Resistance Genes, Plasmids, Multilocus Sequence Typing (MLST), and Phenotypic Resistance of Non-Typhoidal Salmonella (NTS) Isolated from Slaughtered Chickens in Burkina Faso
Abstract
1. Introduction
2. Results
2.1. Serotypes of Isolates from Slaughtered Chickens
2.2. Antimicrobial Resistance Genes and Antibiotic Resistance Phenotypes Detected
2.3. Replicon Types Detected
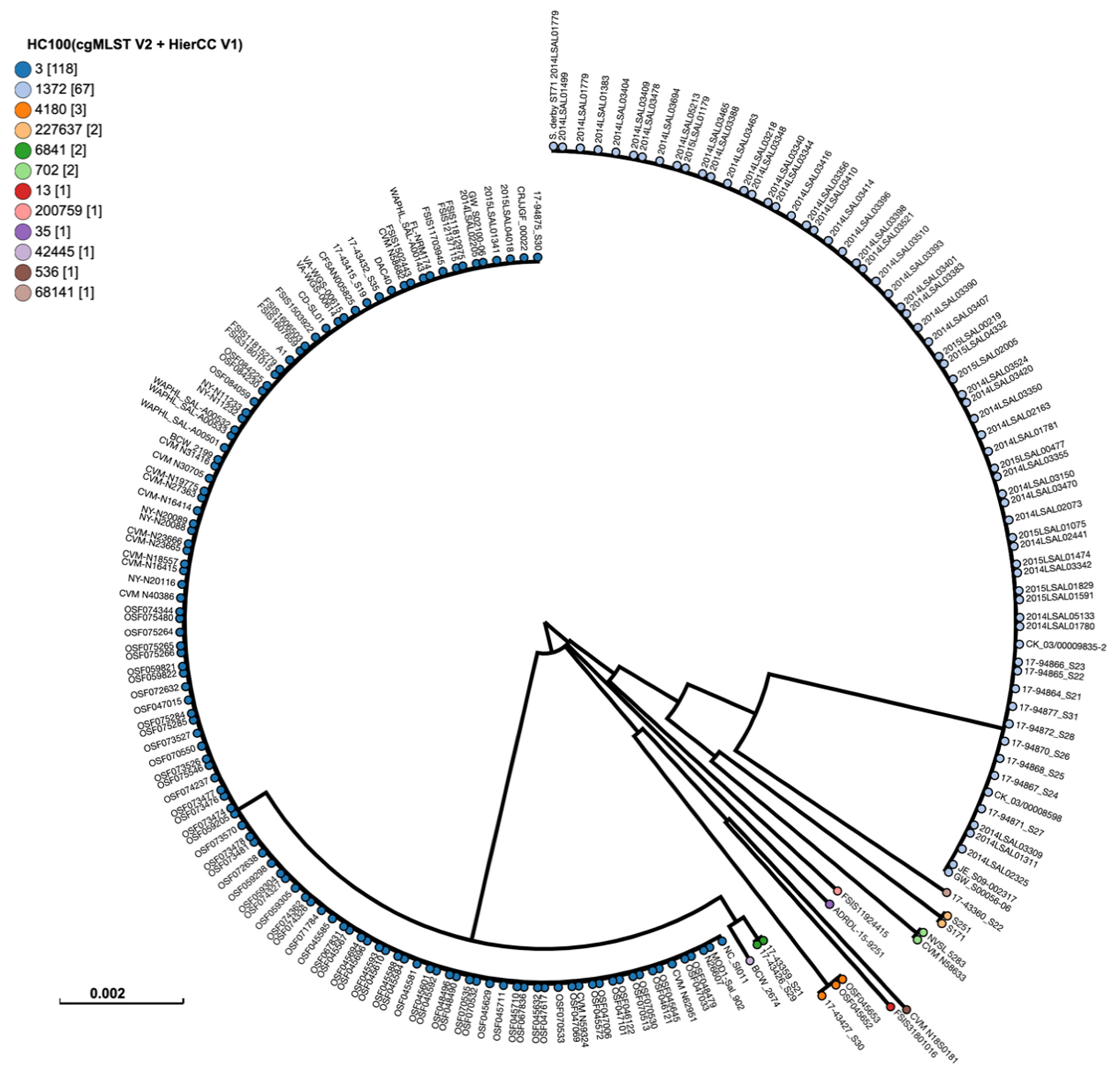
2.4. Phylogenetic Analysis of S. Derby Isolates
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains
4.2. Antimicrobial Susceptibility Testing
4.3. DNA Extraction, Whole-Genome Sequencing, Assembly, Annotation, and Molecular Serotyping
4.4. Phylogenetic Analysis of Salmonella Derby Isolates
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tay, M.Y.F.; Pathirage, S.; Chandrasekaran, L.; Wickramasuriya, U.; Sadeepanie, N.; Waidyarathna, K.D.K.; Liyanage, L.D.C.; Seow, K.L.G.; Hendriksen, R.S.; Takeuchi, M.T.; et al. Whole-Genome Sequencing Analysis of Nontyphoidal Salmonella enterica of Chicken Meat and Human Origin under Surveillance in Sri Lanka. Foodborne Pathog. Dis. 2019, 16, 531–537. [Google Scholar] [CrossRef] [PubMed]
- Eguale, T. Non-typhoidal Salmonella serovars in poultry farms in central Ethiopia: Prevalence and antimicrobial resistance. BMC Vet. Res. 2018, 14, 217. [Google Scholar] [CrossRef] [PubMed]
- Foley, S.L.; Nayak, R.; Hanning, I.B.; Johnson, T.J.; Han, J.; Ricke, S.C. Population dynamics of Salmonella enterica serotypes in commercial egg and poultry production. Appl. Environ. Microbiol. 2011, 77, 4273–4279. [Google Scholar] [CrossRef] [PubMed]
- Kagambèga, A.; Thibodeau, A.; Trinetta, V.; Soro, K.D.; Sama, F.N.; Bako, E.; Bouda, S.C.; Wereme, A.; Fravalo, P.; Barro, N. Salmonella spp. and Campylobacter spp. in poultry feces and carcasses in Ouagadougou, Burkina Faso. Food Sci. Nutr. 2018, 6, 1601–1606. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO Estimates of the Global Burden of Foodborne Diseases: Foodborne Disease Burden Epidemiology Reference Group 2007–2015; World Health Organization: Geneva, Switzerland, 2015; ISBN 9789241565165. Available online: https://www.who.int/ (accessed on 22 March 2022).
