Genomic Changes within a Subset of IncI2 Plasmids Associated with Dissemination of mcr-1 Genes and Other Important Antimicrobial Resistance Determinants
Abstract
1. Introduction
2. Results
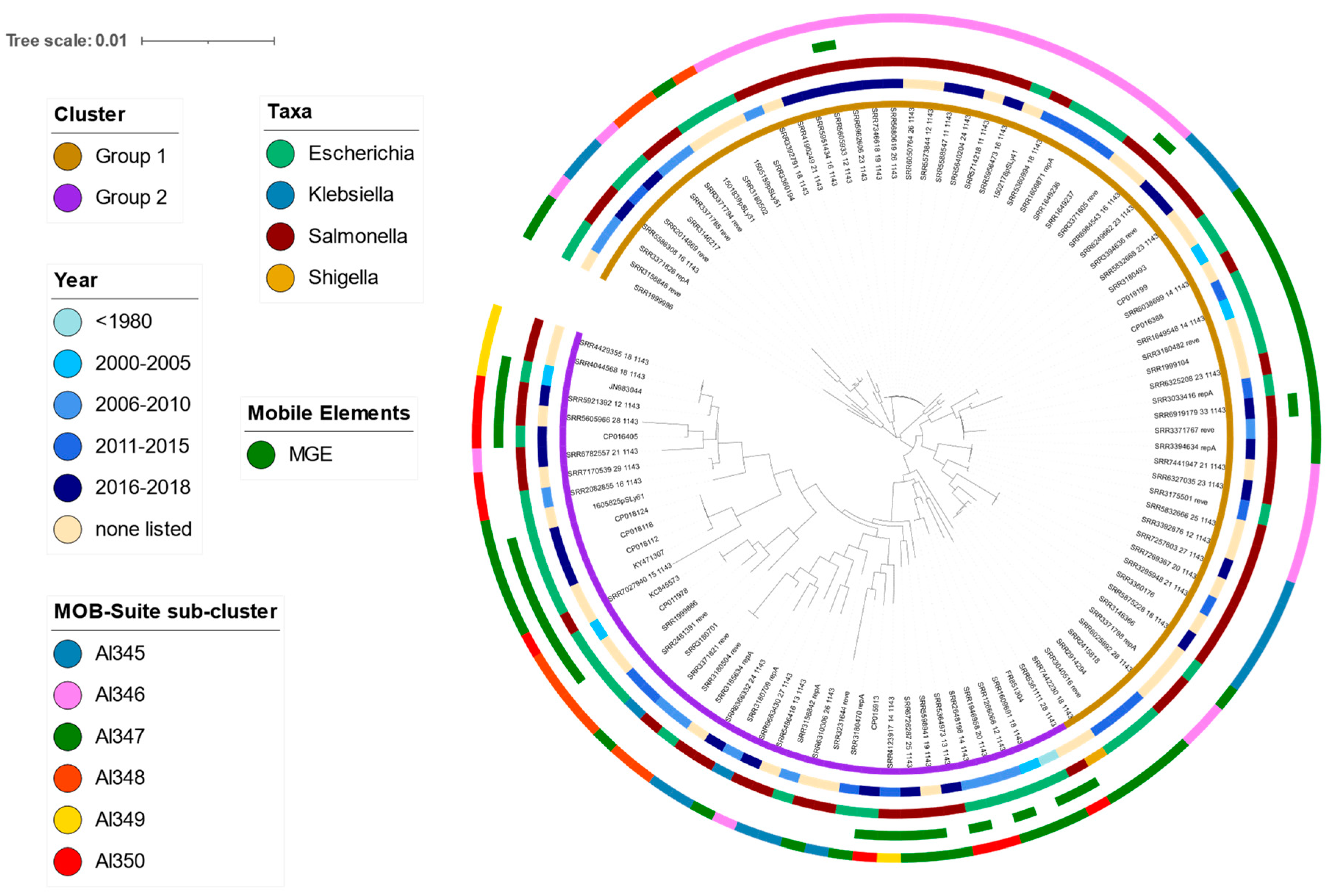
2.1. Global IncI2 Diversity
2.2. Comparison of IncI2 Plasmids from USA Surveillance
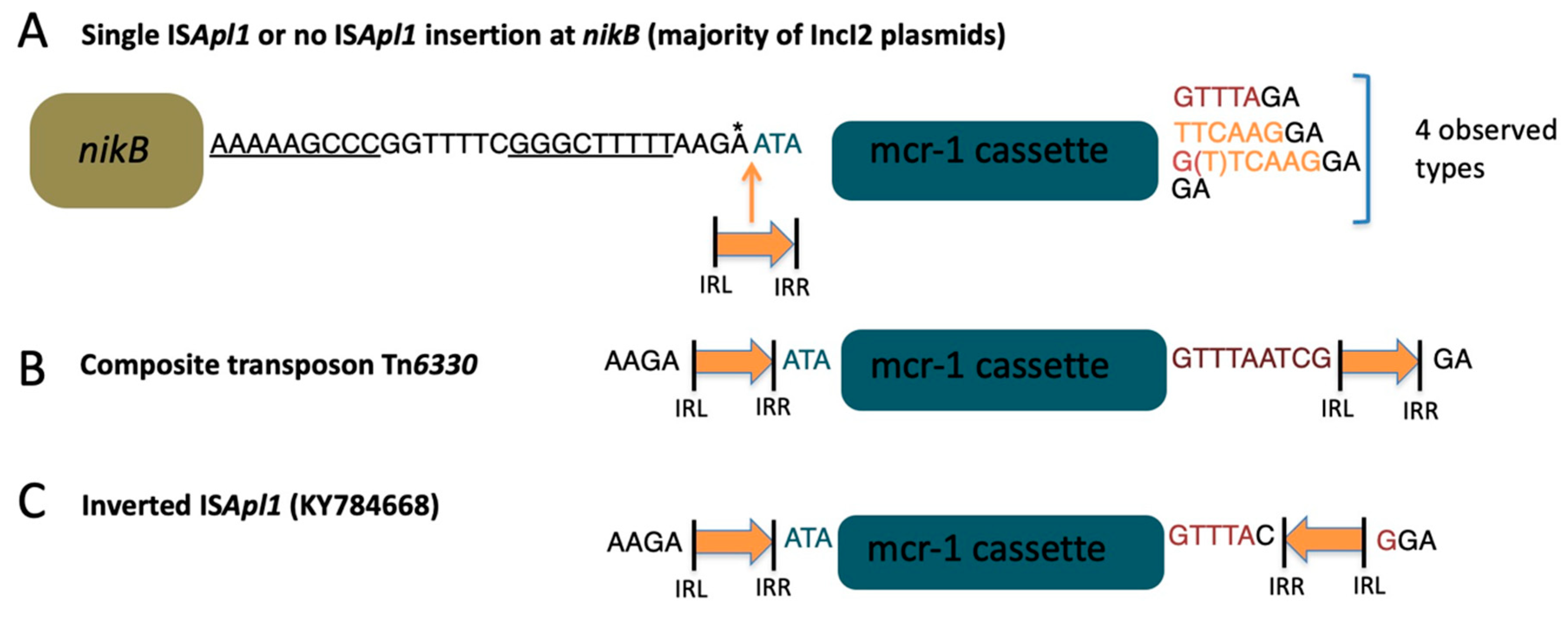
3. Discussion
4. Materials and Methods
4.1. Phylogenetic Reconstruction of IncI2 Plasmids
4.2. Analysis of USA Plasmids
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Douarre, P.E.; Mallet, L.; Radomski, N.; Felten, A.; Mistou, M.Y. Analysis of COMPASS, a New Comprehensive Plasmid Database Revealed Prevalence of Multireplicon and Extensive Diversity of IncF Plasmids. Front. Microbiol. 2020, 11, 483. [Google Scholar] [CrossRef] [PubMed]
- Wong, M.H.; Liu, L.; Yan, M.; Chan, E.W.; Chen, S. Dissemination of IncI2 Plasmids That Harbor the blaCTX-M Element among Clinical Salmonella Isolates. Antimicrob. Agents Chemother. 2015, 59, 5026–5028. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bortolaia, V.; Hansen, K.H.; Nielsen, C.A.; Fritsche, T.R.; Guardabassi, L. High diversity of plasmids harbouring bla(CMY-2) among clinical Escherichia coli isolates from humans and companion animals in the upper Midwestern USA. J. Antimicrob. Chemother. 2014, 69, 1492–1496. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.Y.; Wang, Y.; Walsh, T.R.; Yi, L.X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Rhouma, M.; Letellier, A. Extended-spectrum β-lactamases, carbapenemases and the mcr-1 gene: Is there a historical link? Int. J. Antimicrob. Agents 2017, 49, 269–271. [Google Scholar] [CrossRef]
