Biosynthetic Potential of Streptomyces Rationalizes Genome-Based Bioprospecting
Abstract
:1. Introduction
2. Deluge of Genome Data
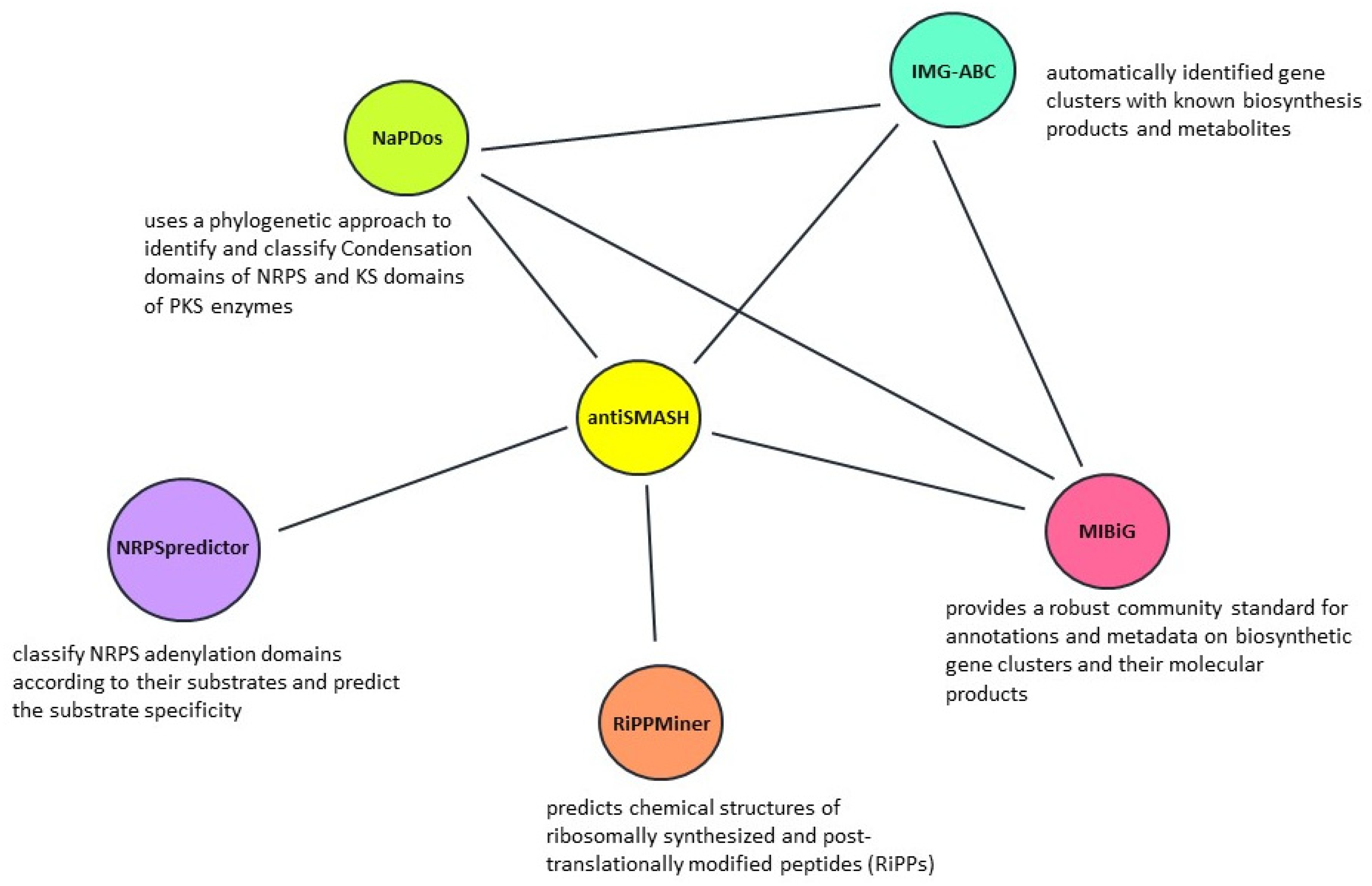
3. Genome Inspired Mining Tools
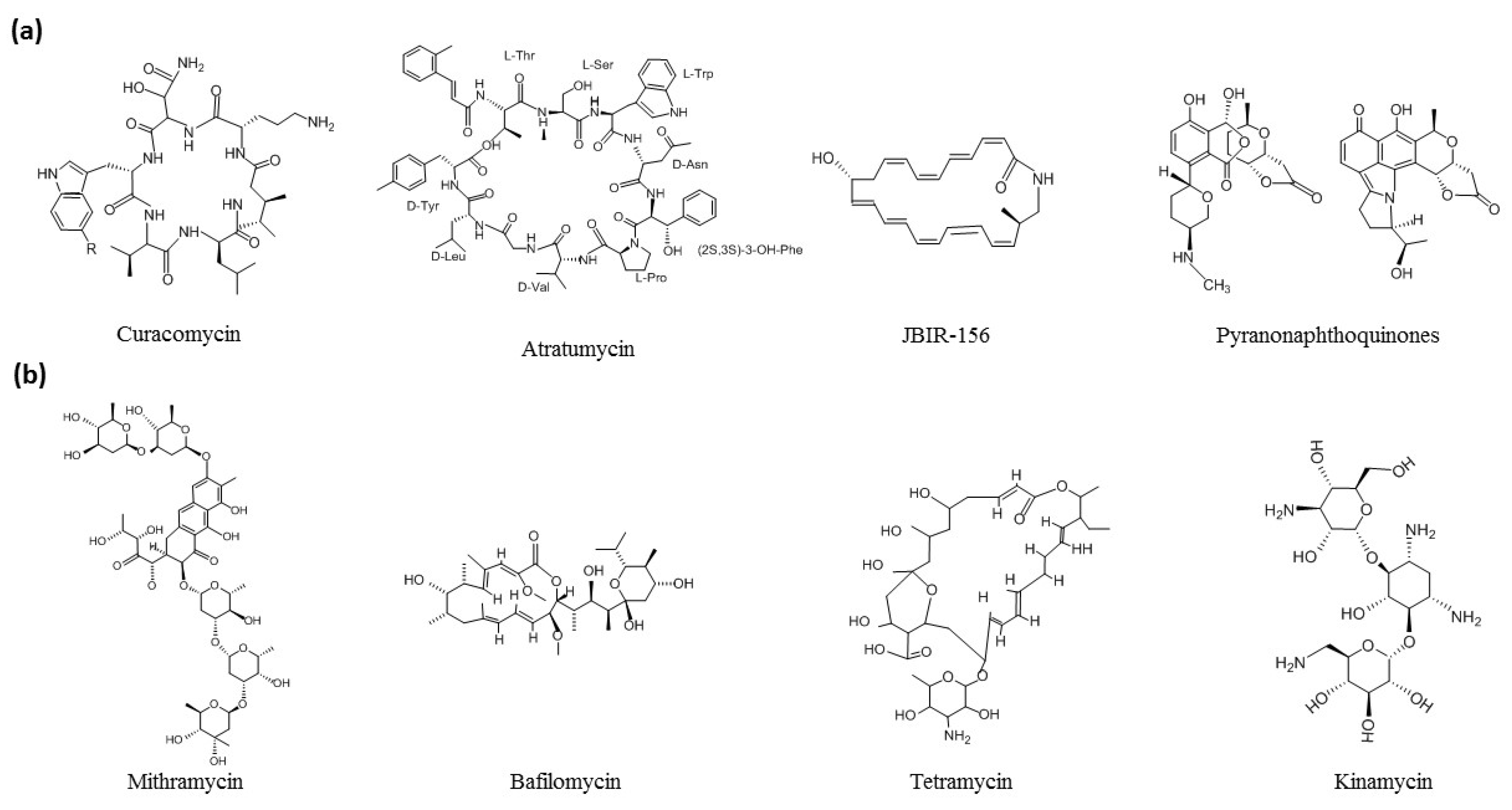
4. Genome Inspired Efforts Nourish Natural Products Research
5. Integrated Genome Mining and Metabolomics Approach
6. Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baltz, R.H. Natural product drug discovery in the genomic era: Realities, conjectures, misconceptions, and opportunities. J. Ind. Microbiol. Biotechnol. 2019, 46, 281–299. [Google Scholar] [CrossRef] [PubMed]
- Jose, P.A.; Jha, B. New dimensions of research on actinomycetes: Quest for next generation antibiotics. Front. Microbiol. 2016, 7, 1295. [Google Scholar] [CrossRef] [PubMed]
- Jose, P.A.; Maharshi, A.; Jha, B. Actinobacteria in natural products research: Progress and prospects. Microbiol. Res. 2021, 246, 126708. [Google Scholar] [CrossRef] [PubMed]
- Katz, L. The DEBS paradigm for type I modular polyketide synthases and beyond. Methods Enzymol. 2009, 459, 113–142. [Google Scholar] [PubMed]
- Katz, L.; Baltz, R.H. Natural product discovery: Past, present, and future. J. Ind. Microbiol. Biotechnol. 2016, 43, 155–176. [Google Scholar] [CrossRef]
- Bode, H.B.; Bethe, B.; Höfs, R.; Zeeck, A. Big effects from small changes: Possible ways to explore nature’s chemical diversity. ChemBioChem 2002, 3, 619–627. [Google Scholar] [CrossRef]
- Baltz, R.H. Microbial genome mining for natural product drug discovery. In Chemical Biology of Natural Products; CRC Press: Boca Raton, FL, USA, 2017. [Google Scholar]
- Mills, M.C.; Rahal, C. A scientometric review of genome-wide association studies. Commun. Biol. 2019, 2, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Bentley, S.D.; Chater, K.F.; Cerdeño-Tárraga, A.-M.; Challis, G.L.; Thomson, N.; James, K.D.; Harris, D.E.; Quail, M.A.; Kieser, H.; Harper, D. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3 (2). Nature 2002, 417, 141–147. [Google Scholar] [CrossRef]
- Ikeda, H.; Ishikawa, J.; Hanamoto, A.; Shinose, M.; Kikuchi, H.; Shiba, T.; Sakaki, Y.; Hattori, M.; Ōmura, S. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 2003, 21, 526. [Google Scholar] [CrossRef]
- Blin, K.; Medema, M.H.; Kazempour, D.; Fischbach, M.A.; Breitling, R.; Takano, E.; Weber, T. antiSMASH 2.0—A versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res. 2013, 41, W204–W212. [Google Scholar] [CrossRef] [Green Version]