- Ministry of Animal Resources. Yearbooks of Livestock Statistics; Ministry of Animal Resources: Ouagadougou, Burkina Faso, 2021; 177p. [Google Scholar]
- Kagambèga, A.; Haukka, K.; Siitonen, A.; Traoré, A.S.; Barro, N. Prevalence of Salmonella enterica and the hygienic indicator Escherichia coli in raw meat at markets in Ouagadougou, Burkina Faso. J. Food Prot. 2011, 74, 1547–1551. [Google Scholar] [CrossRef]
- Lee, L.E.; Niode, O.; Simonne, A.H.; Bruhn, C.M. Consumer perceptions on food safety in Asian and Mexican restaurants. Food Control 2012, 26, 531–538. [Google Scholar] [CrossRef]
- Kagambèga, A.; Lienemann, T.; Aulu, L.; Traoré, A.S.; Barro, N.; Siitonen, A.; Haukka, K. Prevalence and characterization of Salmonella enterica from the intestines of cattle, poultry, swine and hedgehogs in Burkina Faso and their comparison to human Salmonella isolate. Salmonella. BMC Microbiol. 2013, 13, 253. [Google Scholar] [CrossRef]
- Bouda, S.C.; Kagambèga, A.; Bonifait, L.; Le Gall, F.; Ibrahim, H.B.; Bako, E.; Bagre, T.S.; Zongo, C.; N’diaye, W.A.; Traore, A.S.; et al. Prevalence and Antimicrobial Resistance of Salmonella enterica Isolated from Chicken and Guinea Fowl in Burkina Faso. Int. J. Microbiol. Biotechnol. 2019, 4, 64–71. [Google Scholar] [CrossRef]
- Kagambèga, A.; Hiott, L.M.; Boyle, D.S.; McMillan, E.A.; Sharma, P.; Gupta, S.K.; Ramadan, H.; Cho, S.; Humayoun, S.B.; Woodley, T.A.; et al. Serotyping of sub-Saharan Africa Salmonella strains isolated from poultry feces using multiplex PCR and whole genome sequencing. BMC Microbiol. 2021, 21, 29. [Google Scholar] [CrossRef]
- Colavecchio, A.; Jeukens, J.; Freschi, L.; Edmond Rheault, J.G.; Kukavica-Ibrulj, I.; Levesque, R.; Goodridge, L. AnCo3, a new member of the emerging family of Phage-Like Plasmids. Genome Announc. 2017, 5, e00110-17. [Google Scholar] [CrossRef]
- Bonkoungou, I.J.O.; Haukka, K.; Österblad, M.; Hakanen, A.J.; Traoré, A.S.; Barro, N.; Siitonen, A. Bacterial and viral aetiology of childhood diarrhea in Ouagadougou, Burkina Faso. BMC Pediatrics 2013, 13, 36. [Google Scholar] [CrossRef] [PubMed]
- Simporé, J.; Ouermi, D.; Ilboudo, D.; Kabre, A.; Zeba, B.; Pietra, V.; Pignatelli, S.; Nikiema, J.B.; Kabre, G.B.; Caligaris, S.; et al. Aetiology of acute gastro-enteritis in children at Saint Camille Medical Centre, Ouagadougou, Burkina Faso. Pak. J. Biol. Sci. 2009, 12, 258–263. [Google Scholar] [CrossRef] [PubMed]
- Sajid, S.U.; Sajid, M.; Hashmi, R.I. Isolation studies on the prevalence of salmonellae in chicken organs, eggs and feed components. J. Ayub Med. Coll. Abbottabad 2015, 27, 530–533. [Google Scholar] [PubMed]
