The Urinary Resistome of Clinically Healthy Companion Dogs: Potential One Health Implications
Abstract
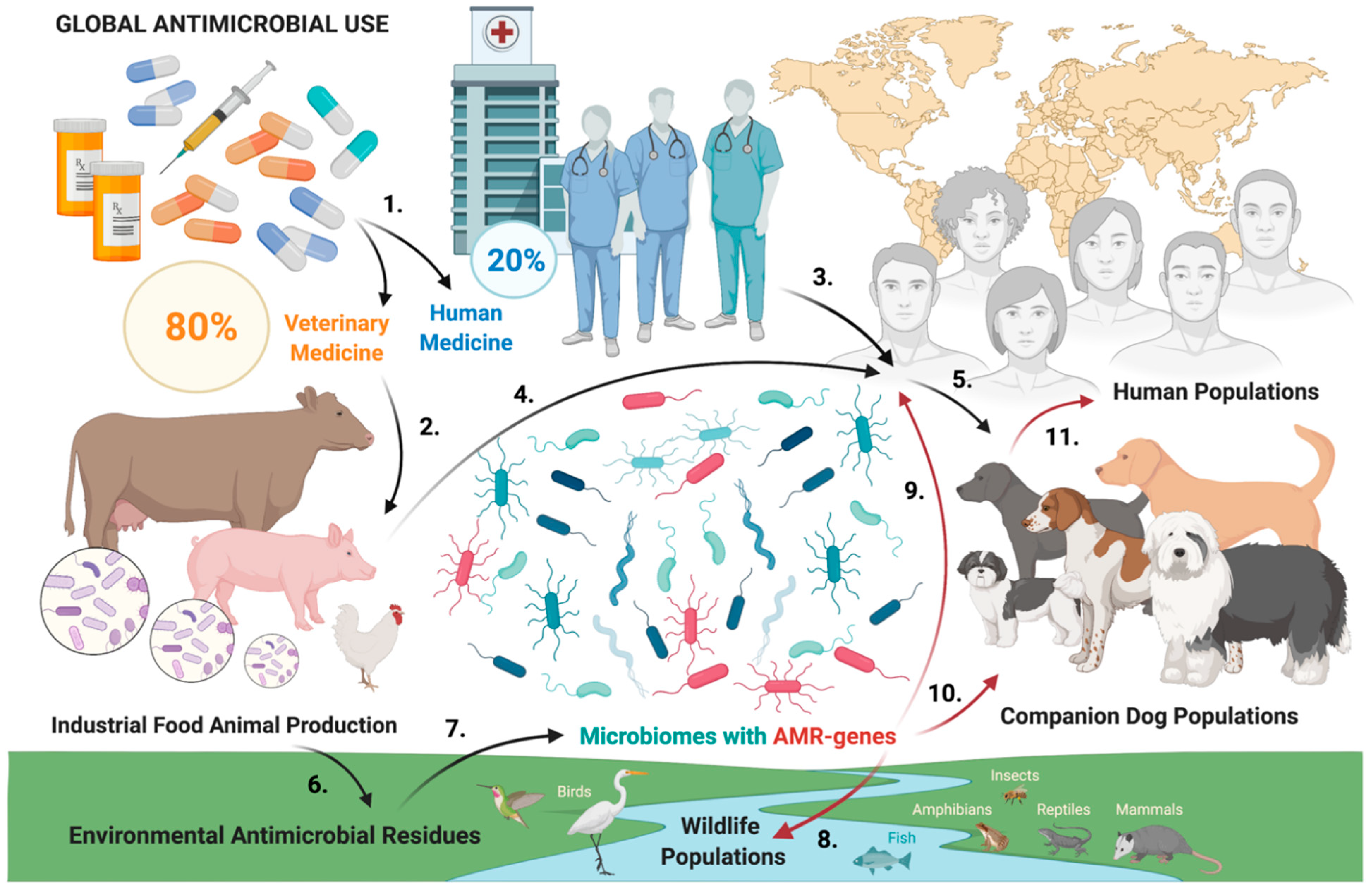
:1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Subjects included, Urine Sample Collection, and Analysis of Urine (UA)
4.2. DNA Extraction and Urine Microbiota 16S Analysis
4.3. Bioinformatics and Statistical Analysis
4.4. Detection of the Urinary Resistome
5. Conclusions
Author Contributions
Funding
Institutional Animal Care and Use Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AMR | antimicrobial resistance |
| CDC | Centers for Disease Control and Prevention |
| IFAP | industrial food animal production |
| MLS | macrolides, lincosamides, and streptogramins |
| WHO | World Health Organization |
References
- O’Neil, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. The Review on Antimicrobial Resistance. United Kingdom. 2016. Available online: https://amr-review.org/sites/default/files/160518_Final%20paper_with%20cover.pdf (accessed on 10 August 2021).
- Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States; U.S. Department of Health and Human Services, CDC: Atlanta, GA, USA, 2019. Available online: http://dx.doi.org/10.15620/cdc:82532 (accessed on 10 August 2021).
- Van Boeckel, T.P.; Brower, C.; Gilbert, M.; Grenfell, B.T.; Levin, S.A.; Robinson, T.P.; Teillant, A.; Laxminarayan, R. Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. USA 2015, 112, 5649–5654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silbergeld, E.; Aidara-Kane, A.; Dailey, J. Agriculture and Food Production as Drivers of the Global Emergence and Dissemination of Antimicrobial Resistance. AMR Control; Johns Hopkins University, Bloomberg School of Public Health: Baltimore, MD, USA, 2017; Available online: http://resistancecontrol.info/2017/agriculture-and-food-production-as-drivers-of-the-global-emergence-and-dissemination-of-antimicrobial-resistance/ (accessed on 10 August 2021).
- Yamanaka, A.R.; Hayakawa, A.T.; Rocha, I.S.M.; Dutra, V.; Souza, V.R.F.; Cruz, J.N.; Camargo, L.M.; Nakazato, L. The Occurrence of Multidrug Resistant Bacteria in the Urine of Healthy Dogs and Dogs with Cystitis. Animals 2019, 9, 1087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marques, C.; Belas, A.; Franco, A.; Aboim, C.; Gama, L.T.; Pomba, C. Increase in antimicrobial resistance and emergence of major international high-risk clonal lineages in dogs and cats with urinary tract infection: 16 year retrospective study. J. Antimicrob. Chemother. 2018, 73, 377–384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melgarejo, T.; Oakley, B.B.; Krumbeck, J.A.; Tang, S.; Krantz, A.; Linde, A. Assessment of bacterial and fungal populations in urine from clinically healthy dogs using next-generation sequencing. J. Vet. Intern. Med. 2021, 35, 1416–1426, Epub 19 March 2021. [Google Scholar] [CrossRef] [PubMed]
- Abadi, A.T.B.; Rizvanov, A.A.; Haertlé, T.; Blatt, N.L. World Health Organization Report: Current Crisis of Antibiotic Resistance. BioNanoScience 2019, 9, 778–788. [Google Scholar] [CrossRef]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin. Microbiol. Rev. 2018, 31, e00088-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.W.; Cha, C.J. Antibiotic resistome from the One-Health perspective: Understanding and controlling antimicrobial resistance transmission. Exp. Mol. Med. 2021, 53, 301–309. [Google Scholar] [CrossRef] [PubMed]
- American Veterinary Medical Association. U.S. pet ownership statistics, 2017–2018. In AVMA Pet Ownership and Demographics Sourcebook; American Veterinary Medical Association: Schaumburg, IL, USA, 1993; Available online: https://www.avma.org/resources-tools/reports-statistics/us-pet-ownership-statistics (accessed on 10 August 2021)ISBN 978–1-882691–53–1 (PDF).
- World Health Organization. Critically Important Antimicrobials for Human Medicine: 6th Revision-Ranking of Medically Important Antimicrobials for Risk Management of Antimicrobial Resistance due to Non-Human Use; WHO Advisory Group on Integrated Surveillance of Antimicrobial Resistance (AGISAR): Geneva, Switzerland, 2018; ISBN 978–92–4-151552–8. Published 20 March 2019; Available online: https://www.who.int/publications/i/item/9789241515528 (accessed on 10 August 2021).
- Shaw, K.J.; Rather, P.N.; Hare, R.S.; Miller, G.H. Molecular Genetics of Aminoglycoside Resistance Genes and Familial Relationships of the Aminoglycoside-Modifying Enzymes. Microbiol. Rev. 1993, 57, 138–163. [Google Scholar] [CrossRef] [PubMed]
- FDA Releases Annual Summary Report on Antimicrobials Sold or Distributed in 2019 for Use in Food-Producing Animals. Available online: https://www.fda.gov/animal-veterinary/cvm-updates/fda-releases-annual-summary-report-antimicrobials-sold-or-distributed-2019-use-food-producing (accessed on 9 December 2021).