- Ziemert, N.; Podell, S.; Penn, K.; Badger, J.H.; Allen, E.; Jensen, P.R. The natural product domain seeker NaPDoS: A phylogeny based bioinformatic tool to classify secondary metabolite gene diversity. PLoS ONE 2012, 7, e34064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bachmann, B.O.; Ravel, J. Methods for in silico prediction of microbial polyketide and nonribosomal peptide biosynthetic pathways from DNA sequence data. Methods Enzymol. 2009, 458, 181–217. [Google Scholar] [PubMed]
- Röttig, M.; Medema, M.H.; Blin, K.; Weber, T.; Rausch, C.; Kohlbacher, O. NRPSpredictor2—A web server for predicting NRPS adenylation domain specificity. Nucleic Acids Res. 2011, 39, W362–W367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agrawal, P.; Khater, S.; Gupta, M.; Sain, N.; Mohanty, D. RiPPMiner: A bioinformatics resource for deciphering chemical structures of RiPPs based on prediction of cleavage and cross-links. Nucleic Acids Res. 2017, 45, W80–W88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, S.-S.; Katsuyama, Y.; Bai, L.; Deng, Z.; Ohnishi, Y.; Kim, E.-S. Genome engineering for microbial natural product discovery. Curr. Opin. Microbiol. 2018, 45, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Lage, O.M.; Ramos, M.C.; Calisto, R.; Almeida, E.; Vasconcelos, V.; Vicente, F. Current screening methodologies in drug discovery for selected human diseases. Mar. Drugs 2018, 16, 279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, M.; Wang, Y.; Zhao, Z.; Gao, G.; Huang, S.-X.; Kang, Q.; He, X.; Lin, S.; Pang, X.; Deng, Z. Functional genome mining for metabolites encoded by large gene clusters through heterologous expression of a whole-genome bacterial artificial chromosome library in Streptomyces spp. Appl. Environ. Microbiol. 2016, 82, 5795–5805. [Google Scholar] [CrossRef] [Green Version]
- Myronovskyi, M.; Rosenkränzer, B.; Nadmid, S.; Pujic, P.; Normand, P.; Luzhetskyy, A. Generation of a cluster-free Streptomyces albus chassis strains for improved heterologous expression of secondary metabolite clusters. Metab. Eng. 2018, 49, 316–324. [Google Scholar] [CrossRef] [PubMed]
- Bauermeister, A.; Pereira, F.; Grilo, I.R.; Godinho, C.C.; Paulino, M.; Almeida, V.; Gobbo-Neto, L.; Prieto-Davó, A.; Sobral, R.G.; Lopes, N.P. Intra-clade metabolomic profiling of MAR4 Streptomyces from the Macaronesia Atlantic region reveals a source of anti-biofilm metabolites. Environ. Microbiol. 2019, 21, 1099–1112. [Google Scholar] [CrossRef]