- Fearnley, E.; Raupach, J.; Lagala, F.; Cameron, S. Salmonella in chicken meat, eggs and humans; Adelaide, South Australia. Int. J. Food Microbiol. 2011, 146, 219–227. [Google Scholar] [CrossRef]
- EFSA. The European Union Summary Report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2012. EFSA J. 2014, 12, 3590. [Google Scholar] [CrossRef]
- Abdelkader, A.S.; Oumarou, S.S.; Maârouhi, I.M.; Boubacar, S.A.; Ousseini, M.H.; Yacoubou, B. Diversity and Distribution of Salmonella Isolated from Poultry Offal in Niger (West Africa). Int. J. Microbiol. Biotechnol. 2019, 4, 103–112. [Google Scholar] [CrossRef][Green Version]
- Stephen, J.S.; Barry, G.H. Determining the Limits of the Evolutionary Potential of an Antibiotic Resistance Gene. Mol. Biol. Evol. 2003, 20, 653–659. [Google Scholar] [CrossRef]
- Magnet, S.; Courvalin, P.; Lambert, T. Activation of the Cryptic aac(6′)-IyAminoglycoside Resistance Gene of Salmonella by a Chromosomal Deletion Generating a Transcriptional Fusion. J. Bacteriol. 1999, 181, 6650–6655. [Google Scholar] [CrossRef]
- Chuanchuen, R.; Khemtong, S.; Padungtod, P. Occurrence of qace/qaceδ1 genes and their correlation with class 1 integrons in salmonella enterica isolates from poultry and swine. Southeast Asian J. Trop. Med. Public Health 2007, 38, 855–862. [Google Scholar]
- Randall, L.P.; Cooles, S.W.; Piddock, L.J.; Woodward, M.J. Effect of triclosan or a phenolic farm disinfectant on the selection of antibiotic-resistant Salmonella enterica. J. Antimicrob. Chemother. 2004, 54, 621–627. [Google Scholar] [CrossRef]
- Villa, L.; García-Fernández, A.; Fortini, D.; Carattoli, A. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J. Antimicrob. Chemother. 2010, 65, 2518–2529. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A. Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 2009, 53, 2227–2238. [Google Scholar] [CrossRef] [PubMed]
- Silva, C.; Puente, J.L.; Calva, E. Salmonella virulence plasmid: Pathogenesis and ecology. Pathog. Dis. 2017, 75, ftx070. [Google Scholar] [CrossRef]
- Rozwandowicz, M.; Brouwer, M.S.M.; Fischer, J.; Wagenaar, J.A.; Gonzalez-Zorn, B.; Guerra, B.; Mevius, D.J.; Hordijk, J. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J. Antimicrob. Chemother. 2018, 73, 1121–1137. [Google Scholar] [CrossRef] [PubMed]
- McMillan, E.A.; Gupta, S.K.; Williams, L.E.; Jové, T.; Hiott, L.M.; Woodley, T.A.; Barrett, J.B.; Jackson, C.R.; Wasilenko, J.L.; Simmons, M.; et al. Antimicrobial resistance genes, cassettes, and plasmids present in Salmonella enterica associated with United States food animals. Front. Microbiol. 2019, 10, 832. [Google Scholar] [CrossRef]
- McMillan, E.A.; Jackson, C.R.; Frye, J.G. Transferable Plasmids of Salmonella enterica Associated with Antibiotic Resistance Genes. Front. Microbiol. 2020, 11, 562181. [Google Scholar] [CrossRef]
