Abstract
The use of colistin in food-producing animals favors the emergence and spread of colistin-resistant strains. Here, we investigated the occurrence and molecular mechanisms of colistin resistance among E. coli isolates from a Mexican piglet farm. A collection of 175 cephalosporin-resistant colonies from swine fecal samples were recovered. The colistin resistance phenotype was identified by rapid polymyxin test and the mcr-type genes were screened by PCR. We assessed the colistin-resistant strains by antimicrobial susceptibility test, pulse-field gel electrophoresis, plasmid profile, and mating experiments. Whole-Genome Sequencing data was used to explore the resistome, virulome, and mobilome of colistin-resistant strains. A total of four colistin-resistant E. coli were identified from the cefotaxime-resistant colonies. All harbored the plasmid-borne mcr-1 gene, which was located on conjugative 170-kb IncHI-2 plasmid co-carrying ESBLs genes. Thus, high antimicrobial resistance rates were observed for several antibiotic families. In the RC2-007 strain, the mcr-1 gene was located as part of a prophage carried on non-conjugative 100-kb-plasmid, which upon being transformed into K. variicola strain increased the polymyxin resistance 2-fold. The genomic analysis showed a broad resistome and virulome. Our findings suggest that colistin resistance followed independent acquisition pathways as clonal and non-genetically related mcr-1-harboring strains were identified. These E. coli isolates represent a reservoir of antibiotic resistance and virulence genes in animals for human consumption which could be potentially propagated into other interfaces.
1. Introduction
Colistin represents the last line of therapeutic options against multidrug-resistant strains of Enterobacterales. However, unregulated overuse of colistin in the animal sector as a growth promoter and for preventing infections in food-producing animals has favored the emergence of colistin-resistant strains [1]. Colistin resistance is associated with LPS lipid A modifications, which can be attributed to either chromosomal mutations in regulatory two-component systems or the plasmid-borne mcr gene variants [2].
Plasmid-borne mcr genes have been found in a variety of plasmid types such as IncI2, IncHI2, IncX4, IncP, IncF and the less common IncY [3]. These plasmids are transferable with a broad host range, which could explain the widespread of mcr genes among different bacterial species [4]. Ten variants of the mcr gene have so far been described [5], but the most widespread is the mcr-1 gene [6]. This gene has been identified in isolates from numerous sources including humans, food, farms and wild animals [7,8]. It has been demonstrated that the transfer of the mcr-1 gene in enterobacteria from farm animals to humans through the food chain [9]. Notably, mcr genes are found in association with other antimicrobial resistance genes, usually extended-spectrum-ß-lactamases (ESBL) and carbapenemases, highlighting the emergence of pan-drug resistant Enterobacterales [6].
Moreover, phages also play a role in the dissemination of antimicrobial resistance genes [10]. Some phages have the ability to integrate into plasmids and can be transferred to other bacteria [11]. Particularly, phage-like plasmids carrying the mcr-1 gene have been described in Klebsiella pneumoniae and Escherichia coli [3,12].
In Mexico, polymyxins, as well as other antibiotics, have not been approved as growth promoters since 2012, and there are few reports of isolates carrying the mcr-1 gene [6]; nevertheless, this gene has been identified from clinical [13] and swine farm [14] environments. In addition, it has also been described in cantaloupes [15]. This study provides insight into the molecular mechanisms of colistin resistance circulating in a Mexican piglet farm and by genomic analysis, we explore the resistome, virulome and mobilome of mcr-1-producing E. coli isolates.
2. Results
2.1. Occurrence of Colistin Resistance on a Swine Farm
Of the stool samples plated on agar plates supplemented with cefotaxime, we randomly selected one colony for each plate. The frequency of cefotaxime-resistant colonies was 34.4% (175/508). Among 175 cefotaxime-resistant colonies, four colistin-resistant (C072, C2-033, C2-107 and C2-108) were identified by rapid polymyxin NP test and were confirmed as E. coli according to the biochemical test kit API20E. The plasmid-borne mcr-1 gene was identified in the four E. coli colistin-resistant isolates (Table 1). The C072 isolate was collected in March 2015 and the C2-033, C2-107 and C2-10 isolates, in September 2015. All mcr-1-positive isolates including the RC2-007 isolate showed a colistin MIC of 4 μg/mL. The RC2-007 isolate was previously described from the same farm among 928 E. coli strains [14]. PFGE analysis demonstrated that C2-107 and C2-108 isolates were the same clone and the rest were genetically unrelated. All colistin-resistant isolates were also identified as ESBL-producers and were resistant to ampicillin, ceftazidime, cefotaxime, gentamicin, nalidixic acid and tetracycline but susceptible to imipenem, amikacin and ciprofloxacin. The RC2-007 isolate, however, showed greater resistance to ceftazidime, gentamicin and ciprofloxacin (Table 1).

Table 1.
Molecular characteristics of colistin-resistant E. coli isolates recovered from piglet fecal samples.
2.2. Plasmid Analysis of mcr-1-Harboring E. coli Isolates
The plasmid-borne mcr-1 gene was identified on a ∼170-kb plasmid shared among the C072, C2-033, C2-107 and C2-108 isolates (Table 1). The mating experiments were successful in the isolates that carried the 170-kb plasmid with different conjugation efficiencies (Table 1). In contrast, as the RC2-007 was unable to mobilize the mcr-1 gene, we performed transformation experiments using the E. coli DH10B and K. variicola F2R9 isolates as receptors (to evaluate the mcr-1 gene in a different genetic background). The plasmid that was successfully transferred in both receptors (TpEcoDH10B and TpKvF2R9) was the ~100-kb plasmid (Table 1).