- Ju, K.-S.; Gao, J.; Doroghazi, J.R.; Wang, K.-K.A.; Thibodeaux, C.J.; Li, S.; Metzger, E.; Fudala, J.; Su, J.; Zhang, J.K. Discovery of phosphonic acid natural products by mining the genomes of 10,000 actinomycetes. Proc. Natl. Acad. Sci. USA 2015, 112, 12175–12180. [Google Scholar] [CrossRef] [Green Version]
- Elsayed, S.S.; Trusch, F.; Deng, H.; Raab, A.; Prokes, I.; Busarakam, K.; Asenjo, J.A.; Andrews, B.A.; Van West, P.; Bull, A.T. Chaxapeptin, a lasso peptide from extremotolerant Streptomyces leeuwenhoekii strain C58 from the hyperarid Atacama Desert. J. Org. Chem. 2015, 80, 10252–10260. [Google Scholar] [CrossRef] [PubMed]
- Kaweewan, I.; Komaki, H.; Hemmi, H.; Hoshino, K.; Hosaka, T.; Isokawa, G.; Oyoshi, T.; Kodani, S. Isolation and structure determination of a new cytotoxic peptide, curacozole, from Streptomyces curacoi based on genome mining. J. Antibiot. 2019, 72, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Kaweewan, I.; Komaki, H.; Hemmi, H.; Kodani, S. Isolation and structure determination of new antibacterial peptide curacomycin based on genome mining. Asian J. Org. Chem. 2017, 6, 1838–1844. [Google Scholar] [CrossRef]
- Frattaruolo, L.; Lacret, R.; Cappello, A.R.; Truman, A.W. A genomics-based approach identifies a thioviridamide-like compound with selective anticancer activity. ACS Chem. Biol. 2017, 12, 2815–2822. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Yang, Z.; Zhang, C.; Liu, Z.; He, J.; Liu, Q.; Zhang, T.; Ju, J.; Ma, J. Genome mining of Streptomyces atratus SCSIO ZH16: Discovery of atratumycin and identification of its biosynthetic gene cluster. Org. Lett. 2019, 21, 1453–1457. [Google Scholar] [CrossRef]
- Nguyen, H.T.; Pokhrel, A.R.; Nguyen, C.T.; Dhakal, D.; Lim, H.N.; Jung, H.J.; Kim, T.-S.; Yamaguchi, T.; Sohng, J.K. Streptomyces sp. VN1, a producer of diverse metabolites including non-natural furan-type anticancer compound. Sci. Rep. 2020, 10, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cortés-Albayay, C.; Jarmusch, S.A.; Trusch, F.; Ebel, R.; Andrews, B.A.; Jaspars, M.; Asenjo, J.A. Downsizing class II lasso peptides: Genome mining-guided isolation of huascopeptin containing the first Gly1-Asp7 macrocycle. J. Org. Chem. 2020, 85, 1661–1667. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, T.; Kozone, I.; Hashimoto, J.; Ueoka, R.; Kagaya, N.; Fujie, M.; Sato, N.; Ikeda, H.; Shin-Ya, K. Novel macrolactam compound produced by the heterologous expression of a large cryptic biosynthetic gene cluster of Streptomyces rochei IFO12908. J. Antibiot. 2020, 73, 171–174. [Google Scholar] [CrossRef]
