Abstract
With the increase of drug resistance caused by the improper use and abuse of antibiotics, human beings are facing a global health crisis. Sequencing of Streptomyces genomes revealed the presence of an important reservoir of secondary metabolic gene clusters for previously unsuspected products with potentially valuable bioactivity. It has therefore become necessary to activate these cryptic pathways through various strategies. Here, we used RNA-seq data to perform a comparative transcriptome analysis of Streptomyces ansochromogenes (wild-type, WT) and its global regulatory gene disruption mutant ΔwblA, in which some differentially expressed genes are associated with the abolished nikkomycin biosynthesis and activated tylosin analogue compounds (TACs) production, and also with the oviedomycin production that is induced by the genetic manipulation of two differentially expressed genes (san7324 and san7324L) encoding RsbR. These results provide a significant clue for the discovery of new drug candidates and the activation of cryptic biosynthetic gene clusters.
1. Introduction
Natural products from microbes play an important role in clinical treatments, animal husbandry and plant crop protection [,]. Actinomycetes, especially streptomycetes, are a particularly abundant source of secondary metabolites. The decreased number in clinical antibiotics and the emergence of antibiotic resistance pose new challenges for the discovery of novel antibiotics. Streptomycetes have the potential to produce more new natural products than those previously recognized under laboratory conditions. With the rapid development of genome sequencing technology and analysis tools, more and more cryptic secondary metabolites have been mined and the activated cryptic gene clusters have been identified through various strategies, which provide more opportunities to discover new natural products or drug candidates [,].
Currently used or proposed methods for activating the ‘cryptic’ pathways include the optimization of culture and fermentation conditions, heterologous expression, genetic manipulations of regulatory genes or gene clusters, omic studies, co-culture of different microbes, induction of signaling molecules, ribosomal engineering, reporter-guided strategy, and so on [,,]. Meanwhile, microbial natural product databases, such as ’The Natural Products Atlas’ (https://www.npatlas.org/, accessed on 9 September 2021), antiSMASH database (https://antismash.secondarymetabolites.org, accessed on 9 September 2021), and CH-NMR-NP database (https://www.j-resonance.com/en/nmrdb/, accessed on 9 September 2021) play a key action in the mining of drug candidates [,,]. Various bioinformatics tools have been developed and widely used in genomics or transcriptomics, such as Canu [], Circos [], RSEM (http://deweylab.github.io/RSEM/, accessed on 9 September 2021) [], edgeR (http://www.bioconductor.org/packages/2.12/bioc/html/edgeR.html, accessed on 9 September 2021) [] and DEGs KEGG enrichment analysis (KOBAS 2.0, http://kobas.cbi.pku.edu.cn, accessed on 9 September 2021) [].
There are many successful examples for mining of hidden natural products. For instance, (i) Three natural products, streptothricin, geosmin and strevertene A (polyene), were successfully activated by using the reporter-guided strategy []; (ii) Multiplex activation strategies were implemented to awaken a cryptic biosynthetic gene cluster, resulting in the discovery of eight aromatic polyketides with two types of frameworks, two pentacyclic isomers and six glycosylated tetracyclines []; (iii) In Streptomyces roseosporus, mureidomycin biosynthesis was activated by the introduction of an exogenous regulatory gene ssaA [].
The WhiB-like (Wbl) family of proteins broadly distribute in Actinobacteria, such as Streptomyces, Corynebacteria and Mycobacteria []. Wbls usually play key roles in the differentiation, the biosynthesis of various secondary metabolites, and the involvement in the oxidative stress response [,,]. For instance, wblA plays a crucial role in the repression of daptomycin biosynthesis, which is probably mediated by the regulatory genes atrA, dptR2 and dptR3. Although WblA has been reported to have a DNA-binding HTH domain, there are few examples of direct evidence about its transcriptional regulation from EMSA experiments []. Wbl proteins may change their regulatory function in response to O2 and nitric oxidation (NO) via iron-sulphur clusters essential for nutrient starvation survival []. In addition, some Wbls can interact with their partner proteins to modulate antibiotic resistance [,]. Streptomyces ansochromogenes 7100 was isolated from the northeast soil of China []. It can produce a peptidyl nucleoside compound nikkomycin, a promising antibiotic against phytopathogenic fungi and human pathogens []. Our previous work showed that the biosynthesis of novel cryptic tylosin analogue compounds 1,2,3 (TAC1,2,3 or TACs) with much higher activity than tylosin against Streptococcus pneumonia was activated by the disruption of wblA, meanwhile, nikkomycin was abolished [].