- Baucheron, S.; Chaslus-Dancla, E.; Cloeckaert, A.; Chiu, C.H.; Butaye, P. High-level resistance to fluoroquinolones linked to mutations in gyrA, parC, and parE in Salmonella enterica serovar Schwarzengrund isolates from humans in Taiwan. Antimicrob. Agents Chemother. 2005, 49, 862–863. [Google Scholar] [CrossRef]
- Feng, Y.; Chang, Y.J.; Fang, S.H.; Su, L.H.; Li, H.C.; Yang, H.P.; Yu, M.J.; Chiu, C.H. Emergence and Evolution of High-Level Cephalosporin-Resistant Salmonella Goldcoast in Northern Taiwan. In Open Forum Infectious Diseases; Oxford University Press: Oxford, UK, 2019; Volume 6, p. ofz447. [Google Scholar] [CrossRef]
- M100 Clinical and Laboratory Standards Institute M100 Ed30 Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement; CLSI: Wayne, PA, USA, 2020.
- Coil, D.; Jospin, G.; Darling, A.E. A5-miseq: An updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics 2015, 31, 587–589. [Google Scholar] [CrossRef]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Pontén, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus Sequence Typing of Total Genome Sequenced Bacteria. J. Clin. Micob. 2012, 52, 1501–1510. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Zankari, E.; Garcia-Fernandez, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Aarestrup, F.M.; Hasman, H. PlasmidFinder and pMLST: In silico detection and typing of plasmids. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed]
- Boratyn, G.M.; Camacho, C.; Cooper, P.S.; Coulouris, G.; Fong, A.; Ma, N.; Madden, T.L.; Matten, W.T.; McGinnis, S.D.; Merezhuk, Y.; et al. BLAST: A more efficient report with usability improvements. Nucleic Acids Res. 2013, 41, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef] [PubMed]
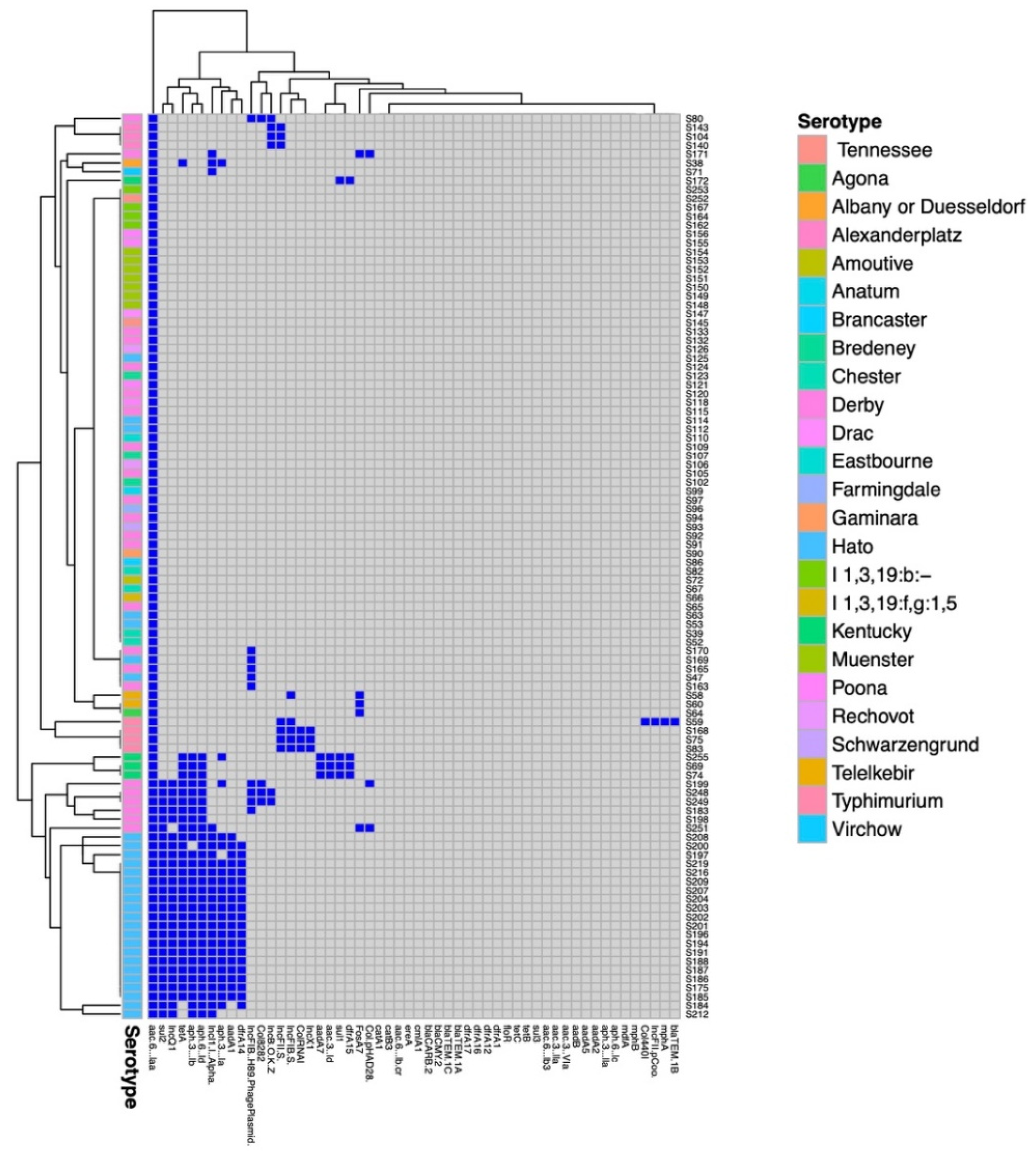
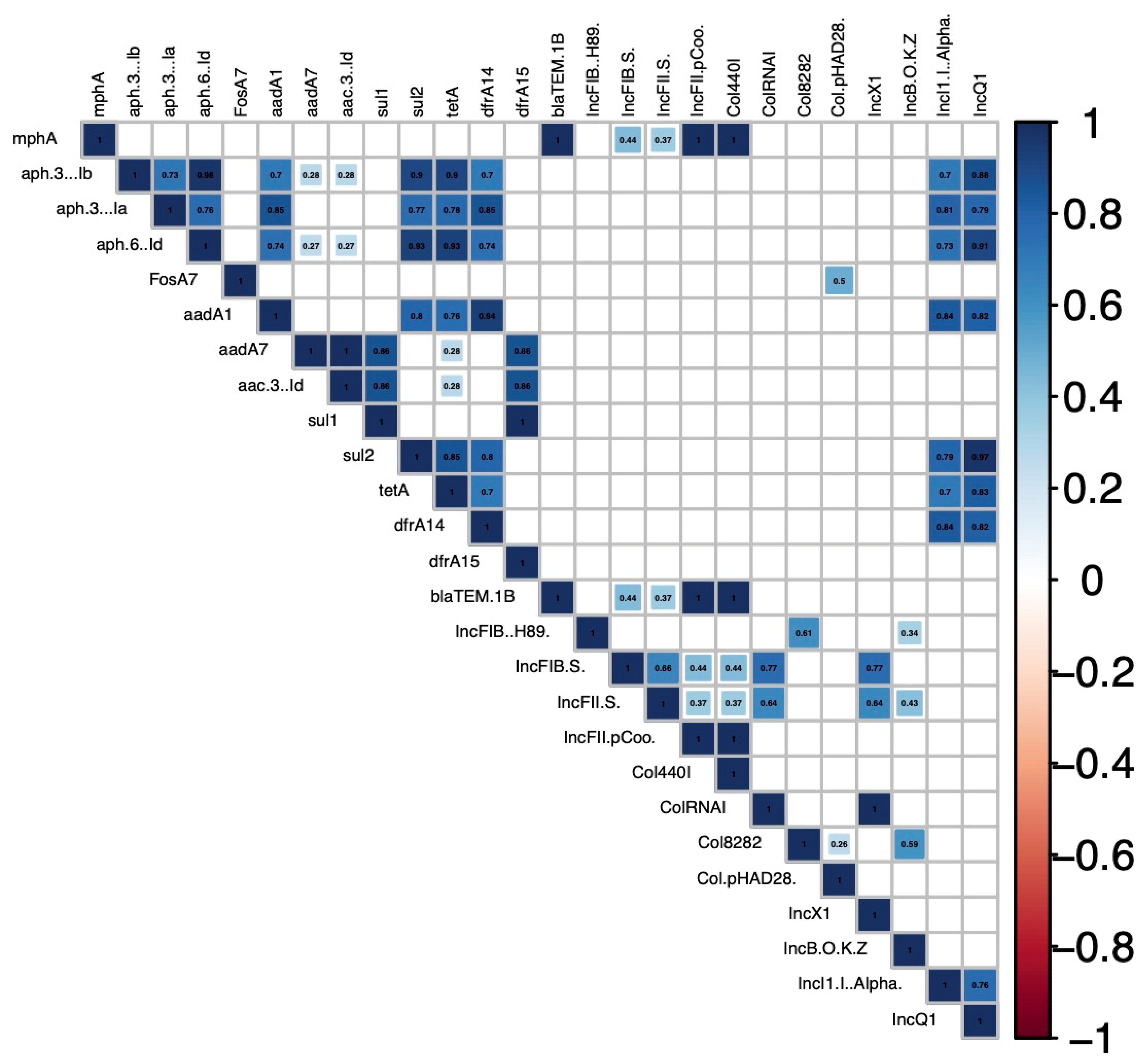
| Sample | Serotype | Antimicrobial Resistances Genes a | Phenotypic Resistance Profile b,c | Plasmid Replicons c | MLST |
|---|---|---|---|---|---|
| S38 | Albany or Dusseldorf | aac(6′)-Iaa; aph(3′)-Ia; tet(A) | TET | IncI1-I (Alpha) | 292 |
| S39 | Chester | aac(6′)-Iaa | ND | ND | 411 |
| S47 | Hato | aac(6′)-Iaa | ND | IncFIB (H89-PhagePlasmid) | Unknown |
| S52 | Chester | aac(6′)-Iaa | ND | ND | 411 |
| S53 | Hato | aac(6′)-Iaa | ND | ND | 3899 |
| S58 | Telelkebir | aac(6′)-Iaa; fosA7 | ND | IncFIB(S) | 2386 |
| S59 | Typhimurium | aac(6′)-Iaa; blaTEM-1B; mph(A) | AMP; AMPSUL (A/S2); PIP; TICCLA(TIM2) | Col440I, IncFIB(S), IncFII(S), IncFII(pCoo) | 313 |
| S60 | Telelkebir | aac(6′)-Iaa; fosA7 | ND | ND | 5494 |
| S63 | Hato | aac(6′)-Iaa | ND | ND | 3899 |
| S64 | Agona | aac(6′)-Iaa; fosA7 | ND | ND | 7876 |