- Tartor, Y.H.; Abd El-Aziz, N.K.; Gharieb, R.M.A.; El Damaty, H.M.; Enany, S.; Soliman, E.A.; Abdellatif, S.S.; Attia, A.S.A.; Bahnass, M.M.; El-Shazly, Y.A.; et al. Whole-Genome Sequencing of Gram-Negative Bacteria Isolated From Bovine Mastitis and Raw Milk: The First Emergence of Colistin mcr-10 and Fosfomycin fosA5 Resistance Genes in Klebsiella pneumoniae in Middle East. Front. Microbiol. 2021, 12, 770813. [Google Scholar] [CrossRef]
- Baron, S.; Hadjadj, L.; Rolain, J.M.; Olaitan, A.O. Molecular mechanisms of polymyxin resistance: Knowns and unknowns. Int. J. Antimicrob. Agents 2016, 48, 583–591. [Google Scholar] [CrossRef]
- Shen, Z.; Wang, Y.; Shen, Y.; Shen, J.; Wu, C. Early emergence of mcr-1 in Escherichia coli from food-producing animals. Lancet Infect. Dis. 2016, 16, 293. [Google Scholar] [CrossRef]
- Li, R.; Xie, M.; Zhang, J.; Yang, Z.; Liu, L.; Liu, X.; Zheng, Z.; Chan, E.W.; Chen, S. Genetic characterization of mcr-1-bearing plasmids to depict molecular mechanisms underlying dissemination of the colistin resistance determinant. J. Antimicrob. Chemother. 2017, 72, 393–401. [Google Scholar] [CrossRef]
- Poirel, L.; Kieffer, N.; Nordmann, P. In Vitro Study of ISApl1-Mediated Mobilization of the Colistin Resistance Gene mcr-1. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef]
- Snesrud, E.; He, S.; Chandler, M.; Dekker, J.P.; Hickman, A.B.; McGann, P.; Dyda, F. A Model for Transposition of the Colistin Resistance Gene mcr-1 by ISApl1. Antimicrob. Agents Chemother. 2016, 60, 6973–6976. [Google Scholar] [CrossRef] [PubMed]