The transconjugants T-C072, T-C2033, T-C2-107, T-C2-108 were confirmed to have decreased colistin susceptibility by MIC determination. Transconjugants exhibited a MIC for colistin of 2 and 1 μg/mL; otherwise, the TpEcoDH10B and TpKvF2R9 transformants exhibited a MIC 2 and 16 μg/mL, respectively (Table 1). In addition, all mcr-1 positive isolates also were ESBL-producers and in all cases the ESBLs CTX-M-type were identified by PCR amplification (Table 1). In the C072, C2-033, C2-107 and C2-108 isolates the CTX-M-type genes are contained in the same plasmid as the mcr-1 gene. In contrast, the colistin and cephalosporin resistance exhibited by the RC2-007 isolate were encoded on different plasmids (Table 1).
2.3. Whole Genome Analysis of mcr-1-Producing E. coli Isolates
Genome features of C072, C2-033, C2-107 and C2-108 isolates are described in Supplementary Table S1. The C072 and C2-033 isolates belonged to distinct sequence types (ST), and the isolates C2-107 and C2-108 were ST1286. In the same way, serotypes varied across the strains. ClermonTyper determined that the majority of the mcr-1-positive strains were grouped into phylogroups from commensal strains (group A or B1) but C2-033 belonged to group F (Table 2).

Table 2.
Typing results of E. coli isolates collected from piglets.
In order to explore the genetic relationship of the five mcr-1 positive isolates from this study, we compared them with E. coli isolates from other sources and/or isolates carrying the mcr-1 gene that have the same ST. For this purpose, we performed a minimum spanning tree (MSP) based on cgMLST (Figure 1). Using a threshold of less than 25 allele differences, only the C072 isolate was found to be genetically related to the VREC0606 isolate derived from turkey feces: this isolate was not mcr-1-producing E. coli. Then, by using a threshold of 50 and 100 allele differences, we identified that C2-107 and C2-108 demonstrated a 33-allele difference to the closest EC_13 isolate, derived from chicken infection and the RC2-007 isolate was related with isolates from pig rectum which were mcr-1-producing. No close genetic relationship was found for the C2-033 isolate.
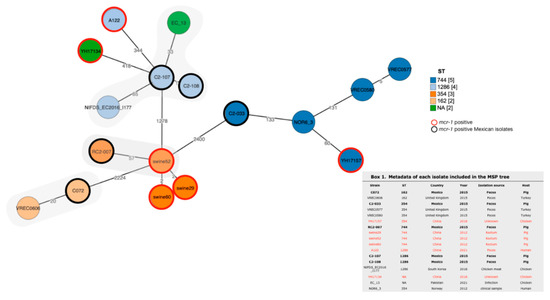
Figure 1.
Minimum spanning tree based on cgMLST comprising 2513 genes of E. coli. Each group is formed by isolates with the same ST as marked by color code. This analysis includes both mcr-1-producing and -non-producing but genetically related to C072, C2-107, C2-108, C2-033 and RC2-007. Numbers between lines indicate allele differences between isolates. Gray shadows represent closely related isolates. An information box containing metadata for each isolate is included.
Consistent with their multi-drug resistant phenotype, the isolates showed an extensive resistome, with genes and chromosomal mutations that confer resistance to several antibiotic families as aminoglycosides, tetracyclines, florfenicol, chloramphenicol, trimethoprim, fosfomycin, macrolides, sulfonamides, lincosamide, β-lactam, quinolones and fluoroquinolones (Figure 2). The main ESBLs corresponded to CTX-M-14, followed by CTX-M-55, and TEM-116. In all isolates the chromosomal mutations in the quinolone resistance-determining region (QRDR) in GyrA and ParC proteins were identified (Supplementary Table S2). Genes encoding for resistance to some heavy metals were found except for copper and lead (Figure 2). Replicon typing showed diverse incompatibility groups (Table 2), the IncHI2 and IncN were shared among C072, C2-033, C2-107 and C2-108 isolates; however, the Incp0111 was identified in the RC2-007 isolate (Table 2).
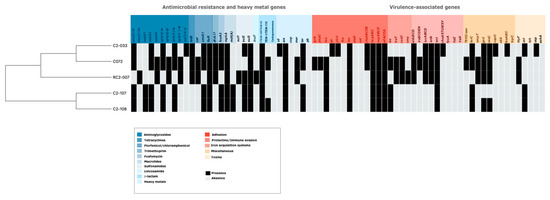
Figure 2.
Repertoire of antimicrobial resistance and virulence genes among mcr-1-producing E. coli isolates from fecal piglet samples. Blue colors represent different antibiotic families and red colors are indicative of virulence-associated genes involved in pathogenesis. Abbreviations for heavy metals correspond to sil: silver, ars: arsenate, cop: copper, mer: mercury, ter: tellurium, and pb: lead.
The genetic context of the mcr-1 gene was analyzed; however, the ISApI1 upstream of mcr-1-PAP2 genes could not be resolved by whole genome sequencing (except for RC2-007), thus we verified its presence by PCR and sequencing (Table 2). The whole-genome analysis revealed that the ISApI1-mcr-1-PAP2 genetic structure in the 170-kb plasmid from C072, C2033, C2-107 and C2-108 is conserved across these isolates (Figure 3). For these isolates, downstream from the ISApI1-mcr-1-PAP2 region we found a Cys-tRNA, the tellurium resistance cluster, and the HigAB toxin-antitoxin system (Figure 3). No other antimicrobial resistance genes were found in this region. However, we detected genes conferring resistance to aminoglycosides, chloramphenicol, florfenicol and quaternary ammonium located in class I integron (data not shown). In the case of the RC2-007 isolate the mcr-1 gene was contained in an intact phage of 79,052 bp identified in a contig of 80,111 bp. In this contig, we also found the pdh/doc toxin-antitoxin system (data not shown).