In this study, we describe the comparative transcriptome analysis, in which some differentially expressed genes are related to the gene cluster’s activation/inactivation. This would provide an efficient approach for the activation of secondary metabolites guided by RNA-seq data.
2. Results and Discussion
2.1. Sequencing and AntiSMASH Analysis of S. Ansochromogenes 7100 Genome
In order to obtain a complete genome sequence of S. ansochromogenes 7100, the third generation sequencing was performed on a PacBio RSII platform. A total of 157,549 quality subreads were obtained in genome sequencing with an approximate coverage fold of more than 100× (Table S1, Supplementary Materials).
Then the raw data were filtered and assembled into a nearly complete genome of 9.56 Mb (Table S2) with software Canu. The main features of the S. ansochromogenes 7100 chromosome are shown in Figure 1. The genome of S. ansochromogenes 7100 is a linear chromosome with a repetitive sequence content of 2.37% (Table S3), which is composed of 8235 protein-coding genes (Table S4), 72 tRNAs, 18 rRNA and 50 nc RNAs genes (Table S5). Of the total gene set, 98% were classified into functional categories according to NR, COG, Pfam, KEGG pathway and Gene Ontology database (Table S6). The average G + C content of the genome is 72.41%.
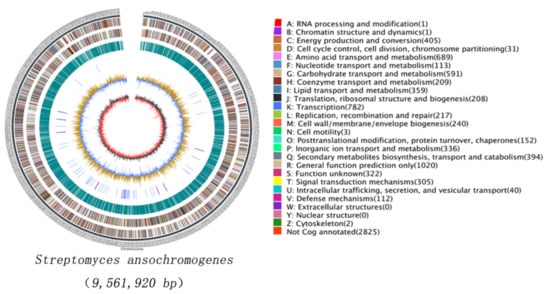
Figure 1.
Circular representation of the Streptomyces ansochromogenes chromosome. The circle 1 (outer circle) is a marker of genome total size with a scale of 5 kb each; circles 2 and 3 (from the outside in) are genes on the forward and reverse strand respectively, which are color-coded by function; circle 4 indicates the distributed position of repeat sequence; circle 5 represents tRNAs and rRNAs; circle 6 shows GC content (the light yellow part indicates that the GC content in this region is higher than the average GC content in the genome, the blue part indicates that the GC content in this region is lower than the average GC content in the genome), circle 7 (inner circle) is GC-skew (Black represents the area where G content is greater than C, and red represents the area where C content is greater than G).
The potential secondary metabolites biosynthetic gene clusters in S. ansochromogenes 7100 were analyzed by the antiSMASH program [,]. A total of 41 gene clusters were identified, which include nikkomycin gene cluster, TACs gene cluster and 39 uncharacterized gene clusters that are predicted to be involved in the biosynthesis of polyketides (PKSs), lassopeptide, terpene, lanthipeptide, bacteriocin, non-ribosomal peptides (NRPSs), terpene, butyrolactone, betalactone, siderophore, ectoine, melanin and so on (Table 1). Although only a few secondary metabolites have been identified, the abundant ‘cryptic’ gene clusters in S. ansochromogenes 7100 implied the potential for discovery of novel bioactive products.

Table 1.
AntiSMASH analysis of genome sequence of the S. ansochromogenes 7100.
2.2. Genome-Wide Differentially Expressed Genes in ΔwblA
wblA is an important regulatory gene usually involved in the sporulation, biosynthesis of secondary metabolites and oxidative stress in Streptomyces []. The disruption of wblA in S. ansochromogenes 7100 caused the failure of sporulation and the loss of nikkomycins production []. Instead, three novel 16-membered tylosin-like macrolides were accumulated in the resulting mutant strain (ΔwblA), arousing much interest in how the regulatory pathway works.
To elucidate the landscape of transcriptomic changes of S. ansochromogenes 7100 and ΔwblA, RNA-seq transcriptional profiles at 24 h and 48 h were analyzed. The production of TACs was not detectable at 24 h but appeared at 48 h with the subsequent expression of the biosynthetic genes. RNAs were isolated from the WT and ΔwblA strains at the two time points for cDNA synthesis, library construction and sequencing. The transcriptome sequencing of four libraries resulted in a total unique map of 76,486,734 sequence reads, which were aligned to the reference genome of S. ansochromogenes 7100 (Table S7). More than 95% of unique mapped reads ratio in each library mapped to the S. ansochromogenes genome. The data reached saturation (Figure S1).