- Wu, C.; Ichinose, K.; Choi, Y.H.; van Wezel, G.P. Aromatic polyketide GTRI-02 is a previously unidentified product of the act gene cluster in Streptomyces coelicolor A3 (2). ChemBioChem 2017, 18, 1428–1434. [Google Scholar] [CrossRef]
- Jose, P.A.; Sivakala, K.K.; Jebakumar, S.R.D. Formulation and statistical optimization of culture medium for improved production of antimicrobial compound by Streptomyces sp. JAJ06. Int. J. Microbiol. 2013, 2013, 526260. [Google Scholar]
- Rajeswari, P.; Jose, P.A.; Amiya, R.; Jebakumar, S.R.D. Characterization of saltern based Streptomyces sp. and statistical media optimization for its improved antibacterial activity. Front. Microbiol. 2015, 5, 753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turło, J.; Gajzlerska, W.; Klimaszewska, M.; Król, M.; Dawidowski, M.; Gutkowska, B. Enhancement of tacrolimus productivity in Streptomyces tsukubaensis by the use of novel precursors for biosynthesis. Enzym. Microb. Technol. 2012, 51, 388–395. [Google Scholar] [CrossRef]
- Yi, J.S.; Kim, M.-S.; Kim, S.-J.; Kim, B.-G. Effects of sucrose, phosphate, and calcium carbonate on the production of pikromycin from Streptomyces venezuelae. J. Microbiol. Biotechnol. 2015, 25, 496–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zabala, D.; Braña, A.F.; Flórez, A.B.; Salas, J.A.; Méndez, C. Engineering precursor metabolite pools for increasing production of antitumor mithramycins in Streptomyces argillaceus. Metab. Eng. 2013, 20, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Jung, W.S.; Kim, E.; Yoo, Y.J.; Ban, Y.H.; Kim, E.J.; Yoon, Y.J. Characterization and engineering of the ethylmalonyl-CoA pathway towards the improved heterologous production of polyketides in Streptomyces venezuelae. Appl. Microbiol. Biotechnol. 2014, 98, 3701–3713. [Google Scholar] [CrossRef]
- Thykaer, J.; Nielsen, J.; Wohlleben, W.; Weber, T.; Gutknecht, M.; Lantz, A.E.; Stegmann, E. Increased glycopeptide production after overexpression of shikimate pathway genes being part of the balhimycin biosynthetic gene cluster. Metab. Eng. 2010, 12, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Liao, G.; Wang, L.; Liu, Q.; Guan, F.; Huang, Y.; Hu, C. Manipulation of kynurenine pathway for enhanced daptomycin production in Streptomyces roseosporus. Biotechnol. Prog. 2013, 29, 847–852. [Google Scholar] [CrossRef]
- Lee, D.W.; Ng, B.G.; Kim, B.S. Increased valinomycin production in mutants of Streptomyces sp. M10 defective in bafilomycin biosynthesis and branched-chain α-keto acid dehydrogenase complex expression. J. Ind. Microbiol. Biotechnol. 2015, 42, 1507–1517. [Google Scholar] [CrossRef] [PubMed]