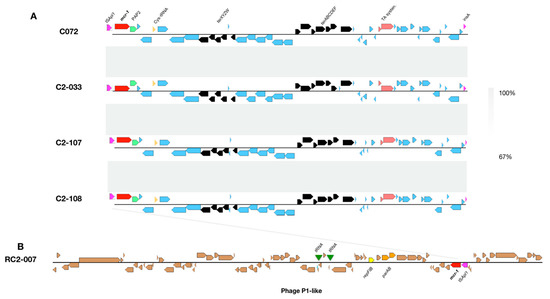
Figure 3.
(A). Genetic contexts of the mcr-1 gene among E. coli from piglet fecal samples. Light grey shadows show BLAST identity percentage. (B). Genetic structure of the phage P1-like carrying the mcr-1 gene. Each CDS is represented as an arrow frame.
Finally, 40 virulence-associated genes that are present in the ExPEC pathotype were screened in the five mcr-1-harboring strains. The highest numbers of ExPEC virulence factors were detected in C2-033 and C072 (Figure 2). However, C072 possessed more virulence genes (22/40) than the remaining strains whereas C2-107 and C2-108 contained less virulence factors (Figure 2). Mobile genetic elements (MGEs) linked with some virulence genes were identified in the C072 and RC2-007 isolates. In the case of C072, we found that ompT, etsC, and hlyF virulence genes were located within the IncFIB plasmid and were associated with the IS629; in contrast, papC, terC and hra were identified in the chromosome possibly located in a genomic island and associated with IS630 and IS609, respectively. Similarly, the RC2-007 contained the virulence genes hlyF, ompT, cvaC, and cma, within an IncFIB plasmid in which the CTX-M-55 was also detected. Other MGEs were detected like the IS629 linked with the Hra adhesin and the ISEc9 linked with protectins.
2.4. In Vitro and In Silico Analysis of Phage Carrying mcr-1
Mitomycin C treatment of the E. coli RC2-007 isolate resulted in phage induction. The phage was purified and analyzed by electronic microscopy (Supplementary Figure S2). However, purified genetic material was negative for the mcr-1 gene by PCR, and consequently the induced phage did not correspond to the phage-like plasmid carrying the mcr-1 gene. The induced phage could correspond to a phage integrated into the chromosome or another plasmid.
The genetic structure of the phage-like plasmid on the pRC2-007 (100-kb) plasmid was compared with other phage-like plasmids carrying mcr-1 genes (Supplementary Figure S3). Two E. coli isolates (WZR78, GD27-62) [16] and one K. pneumoniae (F160070) isolate (MG288678.1) were selected based on BLAST identity. The isolates E. coli WZR78 harboring the pZR78 plasmid and GD27-62 harboring the pGD27-62 plasmid were recovered from a pig farm and chicken gut respectively. The K. pneumoniae F160070 harboring the p160070-MCR plasmid was recovered from food in China. The genetic comparison among these phage-like plasmids carrying the mcr-1 gene showed a >95% nucleotide identity and the pZR78 (91,281 bp), GD27-62 (92,362 bp) and p160070-MCR (97,393 bp) with similar molecular sizes also have a >95% identity. We did not identify additional resistance genes in the pRC2-007 (100-kb) plasmid. However, the E. coli RC2-007 was described as an MDR isolate and an additional conjugative 120-Kb plasmid that transfers the cephalosporin resistance was described. Otherwise, a P7 phage-like plasmid carrying the mcr-1 gene was described in a K. pneumoniae clinical isolate. The mcr-1 was carried by a 97.4-kb pMCR_SCKPLL83 plasmid, which did not carry any additional known antimicrobial resistance genes. The conjugation and transformation of pMCR_SCKPLL83 plasmid were unsuccessful using E. coli as recipient cells.
2.5. Phages and Prophages Carrying mcr-1 Gene in Public Databases
We sought to determine the distribution of phages and prophages harboring the mcr-1 gene from genome assemblies deposited in the GenBank database. A total of 13,557 and 6603 E. coli and K. pneumoniae genomes were obtained from the RefSeq database, respectively (Supplementary Table S3). In E. coli, 689 and 12,868 with the level of complete and draft genomes, respectively, were identified. Meanwhile, in K. pneumoniae 306 complete and 6297 draft genomes were identified. The mcr-1 gene was detected in a total of 868 (6.4%) of E. coli and 45 of K. pneumoniae (0.6%) genomes. The in-silico prediction of phages and prophages carrying the mcr-1 gene in E. coli genomes were 10 intact (Cat-1) and 12 incomplete (Cat-2) phages. Nevertheless, both phages (342) and prophages (214) in the categories Cat-3 and Cat-6 (questionable) were identified. In addition, four incomplete prophages (Cat-5) were also identified in E. coli (Supplementary Table S3). A total of 30 phages and prophages carrying the mcr-1 gene in their different categories were identified in K. pneumoniae (Supplementary Table S3). An intact (Cat-1) and an incomplete (Cat-2) phage were identified, corresponding to SCKP-LL83 and SCKP020138 isolates. Likewise, as in E. coli, the number of questionable phages (25) and prophages (3) were mostly identified in K. pneumoniae. These data showed that at least in E. coli genomes, a high percentage contain the mcr-1 gene. Likewise, the identification of phages or prophages in any of the categories that contain the mcr-1 gene is high (59.6%—518/868). This percentage of positive-mcr-1 genomes decreases in K. pneumoniae, although the percentage of phages and prophages carrying the mcr-1 gene is high (66%—30/45). The replicon containing the phages and prophages carrying the mcr-1 gene in their different categories was not determined.
3. Discussion
In this work, we performed a survey of fecal isolates from a swine farm that is exclusively dedicated to the sale and breeding of piglets. As mcr-1 co-occurs frequently with cephalosporin-resistance phenotype [17], we cultured the fecal samples on a medium supplemented with cefotaxime. Among the cefotaxime-resistant isolates, four E. coli colistin-resistant were identified as being carriers of the plasmid-borne mcr-1 gene. In addition, we included the colistin-resistant and ESBL-producing RC2-007 isolate previously described in the same farm [14]. Our results suggest that the prevalence of E. coli isolates carrying the mcr-1 gene is low, and is higher in ESBL-producing isolates. The mcr-1-positive isolates were obtained only from piglets (Table 1), whereas the adult females and stallions were negative for colistin-resistant E. coli strains. Further, we collected one mcr-1-carrying isolate in march 2015 (C-072) and the other four in September 2015; which suggests that despite the low prevalence, the dissemination of the mcr-1 is maintained in the farm, especially in pigs with a few days old.