RNA-seq analysis revealed a large group of differentially expressed genes, as illustrated in the Heatmap (Figure 2a). Among them, the transcriptional levels of 1162 genes up-regulated 2-fold and 2776 genes down-regulated 2-fold in ΔwblA at 24 h, and 1767 genes up-regulated 2-fold and 2349 genes down-regulated 2-fold in ΔwblA at 48 h. Comparative transcriptome analyses of WT and ΔwblA at both 24 h and 48 h identified 2594 transcripts whose differential expressions were equal or more than 2-fold. Among them, 1433 transcripts were equal or more than 5-fold, and significantly, 1147 transcripts were equal or more than 10-fold (Figure 2b, Table S8). Three blocks were illustrated in the Heatmap (Figure 2a). In block 1, when ΔwblA incubated for 24 h, differentially expressed genes involved in some amino acids and secondary metabolites pathways were sharply decreased compared with those of WT, including the nikkomycin biosynthetic pathway and acarbose, as well as the validamycin biosynthetic pathways (map00525 in KEGG pathway) (Table S9). In block 2, the transcriptional levels of differentially expressed genes increased in the protein biosynthetic pathway, the ribosome biosynthetic pathway (map03010), and the secondary metabolites biosynthetic pathway in ΔwblA at 24 h, such as the TACs biosynthetic pathway, porphyrin and chlorophyll metabolism (map00860). In block 3, when ΔwblA incubated for 48 h, the transcriptional level of differentially expressed genes increased in environmental information processing, signal transducing, quorum sensing pathways and secretion system or transport pathways.
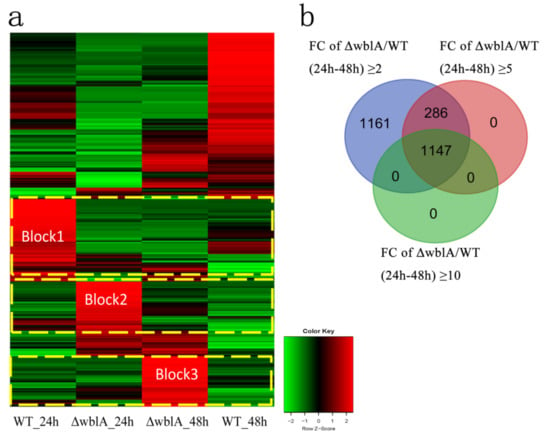
Figure 2.
Heatmap and Venn diagram of gene transcriptional profiles. (a) Heatmap represents all differentially expressed genes whose transcriptional change ≥ 2–fold at 24 h and 48 h in both WT and ΔwblA strains, respectively. padjust < 0.001. Red and green colors represent up-regulated and down-regulated genes, respectively. (b) The Venn diagram represents the numbers of differentially expressed genes in ΔwblA compared with those of WT (fold change ≥ 2-fold, ≥5-fold and ≥10-fold) at both 24 h and 48 h, which was created using DESeq package, a software platform for identifying differentially expressed genes from RNA-seq data. FC indicates fold change.
2.3. The Silent Nikkomycin Pathway and the Activated Tylosin Analogue Compounds (TACs) Biosynthetic Pathway in ΔwblA
Detailed analysis of the transcriptomics revealed a significant up-regulation of TAC biosynthetic genes (from pks1 to orf11) in ΔwblA (Table 2, Figure 3a). Except for orf5 which has a 2.8-fold increase, the transcriptional levels of the TAC biosynthetic genes all showed a 14–192-fold increase at 24 h (Table 2, the column of fold change of ΔwblA_24 h against WT_24 h). Moreover, the transcriptional up-regulation of the genes ctg1_2035 or ctg1_2036 encoding methylmalonyl-CoA mutase and ctg1_2557 or ctg1_4516 encoding acetyl-CoA C-acetyltransferase at 48 h is much higher than that at 24 h; it seems that these genes might be of importance for the biosynthesis of tylosin analogue compounds (TACs). Meanwhile, with the production of the TAC compounds at 48 h, the transcriptional levels of the TACs cluster-situated biosynthetic genes increased approximately 1–10 folds in ΔwblA compared with those of wild-type 7100 at 48 h, even though their transcriptional levels are lower than those at 24 h (Table 2). We speculated that the transcriptional fold change of these genes may be the main factor leading to the production of the the TACs. The decrease of fold change of ΔwblA/WT at 48 h versus fold change of ΔwblA/WT at 24 h may be related to the feedback negative regulation.

Table 2.
Differentially expressed genes in the biosynthesis of tylosin analogue compounds (TACs).
