali, K.; Meheut, T.; Vingadassalon, N.; Ruiz-Ripa, L.; Salmi, D.; Chergui, A.; Chenouf, N.; Hennekinne, J.A.; et al. Emergence of methicillin-resistant Staphylococcus aureus (MRSA) ST8 in raw milk and traditional dairy products in the Tizi Ouzou area of Algeria. J. Dairy Sci. 2019, 102, 6876–6884. [Google Scholar] [CrossRef]
- Alghizzi, M.; Shami, A. The prevalence of Staphylococcus aureus and methicillin resistant Staphylococcus aureus in milk and dairy products in Riyadh, Saudi Arabia. Saudi J. Biol. Sci. 2021, 28, 7098–7104. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, A.; Bhattarai, R.K.; Luitel, H.; Karki, S.; Basnet, H.B. Prevalence of methicillin-resistant Staphylococcus aureus and pattern of antimicrobial resistance in mastitis milk of cattle in Chitwan, Nepal. BMC Vet. Res. 2021, 17, 239. [Google Scholar] [CrossRef] [PubMed]
- Neelam; Jain, V.K.; Singh, M.; Joshi, V.G.; Chhabra, R.; Singh, K.; Rana, Y.S. Virulence and antimicrobial resistance gene profiles of Staphylococcus aureus associated with clinical mastitis in cattle. PLoS ONE 2022, 17, e0264762. [Google Scholar]
- Caruso, M.; Latorre, L.; Santagada, G.; Fraccalvieri, R.; Miccolupo, A.; Sottili, R.; Palazzo, L.; Parisi, A. Methicillin-resistant Staphylococcus aureus (MRSA) in sheep and goat bulk tank milk from Southern Italy. Small Rumin. Res. 2016, 135, 26–31. [Google Scholar] [CrossRef]
- Pardo, L.; Giudice, G.; Mota, M.I.; Gutiérrez, C.; Varela, A.; Algorta, G.; Seija, V.; Galiana, A.; Aguerrebere, P.; Klein, M.; et al. Phenotypic and genotypic characterization of oxacillin-susceptible and mecA positive Staphylococcus aureus strains isolated in Uruguay. Rev. Argent Microbiol. 2022, 54, 293–298. [Google Scholar] [PubMed]
- Naranjo-Lucena, A.; Slowey, R. Invited review: Antimicrobial resistance in bovine mastitis pathogens: A review of genetic determinants and prevalence of resistance in European countries. J. Dairy Sci. 2023, 106, 1–23. [Google Scholar] [CrossRef]
- Argudín, M.A.; Roisin, S.; Nienhaus, L.; Dodémont, M.; de Mendonça, R.; Nonhoff, C.; Deplano, A.; Denis, O. Genetic Diversity among Staphylococcus aureus Isolates Showing Oxacillin and/or Cefoxitin Resistance Not Linked to the Presence of mec Genes. Antimicrob Agents Chemother. 2018, 62, e00091-30. [Google Scholar] [CrossRef]
- Velasco, V.; Mallea, A.; Bonilla, A.M.; Campos, J.; Rojas-García, P. Antibiotic-resistance profile of Staphylococcus aureus strains in the pork supply chain. Chil. J. Agric. Anim. Sc. 2022, 38, 234–240. [Google Scholar] [CrossRef]