Colistin has been widely used in poultry and pig farms and its use is linked with high rates of resistance [6]. However, in this work the occurrence of colistin resistance was low; this may imply that colistin is not used on the animals as an antimicrobial treatment or growth promoter. As we could not find mcr-1-colonizing strains from the mother, this may suggest that the acquisition of colistin-resistant strains comes from other sources in the environment. For the previously described RC2-007 isolate, the genetic context of the mcr-1 gene was found in a phage contained on a non-conjugative plasmid; however, by mitomycin C treatment we could not induce this phage. This plasmid could transfer the colistin resistance phenotype by transformation; moreover, transformants exhibited a high MIC for K. variicola (Table 1). This data suggest that the mcr-1 gene could confer greater resistance to colistin in this bacterial species. Colistin-resistant K. variicola isolates have been described [18], and recently one isolate of K. variicola positive for the mcr-1 gene was identified in China [19].
Here, three mcr-1 positive isolates were genetically unrelated and two were clonally related as demonstrated by PFGE analysis and the MSP tree (Figure 1 and Table 1); thus, colistin resistance followed independent acquisition pathways: plasmid-mediated, clonal dissemination, and phage-mediated (Figure 1 and Table 1). However, the self-transmissible 170-kb IncHI2 plasmid had a major distribution in the mcr-1 positive isolates (Table 1). This is consistent with the fact that the spread of the mcr-1 gene in Latin America seems to be more related to the spread of mcr-1 carrying plasmids among E. coli isolates than to the clonal expansion of MCR-1 producing isolates [6]. The most prevalent mcr-1 carrying plasmids in Latin America are the IncX4- and IncI2-types. These Inc-types were not identified in this study. Nevertheless, the IncI2 was reported in a clinical isolate from Mexico [13]. The spreading of the mcr-1 gene in E. coli from the veterinary sector in Mexico could be determined by the dissemination of plasmids different from those in the clinical setting, such as the IncHI2-type plasmids [6]. Interestingly, we identified toxin-antitoxin systems in the genetic contexts of all mcr-1 positive isolates, suggesting that these plasmids could be maintained even in the absence of selection pressure.
To place in a global context, the five mcr-1-positive isolates were compared with E. coli isolates from other regions. The results support the role of the mcr-1-carrying plasmid, as we identified a genetic relationship with non mcr-1-producing isolates from turkey and chicken (Figure 1). Only one E. coli isolate (RC2-007) was closely related to an mcr-1-producing E. coli isolate (swine52), but its mechanism may not be associated with a phage-like plasmid (data not shown)
Unfortunately, we were not provided information about antibiotics administered, age of piglet sale, strategies of waste management or cleaning, but piglets are fed with food enriched with enrofloxacin probably to prevent urinary or intestinal infections. This seems to be related to resistance to quinolone and fluoroquinolones (Table 1 and Figure 2).
The presence of virulence-associated genes such as those coding for colonizing factors (fimbriae and adhesins), survival of bacterial cells (protectins and siderophores) or causing the host inflammatory response, e.g., toxin production [20] were determined in the five mcr-1-harboring E. coli (Figure 2). The isolates C072 and C2-033 exhibited more virulence-associated genes; this is consistent with their phylogroups, B1 and F, respectively. The strains from phylogroup F are considered highly virulent pathotypes and phylogroup B1 are intestinal pathogenic E. coli [21,22]. Moreover, the C072 isolate showed genes coding for a TSS6, which has an important role on virulence (Figure 2) [23].
The RC2-007 also possessed a vast distribution of ExPEC virulence factors (Figure 2). Although phylogroup A is associated with commensal strains, some pathogenic E. coli fall into group A, and this might be the case for RC2-007. Moreover, some virulence factors were found on IncFIB plasmids in some cases associated with insertion sequences; this shows that IncFIB plasmids are disseminating antibiotic resistance such as CTX-M-type and virulence genes. Correlation between virulence and antimicrobial resistance has been observed in mcr-1 positive strains [24]. Finally, our results showed that phages carrying the mcr-1 gene are more common in E. coli than in K. pneumoniae. A deeper analysis of the genomic structure of the phages as important vehicles for disseminating colistin resistance deserves study.
Although we found a few colistin-resistant strains probably from environmental contamination by mcr-1-harboring E. coli, this represents a reservoir of antibiotic resistance and virulence genes in animals for human consumption that could be potentially propagated into other interfaces. Finally, the mcr-1-producing E. coli strains fall into virulent pathotypes which could represent harm to human health.
4. Material and Methods
4.1. Sampling and Bacterial Selection
A semi-tech swine farm located in Jiutepec, Morelos at 90 km from Mexico City was selected to carry out this study. The sampling was performed in the total swine population at two stages, stage 1 in March 2015 and stage 2 in September 2015. Overall, 280 swine from stage 1 (163 piglets, 112 females and five stallions) and 228 from stage 2 (123 piglets, 99 females and six stallions) were sampled. In each stage the swine stools were collected and transported in Cary-Blair medium (DELTALAB, Barcelona, Spain) at 4 °C. All stool samples (508) were cultured on MacConkey agar plates supplemented with 1 µg/mL of cefotaxime at 37 °C for 24 h.
The RC2-007 isolate was recovered from the same farm but it was part of a different bacterial collection [14]. At that time (2016), phenotypic tests for colistin resistance detection were not available, and so screening of potential colistin-resistant strains in that particular collection was performed directly by PCR [14]. The RC2-007 isolate was further characterized together with the colistin-resistant isolates found in the present study.