- Snesrud, E.; McGann, P.; Chandler, M. The Birth and Demise of the ISApl1-mcr-1-ISApl1 Composite Transposon: The Vehicle for Transferable Colistin Resistance. MBio 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Meinersmann, R.J.; Ladely, S.R.; Plumblee, J.R.; Cook, K.L.; Thacker, E. Prevalence of mcr-1 in the Cecal Contents of Food Animals in the United States. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef] [PubMed]
- Meinersmann, R.J.; Ladely, S.R.; Bono, J.L.; Plumblee, J.R.; Hall, M.C.; Genzlinger, L.L.; Cook, K.L. Complete Genome Sequence of a Colistin Resistance Gene (mcr-1)-Bearing Isolate of Escherichia coli from the United States. Genome Announc. 2016, 4, e01283-16. [Google Scholar] [CrossRef] [PubMed]
- Meinersmann, R.J.; Ladely, S.R.; Plumblee, J.R.; Hall, M.C.; Simpson, S.A.; Ballard, L.L.; Scheffler, B.E.; Genzlinger, L.L.; Cook, K.L. Colistin Resistance mcr-1-Gene-Bearing Escherichia coli Strain from the United States. Genome Announc. 2016, 4, e00898-16. [Google Scholar] [CrossRef] [PubMed]
- Meinersmann, R.J. The biology of IncI2 plasmids shown by whole-plasmid multi-locus sequence typing. Plasmid 2019, 106, 102444. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.Q.; Li, Y.X.; Lei, C.W.; Zhang, A.Y.; Wang, H.N. Novel plasmid-mediated colistin resistance gene mcr-7.1 in Klebsiella pneumoniae. J. Antimicrob. Chemother. 2018, 73, 1791–1795. [Google Scholar] [CrossRef] [PubMed]
- Qiao, M.; Ying, G.G.; Singer, A.C.; Zhu, Y.G. Review of antibiotic resistance in China and its environment. Environ. Int. 2018, 110, 160–172. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2007, 23, 127–128. [Google Scholar] [CrossRef] [PubMed]
- Valiakos, G.; Kapna, I. Colistin Resistant mcr Genes Prevalence in Livestock Animals (Swine, Bovine, Poultry) from a Multinational Perspective. A Systematic Review. Vet. Sci. 2021, 8, 265. [Google Scholar] [CrossRef]