- Ba, X.; Kalmar, L.; Hadjirin, N.F.; Kerschner, H.; Apfalter, P.; Morgan, F.J.; Paterson, G.K.; Girvan, S.L.; Zhou, R.; Harrison, E.M.; et al. Truncation of GdpP mediates β-lactam resistance in clinical isolates of Staphylococcus aureus. J. Antimicrob Chemother. 2019, 74, 1182–1191. [Google Scholar] [CrossRef]
- Sommer, A.; Fuchs, S.; Layer, F.; Schaudinn, C.; Weber, R.E.; Richard, H.; Erdmann, M.B.; Laue, M.; Schuster, C.F.; Werner, G.; et al. Mutations in the gdpP gene are a clinically relevant mechanism for β-lactam resistance in meticillin-resistant Staphylococcus aureus lacking mec determinants. Microb. Genom. 2021, 7, 000623. [Google Scholar] [CrossRef]
- Sasaki, H.; Ishikawa, H.; Itoh, T.; Arano, M.; Hirata, K.; Ueshiba, H. Penicillin-Binding Proteins and Associated Protein Mutations Confer Oxacillin/Cefoxitin Tolerance in Borderline Oxacillin-Resistant Staphylococcus aureus. Microb Drug Resist. 2021, 27, 590–595. [Google Scholar] [CrossRef] [PubMed]
- Molineri, A.I.; Camussone, C.; Zbrun, M.V.; Suárez Archilla, G.; Cristiani, M.; Neder, V.; Calvinho, L.; Signorini, M. Antimicrobial resistance of Staphylococcus aureus isolated from bovine mastitis: Systematic review and meta-analysis. Prev. Vet. Med. 2021, 188, 105261. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, J.; Jin, J.; Li, X.; Zhang, H.; Shi, X.; Zhao, C. Prevalence, antibiotic resistance, and enterotoxin genes of Staphylococcus aureus isolated from milk and dairy products worldwide: A systematic review and meta-analysis. Food Res. Int. 2022, 162, 111969. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Cha, J.; Liu, K.; Deng, J.; Yang, B.; Xu, H.; Wang, J.; Zhang, L.; Gu, X.; Huang, C.; et al. The prevalence of bovine mastitis-associated Staphylococcus aureus in China and its antimicrobial resistance rate: A meta-analysis. Front. Vet. Sci. 2022, 9, 1006676. [Google Scholar] [CrossRef] [PubMed]
- Karatuna, O. Phenotypic Methods Used in Antimicrobial Susceptibility Testing. ICARS—ILRI Webinar Series. 1 December 2021. Available online: https://cgspace.cgiar.org/bitstream/handle/10568/119836/Phenotypic%20AST.pdf?sequence=1&isAllowed=y (accessed on 27 December 2022).
- Poli, A.; Guglielmini, E.; Sembeni, S.; Spiazzi, M.; Dellaglio, F.; Rossi, F.; Torriani, S. Detection of Staphylococcus aureus and enterotoxin genotype diversity in Monte Veronese, a Protected Designation of Origin Italian cheese. Lett. Appl. Microbiol. 2007, 45, 529–534. [Google Scholar] [CrossRef]
| Primer and Probe Labels and Sequences (5′-3′) | Target Gene | Amplicon Size (bp) |
|---|---|---|
| AadA12f: CCTGGAGAGAGCGAGA AadA12p: FAM-TTTGGAGAATGGCAGCGCAATGAC-BHQ1 AadA12r: CTATGTTCTCTTGCTTTTGT | aadA12 | 197 |
| AadA-aph2f: GGTAGTGGTTATGATAGTG AadA-aph2p: FAM-TAGAAACTAATGTAAAAATTCCTAA-MGBEQ AadA-aph2r: TTCTGGTGTTAAAAAAGTTCC | aadA-aph2 | 231 |