4.2. Identification of Colistin Resistant and Screening of Colistin and Cephalosporin Resistant Genes
All E. coli phenotype colonies were analyzed using the Rapid Polymyxin NP assay [25] by determining the colistin-resistant phenotype. The plasmid-borne mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 genes and the ß-lactamases of the family CTX-M-type genes were screened by PCR using generic primers (Supplementary Table S4). The colistin-resistant isolates were analyzed by API20E.
4.3. Antimicrobial Susceptibility Testing
For colistin-resistant isolates, the minimal inhibitory concentration (MIC) for colistin using the broth microdilution procedure according to the EUCAST breakpoints was determined [26]. The ESBL-producer isolates were determined by double disc synergy according to CLSI [27]. Additionally, the MIC of ampicillin (AMP), ceftazidime (CAZ), cefotaxime (CTX), imipenem (IMP), amikacin (AK), gentamicin (GEN), ciprofloxacin (CIP), nalidixic acid (NAL) and tetracycline (TET) were determined according to CLSI standards [27].
4.4. PFGE Analysis
The genetic relatedness of isolates was examined by pulsed-field gel electrophoresis (PFGE) [28]. The results were analyzed using GelCompar II software (Applied Maths, Kortrijk, Belgium).
4.5. Plasmid Profile Determination, Mating and Transformation Experiments
Plasmid DNA profiles were obtained from the isolates according to the method described by Kieser [29] E. coli NCTC 50192 plasmids 154-, 66-, 48- and 7-kb were used as molecular size markers [30]. The linear regression equation was used for molecular weight plasmid calculation. Colistin resistance transfer was evaluated by mating and transformation experiments [31,32]. The conjugation assays were performed using the E. coli strain J53-2 and co-selected with 100 μg/mL rifampicin and 32 μg/mL of potassium tellurite. Since the ter operon was identified in the genetic context of mcr-1 (Supplementary Figure S1) and considering that colistin has a low diffusion in solid media, we decided to use tellurite resistance as a phenotypic selection marker in conjugation assays. On the other hand, the pRC2-007 plasmid was extracted using ion-exchange columns (Qiagen, Hilden, Germany). The plasmid was electroporated to E. coli DH10B (Amersham, Darmstadt, Germany) and Klebsiella variicola F2R9 [33]. Transformants were selected on agar plates supplemented with 8 µg/mL of colistin.
4.6. Whole Genome Sequencing and In Silico Analysis
Total genomic DNA was extracted and purified using the DNeasy Kit (Qiagen, Hilden, Germany). Whole-genome sequencing of bacterial and phage samples was generated using the Illumina (MiSeq) platform. Quality-based trimming was performed with the SolexaQA software and de novo assembly was done with SPAdes v3.1.1. The contigs were subjected to a scaffolding process with SSpace v2.0. Gene prediction and annotation were carried out using the bioinformatic MicroScope platform [34].
The Multilocus Sequence Typing (MLST), serotype and incompatibility group (Inc) of plasmids were determined in silico (http://www.genomicepidemiology.org, accessed on 15 October 2021) and prediction of closely related plasmids were obtained by BacWGSTdb (http://bacdb.cn/BacWGSTdb/, accessed on 5 July 2021) Virulence genes were determined by three platforms: VirulenceFinder 2.0 [35], VFDB [36] and BacWGSTdb [37] (http://bacdb.cn/BacWGSTdb/, accessed on 7 July 2021). Antimicrobial resistance genes were determined by ResFinder (http://www.genomicepidemiology.org, accessed on 15 July 2021) and BacWGSTdb (http://bacdb.cn/BacWGSTdb/, accessed on 12 October 2021). Phylogroup assignation was performed by the ClermonTyper web platform (http://clermontyping.iame-research.center, accessed on 17 October 2021) [38]. MobileElementFinder (http://www.genomicepidemiology.org, accessed on 25 October 2021) was used to detect mobile genetic elements associated with antibiotic resistance and virulence genes. The toxin-antitoxin systems HigAB and pdh/doc were screened by BLASTn search (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 25 October 2021). Finally, the prophage structure was determined by the PHASTER program (https://phaster.ca, accessed on 25 October 2021). Plasmids and prophage with similar structures to the pRC2-007 were searched by BLASTn using >90% of nucleotide identity. Alignment of plasmids that contained the mcr-1 gene inside a prophage and genetic structures of mcr-1 was generated by Easyfig win_2.2 [39].
4.7. Comparative Analysis of mcr-1 E. coli Isolates
To explore the genetic relatedness between Mexican mcr-1 positive isolates and isolates from other sources, we performed a core genome MLST (cgMLST) analysis. A minimum spanning tree (MST) was constructed including isolates with the same sequence type, closely related and positive to the mcr-1 positive isolates which were identified by the BacWGSTdb 2.0 [37]. The threshold for cluster determination was 25 alleles according to BacWGSTdb 2.0; however, this cut-off point was not stringent when we could not find close relatives.
4.8. In Silico Identification of Phages and Prophages Carrying mcr-1 Genes
The E. coli and K. pneumoniae genomes were obtained from the RefSeq Database (https://www.ncbi.nlm.nih.gov/refseq/) accessed on 1 November 2020. The mcr-1 gene was screened by the ggsearch36 program in all E. coli and K. pneumoniae genomes obtained from RefSeq Database (https://www.ncbi.nlm.nih.gov/refseq/) accessed on 5 November 2019. Subsequently, the prediction of phages and prophages was made in those genomes positive for mcr-1 using the VirSorter program and the Refseqdb and Viromedb databases [40]. Finally, those genomes having the mcr-1 gene within the phage or prophage prediction were selected and classified according to six categories (Cat). Cat-1 and Cat-2 correspond to intact and incomplete phages, respectively. Cat-3 and Cat-6 correspond to questionable phages or prophages and Cat-5 corresponds to incomplete prophages.