- Rhouma, M.; Fairbrother, J.M.; Beaudry, F.; Letellier, A. Post weaning diarrhea in pigs: Risk factors and non-colistin-based control strategies. Acta Vet. Scand. 2017, 59, 31. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Abbas, M.; Rehman, M.U.; Wang, M.; Jia, R.; Chen, S.; Liu, M.; Zhu, D.; Zhao, X.; Gao, Q.; et al. Updates on the global dissemination of colistin-resistant Escherichia coli: An emerging threat to public health. Sci. Total Environ. 2021, 799, 149280. [Google Scholar] [CrossRef]
- Li, X.P.; Sun, R.Y.; Song, J.Q.; Fang, L.X.; Zhang, R.M.; Lian, X.L.; Liao, X.P.; Liu, Y.H.; Lin, J.; Sun, J. Within-host heterogeneity and flexibility of mcr-1 transmission in chicken gut. Int. J. Antimicrob. Agents 2020, 55, 105806. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef] [PubMed]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef] [PubMed]
- Hadfield, J.; Croucher, N.J.; Goater, R.J.; Abudahab, K.; Aanensen, D.M.; Harris, S.R. Phandango: An interactive viewer for bacterial population genomics. Bioinformatics 2018, 34, 292–293. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Lischer, H.E. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Slatkin, M. Linkage disequilibrium in growing and stable populations. Genetics 1994, 137, 331–336. [Google Scholar] [CrossRef]
- Excoffier, L.; Smouse, P.E.; Quattro, J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics 1992, 131, 479–491. [Google Scholar] [CrossRef] [PubMed]
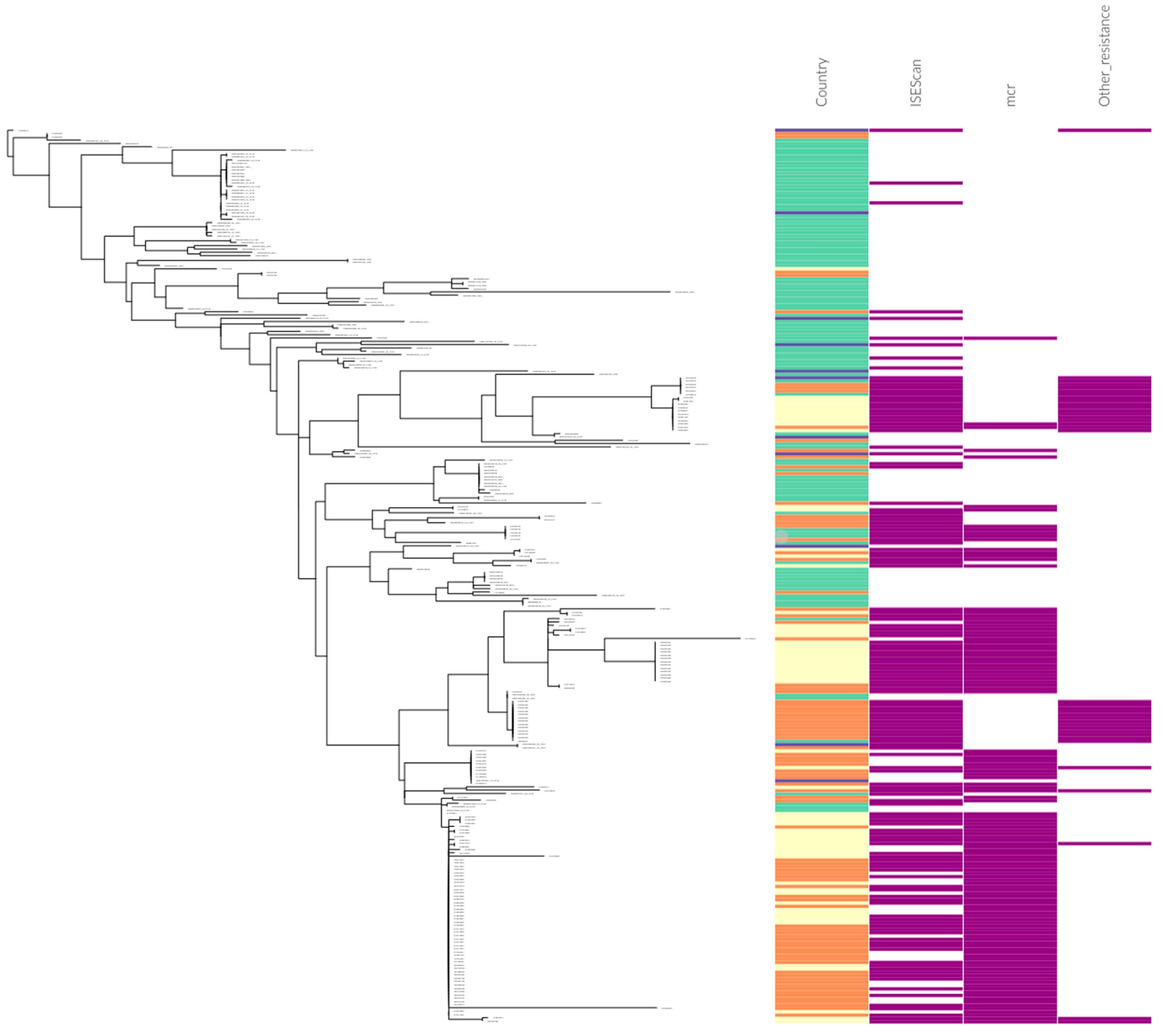
| ISEScan | No AMR | mcr-1 | mcr-7.1_1 | aadA, dfrA | blaCMY-2_1 | blaCTX-M-132_1 | blaCTX-M-15_1 | blaCTX-M-199_1 | blaCTX-M-55_1 | blaCTX-M-64_1 | blaKPC-3, aac(6’)-Ib, aadA, blaOXA-9, blaTEM-1 | blaKPC-3, blaTEM-1 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No IS found | 99 | 32 | - | - | - | - | - | - | - | - | - | - |