| Aac6f: CCTTGCGATGCTCTATG Aac6p: Cy5-CCCGACACTTGCTGACGTACA-MGBEQ Aac6r: TCCCCGCTTCCAAGAG | aac6 b (aac4) | 204 |
| Ant6f: GCGCAAATATTAATATACCTAAA Ant6P: Cy5-TGGGAATATAATAATGATG-MGBEQ Ant6r: GGGCAATAAGGTAAGATCA | ant6 b (aadE) | 157 |
| Aph3f: TGGCTGGAAGGAAAGC Aph3p: FAM-TGATGGCTGGAGCAATCTGCT-BHQ1 Aph3r: TGTCGATGGAGTGAAAGA | aph3-III | 184 |
| BlaZf: AAGGTTGCTGATAAAAGTGG BlaZp: FAM-GTTTATCCTAAGGGCCAATCTGAACCT-BHQ1 BlaZr: AAATTCCTTCATTACACTCTTG | blaZ | 182 |
| Cfrf: AAAACCTAACTGTAGATGAGA Cfrp: Cy5-GATAGCATTTCTTTTATGGGAATGGG-BHQ1 Cfrr: TAAACGAATCAAGAGCATCA | cfr | 138 |
| ErmAf: GGTAAACCCCTCTGAGA ErmAp: Cy5-CATCAGTACGGATATTGTC-MGBEQ ErmAr: CCCTTCTCAACGATAAGA | ermA | 177 |
| ErmBf: TACTCGTGTCACTTTAATTCAC ErmBp: Cy5-CAGTTTCAATTCCCTAACAAACAGAGG-BHQ1 ErmBr: CCCTAGTGTTCGGTGAA | ermB | 205 |
| ErmCTf: AAATGGGTTAACAAAGAATACA ErmCTp: Cy5-GAATTGACGATTTAAACAATATTAGCTTTG-BHQ1 ErmCTr: TATTGAAAAGAGACAAGAATTG | ermC/T a | 123 |
| LnuBf: TAATTCTACCTTATCTAATCG lnuBp: FAM-GTTTAGCCAATTATCAGCAT-MGBEQ LnuBr: CGTTCATTAGAACTCTTATC | lnuB | 113 |
| MecAf: AGAAAAAGAAAAAAGATGGCAAA MecAp: FAM-CAACATGAAAAATGATTATGGCTCAG-BHQ1 MecAr: CTCATGCCATACATAAATGGA | mecA | 184 |
| mecA/Cf: ACWTCACCAGGTTCAAC mecA/Cp: Cy5-ATGGTAARGGTTGGCAAA-MGBEQ mecA/Cr: TCTGATGATTCTATTGCTTG | mecA/C c | 194 |
| Mhpf: GGGACTTACATCCAGG Mphp: FAM-AAGCAAACGTCACAGGTCT-MGBEQ Mhpr: TCGTCGTCGAATACACG | mhp | 134 |
| MsrAF: CTTACCAATTTGAAAAAATAGCA MrsAp: Cy5-GGCAAAACCACATTACTAAATATGATTG-BHQ1 MsrAR: TTCACTCATTAAACTACCGT | mrsA | 240 |
| 2021 | 2022 | ||||
|---|---|---|---|---|---|
| Farm/ Isolate * | AR Phenotype | AR Genotype | Farm/ Isolate * | AR Phenotype | AR Genotype |
| 1 | 1 1 | FOX R | blaZ | ||
| 2 | FOX R | 1 2 | FOX R | blaZ | |
| 3 | FOX R | 2 1 | |||
| 4 1 | FOX R | blaZ | 2 2 | ||
| 4 3 | FOX R | 3 1 | FOX R | blaZ, ermB | |
| 5 | FOX R | aph3, blaZ, ermCT | 3 2 | FOX R | |
| 6 1 | FOX R | 4 | FOX R | blaZ | |
| 6 2 | FOX R | 5 | FOX R | ||
| 7 | FOX R | ant6, aph3, blaZ, ermCT, | 6 | FOX R, CD R | mecA, mph |
| 8 1 | FOX R | blaZ | 7 | blaZ | |
| 8 2 | FOX R | blaZ | 8 1 | ||
| 8 3 | FOX R | blaZ | 8 2 | blaZ | |
| 9 | FOX R | 8 3 | |||
| 10 | FOX R | 8 4 | |||
| 11 | 9 | ||||
| 12 | FOX R | 10 1 | FOX R | blaZ | |
| 13 | blaZ | 10 2 | FOX R | blaZ | |
| 14 1 | FOX R | blaZ | 10 3 | FOX R | blaZ |
| 14 2 | FOX R | blaZ | 11 1 | blaZ | |
| 15 1 | blaZ | 11 2 | blaZ | ||
| 15 2 | blaZ | 11 3 | blaZ | ||
| 15 3 | 12 1 | FOX R | |||
| 16 1 | blaZ | 12 2 | FOX R | blaZ | |
| 16 2 | blaZ | 12 3 | FOX R | ||
| 16 3 | blaZ | 13 1 | FOX R | ||
| 16 4 | blaZ | 13 2 | FOX R | ||
| 16 5 | blaZ | 13 3 | FOX R | ||
| Question | Number of Veterinarians * |
|---|---|
| 1. Antibiotic classes prescribed | |