| S65 | Derby | aac(6′)-Iaa | ND | ND | 7119 |
| S66 | I 1,3,19:f,g:1,5 | aac(6′)-Iaa | ND | ND | Unknown |
| S67 | Chester | aac(6′)-Iaa | ND | ND | 411 |
| S69 | Kentucky | aac(6′)-laa; aac(3)-Id; aadA7; aph(3″)-Ib; aph(6)-Id; dfrA15; sul1; tet(A) | GEN; TET; TRISUL(SXT) | ND | 314 |
| S71 | Virchow | aac(6′)-Iaa | ND | IncI1-I (Alpha) | 181 |
| S72 | Amoutive | aac(6′)-Iaa | ND | ND | Unknown |
| S74 | Kentucky | aac(6′)-Iaa; aac(3)-Id; aadA7; aph(3″)-Ib; aph(6)-Id; dfrA15; sul1; tet(A) | GEN; TET; TRISUL(SXT) | ND | 314 |
| S75 | Typhimurium | aac(6′)-Iaa | GEN; TET; TRISUL(SXT) | ColRNAI, IncFIB(S), IncFII(S), IncX1 | 19 |
| S80 | Derby | aac(6′)-Iaa | ND | Col8282, IncFIB (H89-PhagePlasmid) | 5421 |
| S82 | Chester | aac(6′)-Iaa | ND | ND | 411 |
| S83 | Typhimurium | aac(6′)-Iaa | ND | ColRNAI, IncFIB(S), IncFII(S), IncX1 | 19 |
| S86 | Brancaster | aac(6′)-Iaa | ND | ND | Unknown |
| S90 | Gaminara | aac(6′)-Iaa | ND | ND | 2152 |
| S91 | Derby | aac(6′)-Iaa | ND | ND | 7882 |
| S92 | Derby | aac(6′)-Iaa | ND | ND | 7882 |
| S93 | Schwarzengrund | aac(6′)-Iaa | ND | ND | 96 |
| S94 | Derby | aac(6′)-Iaa | ND | ND | 7880 |
| S96 | Farmingdale | aac(6′)-Iaa | ND | ND | Uknown |
| S97 | Derby | aac(6′)-Iaa | ND | ND | 7880 |
| S99 | Anatum | aac(6′)-Iaa | ND | ND | 5197 |
| S102 | Bredeney | aac(6′)-Iaa | ND | ND | 306 |
| S104 | Alexanderplatz | aac(6′)-Iaa | ND | IncFII(S) | Unknown |
| S105 | Derby | aac(6′)-Iaa | ND | ND | 7882 |
| S106 | Rechovot | aac(6′)-Iaa | ND | ND | Unknown |
| S107 | Bredeney | aac(6′)-Iaa | ND | ND | 306 |
| S109 | Derby | aac(6′)-Iaa | ND | ND | 7882 |
| S110 | Eastbourne | aac(6′)-Iaa | ND | ND | 414 |
| S112 | Hato | aac(6′)-Iaa | ND | ND | 3997 |
| S114 | Hato | aac(6′)-Iaa | ND | ND | Unknown |
| S115 | Derby | aac(6′)-Iaa | ND | ND | 7880 |
| S118 | Poona | aac(6′)-Iaa | ND | ND | 308 |
| S120 | Derby | aac(6′)-Iaa | ND | ND | 7882 |
| S121 | Poona | aac(6′)-Iaa | ND | ND | 308 |
| S123 | Bredeney | aac(6′)-Iaa | ND | ND | 306 |
| S124 | Derby | aac(6′)-Iaa | ND | ND | 7880 |
| S125 | Hato | aac(6′)-Iaa | ND | ND | Unknown |
| S126 | Rechovot | aac(6′)-Iaa | ND | ND | Unknown |
| S132 | Derby | aac(6′)-Iaa | ND | ND | 7882 |
| S133 | Derby | aac(6′)-Iaa | ND | ND | 7882 |
| S140 | Alexanderplatz | aac(6′)-Iaa | ND | IncFII(S) | Unknown |
| S143 | Alexanderplatz | aac(6′)-Iaa | ND | IncFII(S) | Unknown |
| S145 | Tennessee | aac(6′)-Iaa | ND | ND | 8398 |
| S147 | Drac | aac(6′)-Iaa | ND | ND | 2221 |
| S148 | Muenster | aac(6′)-Iaa | ND | ND | 321 |
| S149 | Muenster | aac(6′)-Iaa | ND | ND | 321 |
| S150 | Muenster | aac(6′)-Iaa | ND | ND | 321 |
| S151 | Muenster | aac(6′)-Iaa | ND | ND | 321 |