- Plumb, D.C. Aminoglycosides. In Plumb’s Veterinary Drug Handbook: Desk, 9th ed.; P Wiley-Blackwell: Hoboken, NJ, USA, 2018; pp. 346–364. [Google Scholar]
- Metlay, J.P.; Waterer, G.W.; Long, A.C.; Anzueto, A.; Brozek, J.; Crothers, K.; Cooley, L.A.; Dean, N.C.; Fine, M.J.; Flanders, S.A.; et al. Diagnosis and Treatment of Adults with Community-acquired Pneumonia. An Official Clinical Practice Guideline of the American Thoracic Society and Infectious Diseases Society of America. Am. J. Respir. Crit. Care Med. 2019, 200, e45–e67. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Outpatient Antibiotics Prescriptions. Available online: https://www.cdc.gov/antibiotic-use/data/report-2020.html (accessed on 25 November 2021).
- Lowy, F.D. Antimicrobial resistance: The example of Staphylococcus aureus. J. Clin. Investig. 2003, 111, 1265. [Google Scholar] [CrossRef] [PubMed]
- Fàbrega, A.; Madurga, S.; Giralt, E.; Vila, J. Mechanism of action of and resistance to quinolones. Microb. Biotechnol. 2009, 2, 40–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- National Institute of Diabetes and Digestive and Kidney Diseases. Liver Tox: Clinical and Research Information on Drug Induced Liver Injury. Bethesda (MD): National Institutes of Health. Fluoroquinolones. 2012. Available online: https://www.ncbi.nlm.nih.gov/books/NBK547840/ (accessed on 10 March 2022).
- Pohanka, M.; Skladal, P. Bacillus anthracis, Francisella tularensis, and Yersinia pestis. The Most Important Bacterial Warfare Agents-review. Folia Microbiol. 2009, 54, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Neu, H.C. The crisis of antimicrobial resistance. Science 1992, 257, 1064–1073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Economou, V.; Gousia, P. Agriculture and food animals as a source of antimicrobial-resistant bacteria. Infect. Drug Resist. 2015, 8, 49–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- World Health Organization Guidelines of use of Medically Important Antibiotics in Food Producing Animals. Available online: http://apps.who.int/iris/bitstream/handle/10665/258970/9789241550130-eng.pdf (accessed on 10 April 2022).
- Levy, S.B. Emergence of Antibiotic-Resistant Bacteria in the Intestinal Flora of Farm Inhabitants. J. Infect. Dis. 1978, 137, 688–690. [Google Scholar] [CrossRef]
- Levy, S.; Fitzgerald, G.; Macone, A. Spread of antibiotic-resistant plasmids from chicken to chicken and from chicken to man. Nature 1976, 260, 40–42. [Google Scholar] [CrossRef] [PubMed]
- Hoelzer, K.; Wong, N.; Thomas, J.; Talkington, K.; Jungman, E.; Coukell, A. Antimicrobial drug use in food-producing animals and associated human health risks: What, and how strong, is the evidence? BMC Vet. Res. 2017, 13, 211. [Google Scholar] [CrossRef] [PubMed]
- Mann, A.; Nehra, K.; Rana, J.S.; Dahiya, T. Antibiotic resistance in agriculture: Perspectives on upcoming strategies to overcome upsurge in resistance. Curr. Res. Microb. Sci. 2021, 2, 100030. [Google Scholar] [CrossRef] [PubMed]
- CDC Antibiotic Resistance Threats in USA 2019. Available online: https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf (accessed on 23 December 2021).
- CARD. The Comprehensive Antibiotic Resistance Database; McMaster University: Hamilton, ON, Canada, 2013; Available online: https://card.mcmaster.ca/ontology/41634 (accessed on 2 June 2021).