| Aminoglycosides (gentamicin, neomicin, kanamycin) | 2 |
| Penicillins (ampicillin, amoxicillin/clavulanic acid, penicillin) | 9 |
| Cephalosporins (cefalexin, cefoperazone) | 5 |
| Lincosamides (lincomycin-spectinomycin) | 2 |
| Fluoroquinolones (enrofloxacin) | 9 |
| Macrolides (spiramycin, tylosin) | 2 |
| 2. Hygiene conditions in farms | |
| Excellent | 0 |
| Good | 7 |
| Acceptable | 9 |
| Inadequate | 2 |
| 3. Milking hygiene | |
| Excellent | 0 |
| Good | 9 |
| Acceptable | 9 |
| Inadequate | 0 |
| 4. Mastitis prevention measures | |
| Excellent | 0 |
| Good | 7 |
| Acceptable | 7 |
| Inadequate | 4 |
| 5. Management of total bacterial and somatic cell counts in bulk tank milk [15] | |
| Excellent | 0 |
| Good | 13 |
| Acceptable | 5 |
| Inadequate | 0 |
| 6. Reason for bacteriological examination and antibiogram request for mastitis cases | |
| Always | 0 |
| In most cases | 2 |
| For severe infections | 0 |
| For recidivating mastitis | 16 |
| After treatment failure | 4 |
| 7. Protocol of antibiotic usage adopted | |
| Always | 0 |
| In most cases | 7 |
| Frequent | 7 |
| Rare | 2 |
| None | 2 |
| 8. Evidences of AR | |
| Frequent | 2 |
| Rare | 16 |
| None | 0 |
| 9. Measures adopted for AR management | |
| Infectious disease expert consultation | 0 |
| Therapy against specific infectious agents | 18 |
| Reduction of antibiotic usage | 9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rossi, F.; Del Matto, I.; Saletti, M.A.; Ricchiuti, L.; Tucci, P.; Marino, L. Recent Trends of Antibiotic Resistance in Staphylococcus aureus Causing Clinical Mastitis in Dairy Herds in Abruzzo and Molise Regions, Italy. Antibiotics 2023, 12, 430. https://doi.org/10.3390/antibiotics12030430
Rossi F, Del Matto I, Saletti MA, Ricchiuti L, Tucci P, Marino L. Recent Trends of Antibiotic Resistance in Staphylococcus aureus Causing Clinical Mastitis in Dairy Herds in Abruzzo and Molise Regions, Italy. Antibiotics. 2023; 12(3):430. https://doi.org/10.3390/antibiotics12030430
Chicago/Turabian StyleRossi, Franca, Ilaria Del Matto, Maria Antonietta Saletti, Luciano Ricchiuti, Patrizia Tucci, and Lucio Marino. 2023. "Recent Trends of Antibiotic Resistance in Staphylococcus aureus Causing Clinical Mastitis in Dairy Herds in Abruzzo and Molise Regions, Italy" Antibiotics 12, no. 3: 430. https://doi.org/10.3390/antibiotics12030430
APA StyleRossi, F., Del Matto, I., Saletti, M. A., Ricchiuti, L., Tucci, P., & Marino, L. (2023). Recent Trends of Antibiotic Resistance in Staphylococcus aureus Causing Clinical Mastitis in Dairy Herds in Abruzzo and Molise Regions, Italy. Antibiotics, 12(3), 430. https://doi.org/10.3390/antibiotics12030430