| IS1 | - | 2 | - | - | - | - | - | - | - | - | - | - |
| IS1, IS200/IS605 | - | 13 | - | - | - | - | - | - | - | - | - | - |
| IS1, IS30, IS91 | - | 2 | - | - | - | - | - | - | - | - | - | - |
| IS1, IS5 | - | 1 | - | - | - | - | - | - | - | - | - | - |
| IS110, IS3, IS30 | - | 1 | - | - | - | - | - | - | - | - | - | - |
| IS1182, IS21 | - | - | - | - | - | - | - | - | - | - | 3 | 2 |
| IS1182, IS21, IS91 | - | - | - | - | - | - | - | - | - | - | 1 | - |
| IS1380 | - | 3 | 1 | - | 13 | 2 | - | 1 | 6 | 3 | - | - |
| IS1380, IS3, IS30 | - | 1 | - | - | - | - | - | - | 1 | - | - | - |
| IS1380, IS3, IS91 | - | - | - | - | - | - | 1 | - | - | - | - | - |
| IS1380, IS30 | - | 2 | - | - | - | - | - | - | 2 | - | - | - |
| IS200/IS605 | 15 | 11 | - | - | - | - | - | - | - | - | - | - |
| IS200/IS605, IS3 | - | 1 | - | - | - | - | - | - | - | - | - | - |
| IS200/IS605, IS30 | 1 | 9 | - | - | - | - | - | - | - | - | - | - |
| IS200/IS605, IS91 | 1 | 1 | - | - | - | - | - | - | - | - | - | - |
| IS3 | 3 | 3 | - | 1 | - | - | - | - | - | - | - | - |
| IS3, IS30 | - | 1 | - | - | - | - | - | - | - | - | - | - |
| IS3, IS91 | - | 1 | - | - | - | - | - | - | - | - | - | - |
| IS30 | 1 | 20 | - | - | - | - | - | - | - | - | - | - |
| IS30, IS5 | - | 1 | - | - | - | - | - | - | - | - | - | - |
| IS30, IS630 | - | 1 | - | - | - | - | - | - | - | - | - | - |
| IS30, IS66 | - | 2 | - | - | - | - | - | - | - | - | - | - |
| IS30, IS91 | - | 1 | - | - | - | - | - | - | - | - | - | - |
| IS4 | - | 4 | - | - | - | - | - | - | - | - | - | - |
| IS66 | 1 | 1 | - | - | - | - | - | - | - | - | - | - |