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles-Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
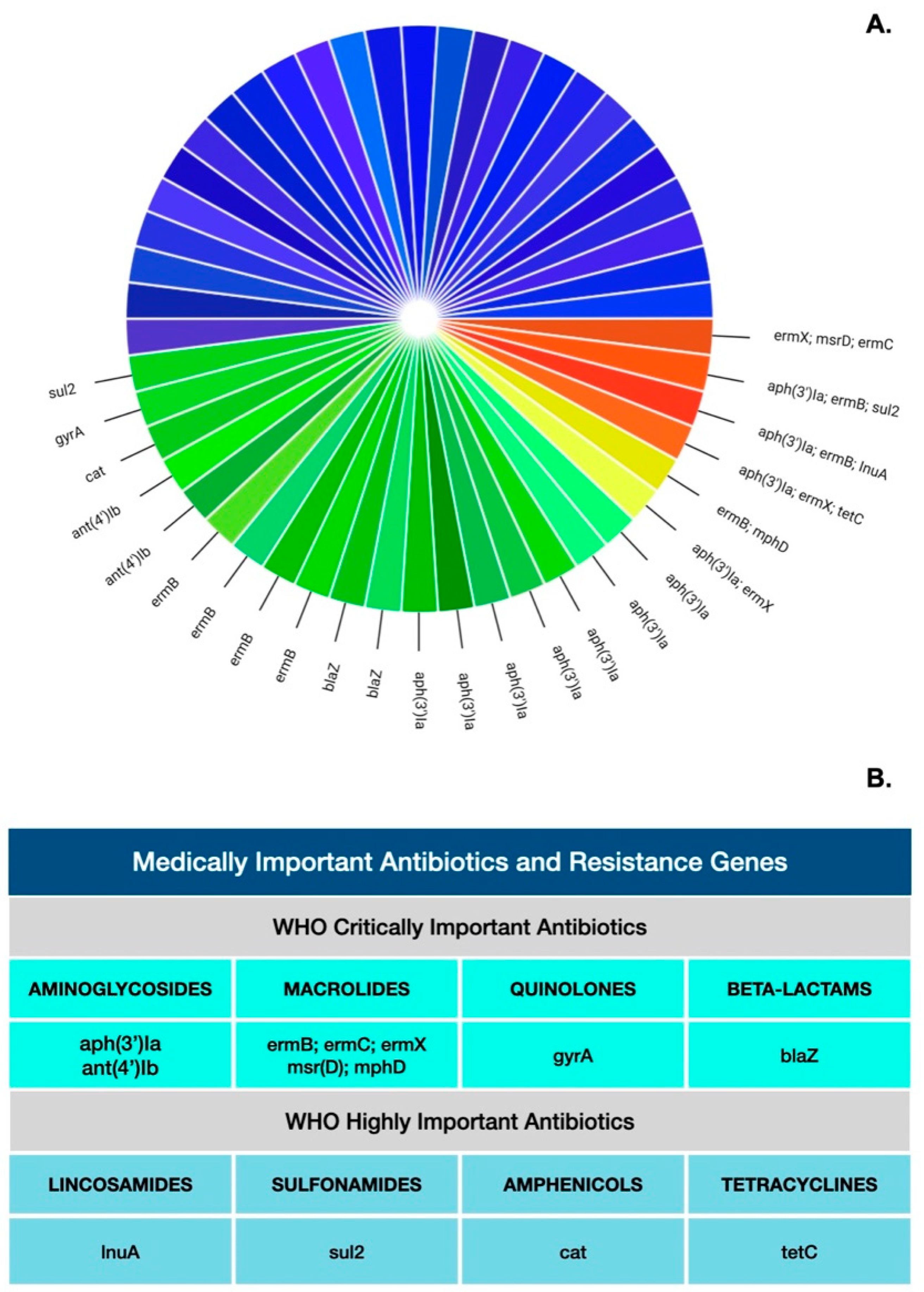
| Antibiotics | AMR Gene | Enzyme Conferring AMR | Positives (%) |
|---|---|---|---|
| Aminoglycosides | aph(3′)Ia | aminoglycoside.phosphotransferase | 11 (22%) |
| ant(4′)Ib | kanamycin.nucleotidyltransferase | 2 (4%) | |
| MLS: Macrolides Lincosamides Streptogramins | ermB | 23S.ribosomal.rna.methyltransferase | 7 (14%) |
| ermX | 23S.ribosomal.rna.methyltransferase | 3 (6%) | |
| mphD | Macrolide.2.phosphotransferase | 1 (2%) | |
| msr(D) | ABC.F.ribosomal.protection.protein | 1 (2%) | |
| ermC | 23S.ribosomal.rna.methyltransferase | 1 (2%) | |
| lnuA | lincosamide.nucleotidyltransferase | 1 (2%) | |
| Beta-lactams | blaZ | staph.blaZ | 2 (4%) |
| Sulfonamides | sul2 | sulfonamide.resistant.dihydropteroate.synthase | 2 (4%) |
| Amphenicols | cat | chloramphenicol.acetyltransferase | 1 (2%) |
| Quinolones | gyrA | staphylococcus.pseudintermedius.250TTG | 1 (2%) |
| Tetracyclines | tetC | tetracycline.efflux.pump | 1 (2%) |
| Bacterial Species (Relative Abundances in Urine Samples) | Genes Detected Against | Intrinsic Resistances Against |
|---|---|---|
| Bifidobacterium longum (3.1%); Escherichia-Shigella coli (6.5%); Lachnoclostridium spp. (1.5%); Rothia mucilaginosa (5.4%) | Clindamycin, Lincomycin, Erythromycin, Azithromycin | |
| Campylobacter upsaliensis (0.1%) | Clindamycin | |
| Corynebacterium auriscanis (45.6%); C. genitalium (4.1%); C. mucifaciens (26.9%); C. pilbarense (9.6%); C. simulans (1.7%); C. pseudogenitalium-tuberculostearicum (0.7%, 3.0%, 4.2%); C. ureicelerivorans (5.9%); Lactobacillus aviarius (2.2%); L. fabifermentans (0.1%); L. vaginalis (1.6%); Staphylococcus cohnii (1.1%); S. capitis-caprae (1.1%); S. epidermidis (1.3%, 2.5%); S. felis (4.6%); S. hominis (8.3%) | Nalidixic acid | |
| Corynebacterium pseudogenitalium-tuberculostearicum (4.2%) | Clindamycin, Lincomycin, Erythromycin, Azithromycin, Neomycin, Amikacin, Gentamicin | Nalidixic acid |
| Corynebacterium ureicelerivorans (1.9%); Enterococcus cecorum (2.06%); Lactobacillus johnsonii (1.6%); L. salivarius (4.5%); Staphylococcus epidermidis (0.5%); S. hominis (0.3%) | Clindamycin, Lincomycin, Erythromycin, Azithromycin | Nalidixic acid |