- Murakami, T.; Burian, J.; Yanai, K.; Bibb, M.J.; Thompson, C.J. A system for the targeted amplification of bacterial gene clusters multiplies antibiotic yield in Streptomyces coelicolor. Proc. Natl. Acad. Sci. USA 2011, 108, 16020–16025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, T.-C.; Kim, B.-G.; Zhong, J.-J. Enhanced production of validamycin A in Streptomyces hygroscopicus 5008 by engineering validamycin biosynthetic gene cluster. Appl. Microbiol. Biotechnol. 2014, 98, 7911–7922. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Cheng, X.; Liu, Y.; Deng, Z.; You, D. Deciphering and engineering of the final step halogenase for improved chlortetracycline biosynthesis in industrial Streptomyces aureofaciens. Metab. Eng. 2013, 19, 69–78. [Google Scholar] [CrossRef]
- Ikeda, H.; Shin-ya, K.; Omura, S. Genome mining of the Streptomycesavermitilis genome and development of genome-minimized hosts for heterologous expression of biosynthetic gene clusters. J. Ind. Microbiol. Biotechnol. 2014, 41, 233–250. [Google Scholar] [CrossRef]
- Horbal, L.; Rebets, Y.; Rabyk, M.; Makitrynskyy, R.; Luzhetskyy, A.; Fedorenko, V.; Bechthold, A. SimReg1 is a master switch for biosynthesis and export of simocyclinone D8 and its precursors. AMB Express 2012, 2, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Yin, M.; Horsman, G.P.; Huang, S.; Shen, B. Manipulation of pathway regulation in Streptomyces globisporus for overproduction of the enediyne antitumor antibiotic C-1027. J. Antibiot. 2010, 63, 482–485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, H.; Ni, X.; Shao, W.; Su, J.; Su, J.; Ren, J.; Xia, H. Functional manipulations of the tetramycin positive regulatory gene ttmRIV to enhance the production of tetramycin A and nystatin A1 in Streptomyces ahygroscopicus. J. Ind. Microbiol. Biotechnol. 2015, 42, 1273–1282. [Google Scholar] [CrossRef] [PubMed]
- Bunet, R.; Song, L.; Mendes, M.V.; Corre, C.; Hotel, L.; Rouhier, N.; Framboisier, X.; Leblond, P.; Challis, G.L.; Aigle, B. Characterization and manipulation of the pathway-specific late regulator AlpW reveals Streptomyces ambofaciens as a new producer of kinamycins. J. Bacteriol. 2011, 193, 1142–1153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, D.; Xia, M.; Li, S.; Wen, J.; Jia, X. Enhancement of FK506 production by engineering secondary pathways of Streptomyces tsukubaensis and exogenous feeding strategies. J. Ind. Microbiol. Biotechnol. 2013, 40, 1023–1037. [Google Scholar] [CrossRef]