| IS91 | - | 6 | - | - | - | - | - | - | - | - | - | - |
| ISEScan Result | No AMR | mcr-1.1 | blaCMY-2 | blaKPC-3, aac(6′)-Ib, aadA, blaOXA-9, blaTEM-1 |
|---|---|---|---|---|
| No IS found | 84 | - | - | - |
| IS1182, IS21 | - | - | - | 2 |
| IS1380 | - | - | 1 | - |
| IS200/IS605 | 8 | 2 | - | - |
| IS200/IS605, IS30 | 1 | 3 | - | - |
| IS200/IS605, IS91 | - | 1 | - | - |
| IS3 | 2 | - | - | - |
| IS30 | 1 | - | - | - |
| IS66 | 1 | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ricker, N.; Chalmers, G.; Whalen, E.; Allen, H.K.; Meinersmann, R.J. Genomic Changes within a Subset of IncI2 Plasmids Associated with Dissemination of mcr-1 Genes and Other Important Antimicrobial Resistance Determinants. Antibiotics 2022, 11, 181. https://doi.org/10.3390/antibiotics11020181
Ricker N, Chalmers G, Whalen E, Allen HK, Meinersmann RJ. Genomic Changes within a Subset of IncI2 Plasmids Associated with Dissemination of mcr-1 Genes and Other Important Antimicrobial Resistance Determinants. Antibiotics. 2022; 11(2):181. https://doi.org/10.3390/antibiotics11020181
Chicago/Turabian StyleRicker, Nicole, Gabhan Chalmers, Elli Whalen, Heather K. Allen, and Richard J. Meinersmann. 2022. "Genomic Changes within a Subset of IncI2 Plasmids Associated with Dissemination of mcr-1 Genes and Other Important Antimicrobial Resistance Determinants" Antibiotics 11, no. 2: 181. https://doi.org/10.3390/antibiotics11020181
APA StyleRicker, N., Chalmers, G., Whalen, E., Allen, H. K., & Meinersmann, R. J. (2022). Genomic Changes within a Subset of IncI2 Plasmids Associated with Dissemination of mcr-1 Genes and Other Important Antimicrobial Resistance Determinants. Antibiotics, 11(2), 181. https://doi.org/10.3390/antibiotics11020181