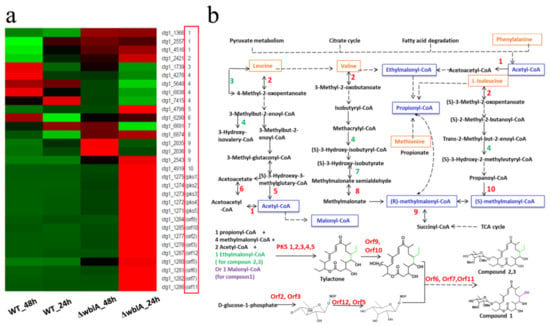
Figure 3.
Differentially expressed genes in tylosin analogue compounds (TACs) biosynthesis in WT and ΔwblA. (a) Analysis of significantly expressed genes in both WT and ΔwblA at 24 h and 48 h. |log2FC| ≥ 1 and padjust < 0.001. Expression variance for each gene was indicated by different colors: red (up-regulated genes), green (down-regulated genes) and black (|log2FC| < 1). The numbers or gene names in the red box are corresponding to the catalytic reaction steps involved in (b). (b) Proposed pathway diagram of TACs biosynthesis and the relevant amino acids catabolism. The blue boxes represent the precursors that incorporated into tylactone, and the orange boxes represent the precursors derived from the catabolism of five amino acids. The numbers 1–10 denote differentially expressed genes in 10 reaction steps of deduced KEGG pathway. Green numbers denote down-regulated genes encoding enzymes responsible for these reaction steps, red numbers denote up-regulated genes encoding enzymes responsible for these reaction steps. Dotted arrows represent multiple genes responsible for these reaction steps.
The transcription of the genes responsible for the TAC precursor biosynthesis was also analyzed at both 24 h and 48 h. Acetyl-CoA, methylmalonyl-CoA, ethylmalonyl-CoA and malonyl-CoA were proposed to be involved in the tylactone’s assembly (Figure 3b). These precursors or intermediates are derived from primary metabolic pathways as amino acid catabolism, fatty acids catabolism, citrate cycle or pyruvate metabolism. The gene (ctg1_4798) responsible for the biosynthesis of acetyl-CoA from leucine metabolism (step 5 in Figure 3b) was transcriptionally up-regulated (6.5-fold increase at 24 h) in ΔwblA. Meanwhile, the genes (ctg1_4276, ctg1_5640, ctg1_6838 and ctg1_7415) catalyzing the branched metabolic pathway in leucine catabolism (step 4 in Figure 3b) were down-regulated, which was proposed to prompt the intermediates’ flow to the formation of acetyl-CoA. The transcriptional levels of the methylmalonyl-CoA biosynthetic genes including ctg1_4919 (step 10 in Figure 3b) and ethylmalonyl-CoA biosynthesis related genes (ctg1_1366, ctg1_2557, ctg1_4516, step 1 in Figure 3b) were all apparently increased to supply more precursors for TAC biosynthesis. Transcriptional analysis of the precursors biosynthetic genes will be helpful for improvement of TACs’ production by optimizing the metabolic pathways.
Nikkomycins, a group of peptidyl nucleoside antibiotics, are a potent competitive inhibitor of fungal chitin synthases, since their chemical structure is similar to the natural substrate of chitin synthase, UDPN-acetylglucosamine []. The abolishment of nikkomycin production in ΔwblA was also approved by analyzing the transcripts. Nearly all the nikkomycin biosynthetic genes were found to be silent at both 24 h and 48 h in ΔwblA (Figure S2a,b and Table S9). Since no direct binding was observed between WblA and the related promoters, the mechanism that wblA disruption activated TACs’ production and simultaneously abolished nikkomycin biosynthesis is still unknown; more investigations need to be performed.
2.4. Production of an Anthracycline Antibiotic Oviedomycin Activated by Disrupting Genes san7324 plus san7324L
In ΔwblA, transcription of an rsbR homologous gene ctg1_705 (named as san7324) in RNA-seq data was found to be dramatically decreased, which was consistent with the RT-PCR results (Figure 4a,b), implying that it was positively regulated by WblA. RsbR encoded by rsbR is a positive regulator modulating sigma factor B activity in Bacillus subtilis [,,]. The disruption of san7324 in S. ansochromogenes 7100 abolished nikkomycin production (Figure S3). However, unlike that in ΔwblA, the production of tylosin analogues in Δsan7324 was not detected []. One hypothesis is that a san7324 homologous gene ctg1_3665 (named as san7324L), which was almost not transcribed in ΔwblA, might play the same function, while san7324 was deficient. In addition, their flanking genes (encoding RsbS, RsbT or sigma factor) also showed low transcriptional levels (Figure 4c,d).