- Moore, S.J.; Lai, H.E.; Needham, H.; Polizzi, K.M.; Freemont, P.S. Streptomyces venezuelae TX-TL–a next generation cell-free synthetic biology tool. Biotechnol. J. 2017, 12, 1600678. [Google Scholar] [CrossRef] [Green Version]
- Yang, X. Applications of CRISPR-Cas9 mediated genome engineering. Mil. Med Res. 2015, 2, 11. [Google Scholar] [CrossRef] [Green Version]
- Cobb, R.E.; Wang, Y.; Zhao, H. High-efficiency multiplex genome editing of Streptomyces species using an engineered CRISPR/Cas system. ACS Synth. Biol. 2015, 4, 723–728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishaque, N.M.; Burgsdorf, I.; Limlingan Malit, J.J.; Saha, S.; Teta, R.; Ewe, D.; Kannabiran, K.; Hrouzek, P.; Steindler, L.; Costantino, V. Isolation, genomic and metabolomic characterization of Streptomyces tendae VITAKN with quorum sensing inhibitory activity from southern India. Microorganisms 2020, 8, 121. [Google Scholar] [CrossRef] [Green Version]
- Sukmarini, L. Recent Advances in Discovery of Lead Structures from Microbial Natural Products: Genomics-and Metabolomics-Guided Acceleration. Molecules 2021, 26, 2542. [Google Scholar] [CrossRef] [PubMed]
- Sivakala, K.K.; Gutiérrez-García, K.; Arul Jose, P.; Thinesh, T.; Anandham, R.; Barona-Gómez, F.; Sivakumar, N. Desert environments facilitate unique evolution of biosynthetic potential in Streptomyces. Molecules 2021, 26, 588. [Google Scholar] [CrossRef] [PubMed]
- Demarque, D.P.; Dusi, R.G.; de Sousa, F.D.; Grossi, S.M.; Silvério, M.R.; Lopes, N.P.; Espindola, L.S. Mass spectrometry-based metabolomics approach in the isolation of bioactive natural products. Sci. Rep. 2020, 10, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gebretsadik, T.; Linert, W.; Thomas, M.; Berhanu, T.; Frew, R. LC–NMR for Natural Product Analysis: A Journey from an Academic Curiosity to a Robust Analytical Tool. Science 2021, 3, 6. [Google Scholar] [CrossRef]
- Navarro-Muñoz, J.C.; Selem-Mojica, N.; Mullowney, M.W.; Kautsar, S.A.; Tryon, J.H.; Parkinson, E.I.; De Los Santos, E.L.; Yeong, M.; Cruz-Morales, P.; Abubucker, S. A computational framework to explore large-scale biosynthetic diversity. Nat. Chem. Biol. 2020, 16, 60–68. [Google Scholar] [CrossRef]
- Kautsar, S.A.; van der Hooft, J.J.; de Ridder, D.; Medema, M.H. BiG-SLiCE: A highly scalable tool maps the diversity of 1.2 million biosynthetic gene clusters. GigaScience 2021, 10, giaa154. [Google Scholar] [CrossRef] [PubMed]