4.9. Bacteriophage Induction
The bacteriophage induction assay was carried out with mitomycin C. In 50 mL of fresh culture (at a cell density corresponding to 2 on the McFarland scale) of strain RC2-007, mitomycin C was added to a concentration of 0.5 μg/mL and the culture was incubated under 37 °C with shaking. The decrease in the absorbance at 600 nm was determined every hour for 6 h. Next, the supernatant was obtained by centrifugation at 3000× g for 12 min at 4 °C, was neutralized to pH 7.0 with 0.1N NaOH, and then filtered through a 0.22 μm pore membrane filter. Finally, supernatant containing bacteriophages was stored at 4 °C.
The assay was performed in triplicate. The phage DNA was extracted and purified using the DNeasy Kit (Qiagen, Hilden, Germany) and the mcr-1 and a bacterial chromosomal control gene (yjaA) were screened by PCR using generic primers (Supplementary Table S4).
In addition, electron microscopy was carried out in a drop of medium containing the phage placed on a 200 mesh copper grid f coated with formvar/carbon (Electron Microscopy Sciences, Washington, PA, USA). The excess liquid was removed and the grid was negatively stained for 5 min with 2.5% uranyl acetate (aqueous solution) and washed several times with ultrapure water. Micrographs were obtained on a transmission electron microscope (JEOL 1011) operated at 80 kV.
4.10. Nucleotide Accession Numbers
The annotated genome sequences are available at the GenBank under the following accession numbers: JAIQXZ000000000 for E. coli C072, JAIQYA000000000 for E. coli C2033, JAIQYB000000000 for E. coli C2-107 and JAIQYC000000000 for E. coli C2-108.
5. Conclusions
In Mexico, there are few studies both in the clinical environment and in the food production chain that monitor resistance to colistin. This study reports the molecular and genomic traits of mcr-1-harboring E. coli from piglets. Our results show that colistin resistance followed independent acquisition pathways, but a common conjugative 170-kb plasmid was identified in the majority of isolates. The genetic relatedness of piglet E. coli isolates determined by cgMLST with non mcr-1-producing isolates derived from avian hosts supports the horizontal transfer of colistin resistance. The phage-like plasmid carrying mcr-1 gene was less common in the farm. The structure of this phage-like plasmid was identified in other plasmids obtained from E. coli and K. pneumoniae recovered from non-clinical samples in China. The phages and prophages carrying mcr-1 in E. coli and K. pneumoniae genomes could be closely related to the dissemination of the colistin resistance. The presence of CTX-M-type genes carried by the mcr-1-producing E. coli isolates implies the potential co-selection of mcr-1 positive isolates with the use of cephalosporins. The co-occurrence of ESBLs and virulence genes in mcr-1-carrying isolates may favor reservoirs of clinically relevant antibiotic-resistant bacteria in the food production chain that influence human health.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/antibiotics11020157/s1, Table S1. Genomic features of E. coli isolates; Table S2. Resistome of colistin-resistant E. coli isolates; Table S3. Phages and prophages carrying mcr-1 identified on E. coli and K. pneumoniae genomes; Table S4. Primers used for mcr-type gene identification; Figure S1. Genome map of prophage and the genetic context of mcr-1 gene. The predicted genes and the functions predicted by PHASTER program [41] are indicated as boxes with the superscript number in each box. Figure S2. Electronic microscopy of phage that contained no detectable mcr-1 gene; Figure S3. Comparison of phage-like plasmid that contains the mcr-1 gene in the pRC2-007 plasmid with other phage-like plasmids. References [42,43,44] are cited in the supplementary materials.
Author Contributions
Conceptualization, J.S.-S., U.G.-R. and C.A.-A.; methodology, J.R.-S., N.R.-M., J.D.-B. and E.M.T.-L.; software, A.A.-V. and N.R.-M.; formal analysis, J.R.-S. and N.R.-M.; investigation, J.T.-S., A.N.L.-V. and J.D.-B.; writing—original draft preparation, J.R.-S. and N.R.-M.; writing—review and editing, U.G.-R., C.A.-A., J.R.-S. and N.R.-M.; visualization, A.N.L.-V.; funding acquisition, U.G.-R. and C.A.-A. All authors have read and agreed to the published version of the manuscript.
Funding
We gratefully acknowledge financial support from the Consejo Nacional de Ciencia y Tecnología (CONACyT) of México, by grant numbers 256988 and 215146.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA762933 and https://www.ncbi.nlm.nih.gov/biosample/SAMEA45274168 (accessed on 18 December 2021).