| S152 | Muenster | aac(6′)-Iaa | ND | ND | 321 |
| S153 | Muenster | aac(6′)-Iaa | ND | ND | 321 |
| S154 | Muenster | aac(6′)-Iaa | ND | ND | 321 |
| S155 | Poona | aac(6′)-Iaa | ND | ND | 608 |
| S156 | Poona | aac(6′)-Iaa | ND | ND | 608 |
| S162 | I 1,3,19:b:- | aac(6′)-Iaa | ND | ND | Unknown |
| S163 | Derby | aac(6′)-Iaa | ND | IncFIB (H89-PhagePlasmid) | 3135 |
| S164 | I 1,3,19:b:- | aac(6′)-Iaa | ND | ND | Unknown |
| S165 | Derby | aac(6′)-Iaa | ND | IncFIB (H89-PhagePlasmid) | 3135 |
| S167 | I 1,3,19:b:- | aac(6′)-Iaa | ND | ND | Unknown |
| S168 | Typhimurium | aac(6′)-Iaa | ND | ColRNAI, IncFIB(S), IncFII(S), IncX1 | 19 |
| S169 | Hato | aac(6′)-Iaa | ND | IncFIB (H89-PhagePlasmid) | 3292 |
| S170 | Derby | aac(6′)-Iaa | ND | IncFIB (H89-PhagePlasmid) | 3135 |
| S171 | Derby | aac(6′)-Iaa; fosA7 | ND | Col(pHAD28), IncI1-I (Alpha) | 7881 |
| S172 | Kentucky | aac(6′)-Iaa; dfrA15; sul1 | TRISUL(SXT) | ND | 314 |
| S175 | Hato | aac(6′)-Iaa; aadA1; sul2; tet(A) | TET; TRISUL(SXT) | IncI1-I (Alpha) | 3899 |
| S183 | Derby | aac(6′)-Iaa; aph(3″)-Ib; aph(6)-Id; sul2; tet(A) | MIN; TET | IncFIB (H89-PhagePlasmid), IncQ1 | 3135 |
| S184 | Hato | aac(6′)-Iaa; aph(3′)-Ia; [aph(3′’)-Ib]; aph(6)-Id; dfrA14; sul2 | TRISUL(SXT) | IncI1-I (Alpha) | 3899 |
| S185 | Hato | aac(6′)-Iaa; aadA1; aph(3′)-Ia; dfrA14; sul2; tet(A) | MIN; TET; TRISUL(SXT) | IncI1-I (Alpha) | 3899 |
| S186 | Hato | aac(6′)-Iaa; aadA1; sul2; tet(A) | TET; TRISUL(SXT) | IncI1-I (Alpha) | 3899 |
| S187 | Hato | aac(6′)-Iaa; aadA1; aph(3′)-Ia; aph(6)-Id; sul2; tet(A) | TET; TRISUL(SXT) | IncI1-I (Alpha) | 3899 |
| S188 | Hato | aac(6′)-Iaa; aadA1; aph(3′)-Ia; [aph(3″)-Ib]; sul2; tet(A) | MIN; TET; TRISUL(SXT) | IncI1-I (Alpha) | 3899 |
| S191 | Hato | aac(6′)-Iaa; aadA1; aph(3′)-Ia; aph(6)-Id; dfrA14; sul2; tet(A) | MIN; TET; TRISUL(SXT) | IncI1-I (Alpha) | 3899 |
| S194 | Hato | aac(6′)-Iaa; aadA1; aph(3′)-Ia; [aph(3″)-Ib]; sul2; tet(A) | TET; TRISUL(SXT) | IncI1-I (Alpha) | 3899 |
| S196 | Hato | aac(6′)-Iaa; aadA1; aph(3′)-Ia; [aph(3″)-Ib]; sul2; tet(A) | TET; TRISUL(SXT) | IncI1-I (Alpha) | 3899 |
| S197 | Hato | aac(6′)-Iaa; aadA1; sul2; tet(A) | TET; TRISUL(SXT) | IncI1-I (Alpha) | 3899 |
| S198 | Derby | aac(6′)-Iaa; aph(3′’)-Ib; aph(6)-Id; sul2; tet(A) | MIN; TET | IncQ1 | 3135 |
| S199 | Derby | aac(6′)-Iaa; aph(3′’)-Ib; aph(6)-Id; sul2; tet(A) | MIN; TET | Col(pHAD28), Col8282, IncFIB (H89-PhagePlasmid), IncQ1 | 3135 |
| S200 | Hato | aac(6′)-Iaa; aadA1; aph(3′)-Ia; aph(6)-Id; dfrA14; sul2; tet(A) | TET; TRISUL(SXT) | IncI1-I (Alpha) | 3899 |
| S201 | Hato | aac(6′)-Iaa; aadA1; aph(3′)-Ia; [aph(3″)-Ib]; aph(6)-Id; dfrA14; sul2; tet(A) | TRISUL(SXT) | IncI1-I (Alpha) | 3899 |