| Escherichia-Shigella boydii-coli-fergusonii (0.9%) | Neomycin, Amikacin, Gentamicin | |
| Haemophilus parainfluenzae (0.6%) | Clindamycin | |
| Pseudomonas spp. (4.0%, 7.1%, 8.2%, 14.4%, 20.4%); P. moraviensis (0.2%) | Cefadroxil, Cefazolin, Cefoxitin, Clindamycin, Erythromycin, Azithromycin, Rifampicin | |
| Staphylococcus hominis (3.8%); S. simulans (6.7%); S. pseudintermedius (17.8%); S. delphini-intermedius-pseudintermedius (100.0%) | Ampicillin, Amoxicillin, Oxacillin, Benzylpenicillin | Nalidixic acid |
| Stenotrophomonas maltophilia (3.0%) Stenotrophomonas maltophilia (5.7%) | Sulfonamide | Ampicillin, Amoxicillin, Cefadroxil, Cefazolin, Cefpodoxime, Ceftiofur, Cefovecin, Cefoxitin, Clindamycin, Erythromycin, Azithromycin, Doxycycline, Tetracycline, Neomycin, Amikacin, Gentamicin, Clavamox, Piperacillin/Tazobactam, Imipenem, Rifampicin |
| Streptococcus alactolyticus (1.4%); S. canis (2.6%, 44.2%); S. gordonii (1.6%); S. oralis (2.0%); S. parasanguinis (16.8%); S. salivarius (0.4%, 6.7%); S. salivarius-thermophilus (1.1%); S. sanguinis (0.4%, 3.4%) | Neomycin, Amikacin, Gentamicin, Nalidixic acid | |
| Streptococcus cristatus (1.3%); S. sanguinis (1.3%) | Clindamycin, Lincomycin, Erythromycin, Azithromycin, | Neomycin, Amikacin, Gentamicin, Nalidixic acid |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Melgarejo, T.; Sharp, N.; Krumbeck, J.A.; Wu, G.; Kim, Y.J.; Linde, A. The Urinary Resistome of Clinically Healthy Companion Dogs: Potential One Health Implications. Antibiotics 2022, 11, 780. https://doi.org/10.3390/antibiotics11060780
Melgarejo T, Sharp N, Krumbeck JA, Wu G, Kim YJ, Linde A. The Urinary Resistome of Clinically Healthy Companion Dogs: Potential One Health Implications. Antibiotics. 2022; 11(6):780. https://doi.org/10.3390/antibiotics11060780
Chicago/Turabian StyleMelgarejo, Tonatiuh, Nathan Sharp, Janina A. Krumbeck, Guangxi Wu, Young J. Kim, and Annika Linde. 2022. "The Urinary Resistome of Clinically Healthy Companion Dogs: Potential One Health Implications" Antibiotics 11, no. 6: 780. https://doi.org/10.3390/antibiotics11060780
APA StyleMelgarejo, T., Sharp, N., Krumbeck, J. A., Wu, G., Kim, Y. J., & Linde, A. (2022). The Urinary Resistome of Clinically Healthy Companion Dogs: Potential One Health Implications. Antibiotics, 11(6), 780. https://doi.org/10.3390/antibiotics11060780