Acknowledgments
We thank Alejandro García Radilla and Bertha Carrillo Quiroz for the excellent technical assistance. We thank Michael Dunn from the Center for Genomic Science-UNAM for reviewing the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Poirel, L.; Jayol, A.; Nordmann, P. Polymyxins: Antibacterial Activity, Susceptibility Testing, and Resistance Mechanisms Encoded by Plasmids or Chromosomes. Clin. Microbiol. Rev. 2017, 30, 557–596. [Google Scholar] [CrossRef] [Green Version]
- Olaitan, A.O.; Morand, S.; Rolain, J.-M. Mechanisms of polymyxin resistance: Acquired and intrinsic resistance in bacteria. Front. Microbiol. 2014, 5, 643. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Feng, Y.; Liu, F.; Jiang, H.; Qu, Z.; Lei, M.; Wang, J.; Zhang, B.; Hu, Y.; Ding, J.; et al. A Phage-Like IncY Plasmid Carrying the mcr-1 Gene in Escherichia coli from a Pig Farm in China. Antimicrob. Agents Chemother. 2017, 61, e02035-16. [Google Scholar] [CrossRef] [Green Version]
- Al-Mir, H.; Osman, M.; Drapeau, A.; Hamze, M.; Madec, J.-Y.; Haenni, M. WGS Analysis of Clonal and Plasmidic Epidemiology of Colistin-Resistance Mediated by mcr Genes in the Poultry Sector in Lebanon. Front. Microbiol. 2021, 12, 624194. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Feng, Y.; Liu, L.; Wei, L.; Kang, M.; Zong, Z. Identification of novel mobile colistin resistance gene mcr-10. Emerg. Microbes Infect. 2020, 9, 508–516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez-Santiago, J.; Cornejo-Juárez, P.; Silva-Sánchez, J.; Garza-Ramos, U. Polymyxin resistance in Enterobacterales: Overview and epidemiology in the Americas. Int. J. Antimicrob. Agents 2021, 58, 106426. [Google Scholar] [CrossRef] [PubMed]
- Anjum, M.F.; Duggett, N.A.; AbuOun, M.; Randall, L.; Nunez-Garcia, J.; Ellis, R.J.; Rogers, J.; Horton, R.; Brena, C.; Williamson, S.; et al. Colistin resistance in Salmonella and Escherichia coli isolates from a pig farm in Great Britain. J. Antimicrob. Chemother. 2016, 71, 2306–2313. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.-Y.; Wang, Y.; Walsh, T.R.; Yi, L.-X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Hu, Y.; Liu, F.; Lin, I.Y.C.; Gao, G.F.; Zhu, B. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect. Dis. 2016, 16, 146–147. [Google Scholar] [CrossRef] [Green Version]
- Touchon, M.; Moura de Sousa, J.A.; Rocha, E.P. Embracing the enemy: The diversification of microbial gene repertoires by phage-mediated horizontal gene transfer. Curr. Opin. Microbiol. 2017, 38, 66–73. [Google Scholar] [CrossRef]
- Oliver, A.; Coque, T.M.; Alonso, D.; Valverde, A.; Baquero, F.; Cantón, R. CTX-M-10 Linked to a Phage-Related Element Is Widely Disseminated among Enterobacteriaceae in a Spanish Hospital. Antimicrob. Agents Chemother. 2005, 49, 1567–1571. [Google Scholar] [CrossRef] [Green Version]
- Zhou, W.; Liu, L.; Feng, Y.; Zong, Z. A P7 Phage-Like Plasmid Carrying mcr-1 in an ST15 Klebsiella pneumoniae Clinical Isolate. Front. Microbiol. 2018, 9, 11. [Google Scholar] [CrossRef] [PubMed]
- Merida-Vieyra, J.; De Colsa-Ranero, A.; Arzate-Barbosa, P.; Arias-de la Garza, E.; Mendez-Tenorio, A.; Murcia-Garzon, J.; Aquino-Andrade, A. First clinical isolate of Escherichia coli harboring mcr-1 gene in Mexico. PLoS ONE 2019, 14, e0214648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garza-Ramos, U.; Tamayo-Legorreta, E.; Arellano-Quintanilla, D.M.; Rodriguez-Medina, N.; Silva-Sanchez, J.; Catalan-Najera, J.; Rocha-Martínez, M.K.; Bravo-Díaz, M.A.; Alpuche-Aranda, C. Draft Genome Sequence of a Multidrug- and Colistin-Resistant mcr-1-Producing Escherichia coli Isolate from a Swine Farm in Mexico. Genome Announc. 2018, 6, e00102-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perez-Garza, J.; Franco-Frias, E.; Garcia-Heredia, A.; Garcia, S.; Leon, J.S.; Jaykus, L.A.; Heredia, N. The Cantaloupe Farm Environment Has a Diverse Genetic Pool of Antibiotic-Resistance and Virulence Genes. Foodborne Pathog. Dis. 2021, 18, 469–476. [Google Scholar] [CrossRef]
- Li, X.-P.; Sun, R.Y.; Song, J.Q.; Fang, L.X.; Zhang, R.M.; Lian, X.L.; Liao, X.P.; Liu, Y.H.; Lin, J.; Sun, J. Within-host heterogeneity and flexibility of mcr-1 transmission in chicken gut. Int. J. Antimicrob. Agents 2020, 2020 55, 105806. [Google Scholar] [CrossRef]
- Mikhayel, M.; Leclercq, S.O.; Sarkis, D.K.; Doublet, B. Occurrence of the Colistin Resistance Gene mcr-1 and Additional Antibiotic Resistance Genes in ESBL/AmpC-Producing Escherichia coli from Poultry in Lebanon: A Nationwide Survey. Microbiol. Spectr. 2021, 9, e0002521. [Google Scholar] [CrossRef]
- Rodríguez-Medina, N.; Barrios-Camacho, H.; Duran-Bedolla, J.; Garza-Ramos, U. Klebsiella variicola: An emerging pathogen in humans. Emerg. Microbes Infect. 2019, 8, 973–988. [Google Scholar] [CrossRef] [Green Version]
- Lu, J.; Dong, N.; Liu, C.; Zeng, Y.; Sun, Q.; Zhou, H.; Hu, Y.; Chen, S.; Shen, Z.; Zhang, R. Prevalence and molecular epidemiology of mcr-1-positive Klebsiella pneumoniae in healthy adults from China. J. Antimicrob. Chemother. 2020, 75, 2485–2494. [Google Scholar] [CrossRef]
- Da Silva, G.J.; Mendonca, N. Association between antimicrobial resistance and virulence in Escherichia coli. Virulence 2012, 3, 18–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Markland, S.M.; LeStrange, K.J.; Sharma, M.; Kniel, K.E. Old Friends in New Places: Exploring the Role of Extraintestinal E. coli in Intestinal Disease and Foodborne Illness. Zoonoses Public Health 2015, 62, 491–496. [Google Scholar] [CrossRef] [PubMed]
- Sarowska, J.; Futoma-Koloch, B.; Jama-Kmiecik, A.; Frej-Madrzak, M.; Ksiazczyk, M.; Bugla-PLoSkonska, G.; Choroszy-Krol, I. Virulence factors, prevalence and potential transmission of extraintestinal pathogenic Escherichia coli isolated from different sources: Recent reports. Gut Pathog. 2019, 11, 10. [Google Scholar] [CrossRef] [Green Version]
- Navarro-Garcia, F.; Ruiz-Perez, F.; Cataldi, A.; Larzabal, M. Type VI Secretion System in Pathogenic Escherichia coli: Structure, Role in Virulence, and Acquisition. Front. Microbiol. 2019, 10, 1965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kubelová, M.; Koláčková, I.; Gelbíčová, T.; Florianová, M.; Kalová, A.; Karpíšková, R. Virulence Properties of mcr-1-Positive Escherichia coli Isolated from Retail Poultry Meat. Microorganisms 2021, 9, 308. [Google Scholar] [CrossRef] [PubMed]
- Nordmann, P.; Jayol, A.; Poirel, L. Rapid Detection of Polymyxin Resistance inEnterobacteriaceae. Emerg. Infect. Dis. 2016, 22, 1038–1043. [Google Scholar] [CrossRef] [Green Version]
- European Committee on Antimicrobial Susceptibility Testing (EUCAST). Recommendations for MIC Determination of Colistin (Polymyxin E) As Recommended by the Joint CLSI-EUCAST Polymyxin Breakpoints Working Group. 2016. Available online: https://eucast.org (accessed on 25 October 2021).