| S202 | Hato | aac(6′)-Iaa; aadA1; aph(3′)-Ia; [aph(3″)-Ib]; aph(6)-Id; dfrA14; sul2; tet(A) | TET; TRISUL(SXT) | IncI1-I (Alpha) | 3899 |
| S203 | Hato | aac(6′)-Iaa; aadA1; aph(3′)-Ia; sul2; tet(A) | TET; TRISUL(SXT) | IncI1-I (Alpha) | 3899 |
| S204 | Hato | aac(6′)-Iaa; aadA1; aph(3′)-Ia; aph(6)-Id; sul2; tet(A) | TET; TRISUL(SXT) | IncI1-I (Alpha) | 3899 |
| S207 | Hato | aac(6′)-Iaa; aadA1; aph(3′)-Ia; [aph(3″)-Ib]; aph(6)-Id; dfrA14; sul2; tet(A) | TET; TRISUL(SXT) | IncI1-I (Alpha) | 3899 |
| S208 | Hato | aac(6′)-Iaa; aadA1; aph(3′)-Ia; sul2; tet(A) | TET; TRISUL(SXT) | IncI1-I (Alpha) | 3899 |
| S209 | Hato | aac(6′)-Iaa; aadA1; aph(3′)-Ia; sul2; tet(A) | TET; TRISUL(SXT) | IncI1-I (Alpha) | 3899 |
| S212 | Hato | aac(6′)-Iaa; aph(3′’)-Ib; aph(6)-Id; sul2 | ND | IncI1-I (Alpha) | 3899 |
| S216 | Hato | aac(6′)-Iaa; aadA1; aph(3′)-Ia; sul2; tet(A) | TET; TRISUL(SXT) | IncI1-I (Alpha) | 3899 |
| S219 | Hato | aac(6′)-Iaa; aadA1; aph(3′)-Ia; [aph(3″)-Ib]; sul2; tet(A) | TET; TRISUL(SXT) | IncI1-I (Alpha) | 3899 |
| S248 | Derby | aac(6′)-Iaa; aph(3′’)-Ib; aph(6)-Id; sul2; tet(A) | TET | Col8282, IncFIB (H89-PhagePlasmid), IncQ1 | Unknown |
| S249 | Derby | aac(6′)-Iaa; aph(3′’)-Ib; aph(6)-Id; sul2; tet(A) | TET | Col8282, IncFIB (H89-PhagePlasmid), IncQ1 | Unknown |
| S251 | Derby | aac(6′)-Iaa; aph(3″)-Ib; aph(6)-Id; fosA7; sul2; tet(A) | MIN; TET | Col(pHAD28), IncI1-I (Alpha) | 7881 |
| S252 | Tennessee | aac(6′)-Iaa | ND | ND | 8398 |
| S253 | I 1,3,19:b:- | aac(6′)-Iaa | ND | ND | Unknown |
| S255 | Kentucky | aac(6′)-Iaa; aac(3)-Id; aadA7; aph(3″)-Ib; aph(6)-Id; dfrA15; sul1; tet(A) | GEN; TET; TRISUL(SXT) | ND | 314 |
| Isolate | Serotypes | Plasmid Replicon | pMLST Type a | Antibiotic Resistance Genes |
|---|---|---|---|---|
| S38 | Albany or Dusseldorf | IncI1 | IncI1 ST 12, CC-12 | aph(3′)-Ia; tet(A) |
| S175 | Hato | IncI1 | IncI1 ST 12, CC-12 | aph(3″)-Ib; aph(6)-Id; dfrA14; sul2 |
| S183 | Derby | IncQ1 | NA | aph(3″)-Ib; aph(6)-Id; sul2; tet(A) |
| S184 | Hato | IncI1 and IncQ1 | IncI1 ST 12, CC-12 | aph(3′’)-Ib; aph(6)-Id; dfrA14; sul2 |
| S185 | Hato | IncI1 | IncI1 ST 12, CC-12 | aph(3″)-Ib; aph(6)-Id |
| S186 | Hato | IncI1 | IncI1 ST 12, CC-12 | aph(3″)-Ib; aph(6)-Id; dfrA14; sul2 |
| S187 | Hato | IncQ1 | NA | aph(3″)-Ib; aph(6)-Id; dfrA14; sul2; |
| S188 | Hato | IncI1 | IncI1 ST 12, CC-12 | aph(3″)-Ib; aph(6)-Id |
| S194 | Hato | IncI1 | IncI1 ST 12, CC-12 | aph(6)-Id |
| S196 | Hato | IncI1 | IncI1 ST 12, CC-12 | aph(3″)-Ib; aph(6)-Id; dfrA14 |
| S197 | Hato | IncI1 | IncI1 ST 12, CC-12 | aph(3″)-Ib; aph(6)-Id; dfrA14; sul2 |
| S198 | Derby | IncQ1 | NA | aph(3′’)-Ib; aph(6)-Id; sul2; tet(A) |