- Mohimani, H.; Kersten, R.D.; Liu, W.-T.; Wang, M.; Purvine, S.O.; Wu, S.; Brewer, H.M.; Pasa-Tolic, L.; Bandeira, N.; Moore, B.S. Automated genome mining of ribosomal peptide natural products. ACS Chem. Biol. 2014, 9, 1545–1551. [Google Scholar] [CrossRef] [PubMed]
- Quinn, R.A.; Nothias, L.-F.; Vining, O.; Meehan, M.; Esquenazi, E.; Dorrestein, P.C. Molecular networking as a drug discovery, drug metabolism, and precision medicine strategy. Trends Pharmacol. Sci. 2017, 38, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whitford, C.M.; Cruz-Morales, P.; Keasling, J.D.; Weber, T. The Design-Build-Test-Learn cycle for metabolic engeenering. of Streptomycetes. Essays Biochem. 2021, EBC20200132. [Google Scholar] [CrossRef] [PubMed]
- Kersten, R.D.; Yang, Y.-L.; Xu, Y.; Cimermancic, P.; Nam, S.-J.; Fenical, W.; Fischbach, M.A.; Moore, B.S.; Dorrestein, P.C. A mass spectrometry–guided genome mining approach for natural product peptidogenomics. Nat. Chem. Biol. 2011, 7, 794–802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.-T.; Lamsa, A.; Wong, W.R.; Boudreau, P.D.; Kersten, R.; Peng, Y.; Moree, W.J.; Duggan, B.M.; Moore, B.S.; Gerwick, W.H. MS/MS-based networking and peptidogenomics guided genome mining revealed the stenothricin gene cluster in Streptomyces roseosporus. J. Antibiot. 2014, 67, 99–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rateb, M.E.; Zhai, Y.; Ehrner, E.; Rath, C.M.; Wang, X.; Tabudravu, J.; Ebel, R.; Bibb, M.; Kyeremeh, K.; Dorrestein, P.C. Legonaridin, a new member of linaridin RiPP from a Ghanaian Streptomyces isolate. Org. Biomol. Chem. 2015, 13, 9585–9592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cibichakravarthy, B.; Jose, P.A. Biosynthetic Potential of Streptomyces Rationalizes Genome-Based Bioprospecting. Antibiotics 2021, 10, 873. https://doi.org/10.3390/antibiotics10070873
Cibichakravarthy B, Jose PA. Biosynthetic Potential of Streptomyces Rationalizes Genome-Based Bioprospecting. Antibiotics. 2021; 10(7):873. https://doi.org/10.3390/antibiotics10070873
Chicago/Turabian StyleCibichakravarthy, Balasubramanian, and Polapass Arul Jose. 2021. "Biosynthetic Potential of Streptomyces Rationalizes Genome-Based Bioprospecting" Antibiotics 10, no. 7: 873. https://doi.org/10.3390/antibiotics10070873
APA StyleCibichakravarthy, B., & Jose, P. A. (2021). Biosynthetic Potential of Streptomyces Rationalizes Genome-Based Bioprospecting. Antibiotics, 10(7), 873. https://doi.org/10.3390/antibiotics10070873