- Clinical Laboratory Standart Institute. M100 Performance Standards for Antimicrobial Susceptibility Testing. 2017. Available online: https://clsi.org (accessed on 25 October 2021).
- Tenover, F.C.; Arbeit, R.D.; Goering, R.V.; Mickelsen, P.A.; Murray, B.E.; Persing, D.H.; Swaminathan, B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: Criteria for bacterial strain typing. J. Clin. Microbiol. 1995, 33, 2233–2239. [Google Scholar] [CrossRef] [Green Version]
- Kieser, T. Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid 1984, 12, 19–36. [Google Scholar] [CrossRef]
- Silva-Sanchez, J.; Barrios, H.; Reyna-Flores, F.; Bello-Diaz, M.; Sanchez-Perez, A.; Rojas, T. Bacterial Resistance Consortium, Garza-Ramos, U. Prevalence and characterization of plasmid-mediated quinolone resistance genes in extended-spectrum β-lactamase-producing Enterobacteriaceae isolates in Mexico. Microb Drug Resist. 2011, 17, 497–505. [Google Scholar] [CrossRef]
- Philippon, L.N.; Naas, T.; Bouthors, A.T.; Barakett, V.; Nordmann, P. OXA-18, a class D clavulanic acid-inhibited extended-spectrum beta-lactamase from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1997, 41, 2188–2195. [Google Scholar] [CrossRef] [Green Version]
- Miller, J.M. Experiments in Molecular Genetics; Cold Spring Harbor Laboratory: New York, NY, USA, 1972. [Google Scholar]
- Garza-Ramos, U.; Rodriguez-Medina, N.; Lozano-Aguirre, L.; Silva-Sanchez, J.; Sanchez-Arias, M.; Rodriguez-Olguin, J.; Martínez-Romero, E. Klebsiella variicola Reference Strain F2R9 (ATCC BAA-830) Genome Sequence. Microbiol. Resour. Announc. 2021, 10, e0032921. [Google Scholar] [CrossRef]
- Vallenet, D.; Belda, E.; Calteau, A.; Cruveiller, S.; Engelen, S.; Lajus, A.; Le Fèvre, F.; Longin, C.; Mornico, D.; Roche, D.; et al. MicroScope--an integrated microbial resource for the curation and comparative analysis of genomic and metabolic data. Nucleic Acids Res. 2013, 41, D636–D647. [Google Scholar] [CrossRef]
- Malberg Tetzschner, A.M.; Johnson, J.R.; Johnston, B.D.; Lund, O.; Scheutz, F. In Silico Genotyping of Escherichia coli Isolates for Extraintestinal Virulence Genes by Use of Whole-Genome Sequencing Data. J. Clin. Microbiol. 2020, 58, e01269-20. [Google Scholar] [CrossRef]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Zou, S.; Chen, H.; Yu, Y.; Ruan, Z. BacWGSTdb 2.0: A one-stop repository for bacterial whole-genome sequence typing and source tracking. Nucleic Acids Res. 2020, 49, D644–D650. [Google Scholar] [CrossRef]
- Beghain, J.; Bridier-Nahmias, A.; Le Nagard, H.; Denamur, E.; Clermont, O. ClermonTyping: An easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Microb. Genom. 2018, 4, e000192. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef] [PubMed]
- Roux, S.; Enault, F.; Hurwitz, B.L.; Sullivan, M.B. VirSorter: Mining viral signal from microbial genomic data. PeerJ 2015, 3, e985. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Liang, Y.; Lynch, K.H.; Dennis, J.J.; Wishart, D.S. PHAST: A fast phage search tool. Nucleic Acids Res. 2011, 39, W347–W352. [Google Scholar] [CrossRef] [PubMed]
- Yin, W.; Li, H.; Shen, Y.; Liu, Z.; Wang, S.; Shen, Z.; Zhang, R.; Walsh, T.R.; Shen, J.; Wang, Y. Novel Plasmid-Mediated Colistin Resistance Gene mcr-3 in Escherichia coli. mBio 2017, 8, e00543-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carattoli, A.; Villa, L.; Feudi, C.; Curcio, L.; Orsini, S.; Luppi, A.; Pezzotti, G.; Magistrali, C.F. Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Eurosurveillance 2017, 22, 30589. [Google Scholar] [CrossRef] [Green Version]
- Borowiak, M.; Fischer, J.; Hammerl, J.A.; Hendriksen, R.S.; Szabo, I.; Malorny, B. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J. Antimicrob. Chemother. 2017, 72, 3317–3324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).