| S199 | Derby | IncQ1 | NA | aph(3′’)-Ib; aph(6)-Id; sul2; tet(A) |
| S201 | Hato | IncI1 and IncQ1 | IncI1 ST 12, CC-12 | aph(3″)-Ib; aph(6)-Id; dfrA14; sul2 |
| S202 | Hato | IncI1 and IncQ1 | IncI1 ST 12, CC-12 | aph(3″)-Ib; aph(6)-Id; dfrA14; sul2 |
| S203 | Hato | IncI1 | IncI1 ST 12, CC-12 | aph(3″)-Ib; aph(6)-Id; dfrA14; sul2 |
| S204 | Hato | IncQ1 | NA | aph(3″)-Ib; aph(6)-Id; dfrA14; sul2 |
| S207 | Hato | IncI1 and IncQ1 | IncI1 ST 12, CC-12 | aph(3″)-Ib; aph(6)-Id; dfrA14; sul2 |
| S208 | Hato | IncI1 | IncI1 ST 12, CC-12 | aph(3″)-Ib; aph(6)-Id; dfrA14; sul2 |
| S209 | Hato | IncI1 | IncI1 ST 12, CC-12 | aph(3″)-Ib; aph(6)-Id; dfrA14; sul2 |
| S212 | Hato | IncI1 and IncQ1 | IncI1 ST 12, CC-12 | aph(3′’)-Ib; aph(6)-Id; sul2 |
| S216 | Hato | IncI1 | IncI1 ST 12, CC-12 | aph(3″)-Ib; aph(6)-Id; dfrA14; sul2 |
| S248 | Derby | IncQ1 | NA | aph(3′’)-Ib; aph(6)-Id; sul2; tet(A) |
| S249 | Derby | IncQ1 | NA | aph(3′’)-Ib; aph(6)-Id; sul2; tet(A) |
| S251 | Derby | Col(pHAD28) | NA | aph(3″)-Ib; aph(6)-Id; sul2; tet(A) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kagambèga, A.; McMillan, E.A.; Bouda, S.C.; Hiott, L.M.; Ramadan, H.; Soro, D.K.; Sharma, P.; Gupta, S.K.; Barro, N.; Jackson, C.R.; et al. Resistance Genes, Plasmids, Multilocus Sequence Typing (MLST), and Phenotypic Resistance of Non-Typhoidal Salmonella (NTS) Isolated from Slaughtered Chickens in Burkina Faso. Antibiotics 2022, 11, 782. https://doi.org/10.3390/antibiotics11060782
Kagambèga A, McMillan EA, Bouda SC, Hiott LM, Ramadan H, Soro DK, Sharma P, Gupta SK, Barro N, Jackson CR, et al. Resistance Genes, Plasmids, Multilocus Sequence Typing (MLST), and Phenotypic Resistance of Non-Typhoidal Salmonella (NTS) Isolated from Slaughtered Chickens in Burkina Faso. Antibiotics. 2022; 11(6):782. https://doi.org/10.3390/antibiotics11060782
Chicago/Turabian StyleKagambèga, Assèta, Elizabeth A. McMillan, Soutongnooma C. Bouda, Lari M. Hiott, Hazem Ramadan, Daniel K. Soro, Poonam Sharma, Sushim K. Gupta, Nicolas Barro, Charlene R. Jackson, and et al. 2022. "Resistance Genes, Plasmids, Multilocus Sequence Typing (MLST), and Phenotypic Resistance of Non-Typhoidal Salmonella (NTS) Isolated from Slaughtered Chickens in Burkina Faso" Antibiotics 11, no. 6: 782. https://doi.org/10.3390/antibiotics11060782
APA StyleKagambèga, A., McMillan, E. A., Bouda, S. C., Hiott, L. M., Ramadan, H., Soro, D. K., Sharma, P., Gupta, S. K., Barro, N., Jackson, C. R., & Frye, J. G. (2022). Resistance Genes, Plasmids, Multilocus Sequence Typing (MLST), and Phenotypic Resistance of Non-Typhoidal Salmonella (NTS) Isolated from Slaughtered Chickens in Burkina Faso. Antibiotics, 11(6), 782. https://doi.org/10.3390/antibiotics11060782