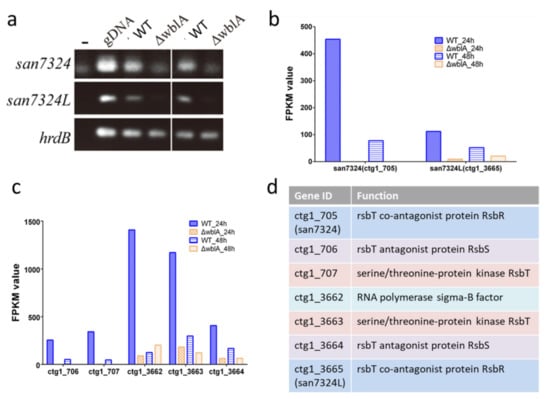
Figure 4.
Transcriptional analysis of san 7324 and san 7324L in both S. ansochromogenes 7100 and ∆wblA. (a) RT-PCR analysis of san7324 and san7324L, -: no DNA template was added. PCR amplifications with cDNA as template were repeated twice. (b) FPKM value of san 7324 and san7324L from RNAseq data. (c) FPKM value of flanking genes adjacent to san 7324 and san 7324L. (d) Gene annotation of ctg1_705–707 and ctg1_3662–3665.
Thus, the double mutation of san7324 and san7324L (Δsan7324/7324L) was constructed and the HPLC results showed TACs were still not be observed. To our surprise, a new peak with maximum absorbance at 280 nm was detected. This compound exhibited an obvious inhibition zone against Bacillus subtilis (Figure 5a,b). Mass spectrometry (MS) analysis showed a [M + H]+ signal at m/z 351.2, which is consistent with oviedomycin (Figure 5c). Oviedomycin is an anthracycline antibiotic which had been previously activated in ΔadpA []. The production of oviedomycin implied that RsbR homologs might negatively regulate oviedomycin biosynthesis.
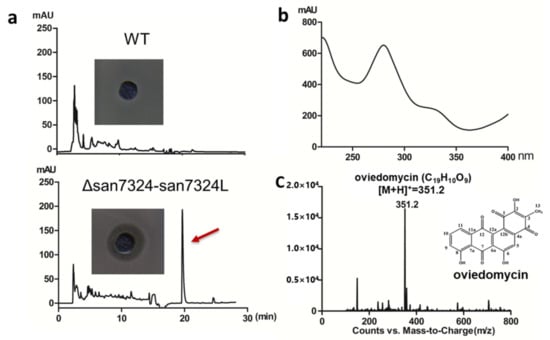
Figure 5.
Identification of oviedomycin in S. ansochromogenes 7100 (WT) and Δsan7324–7324L. (a) HPLC analysis of fermentation supernatant of WT and Δsan7324–7324L. (b) UV absorption spectrum of oviedomycin. (c) Oviedomycin [M + H]+ signal at m/z 351.2 was analyzed by MS and the structure was shown.
2.5. Understanding on Activation of Cryptic Antibiotic Biosynthetic Gene Clusters Guided by RNA-seq Data
The TACs biosynthesis was activated by the disruption of wblA, but the mechanism of the action of WblA is quite complicated because WblA as a global regulator influences a variety of target genes. Furthermore, there is a serious challenge in obtaining sufficient and active WblA protein (an unstable protein) to perform the protein-DNA interaction experiment. However, the RNA-seq data indicated that some sets of genes involved in several precursors’ (Acetyl-CoA, methylmalonyl-CoA etc.) biosynthesis up-regulated in ΔwblA, which may be helpful for the biosynthesis of TACs. In addition to focusing on the role of WblA in secondary metabolites biosynthesis and morphological differentiation, we also noticed that the transcription of wblA (ctg1_3506) itself maintained at a high level in its parent strain, but decreased to 0.35-fold and 0.1-fold in ΔwblA at 24 h and 48 h, respectively. Thus, WblA seems to be a positive regulator for its own transcription, and the detailed mechanism of its action remains to be revealed.
Prior to making a decision on target genes for other products’ biosynthesis, we screened the differentially expressed genes in ΔwblA and found the transcription of san7324 and san7324L was substantially down-regulated more than 10-fold in ΔwblA at 24 h, and also their deduced products (enzymes) are probably involved in biosynthetic pathways, therefore the two genes were selected for further experiment. The results demonstrated that oviedomycin production can be triggered by the disruption of san7324 and san7324L, but further investigations on their specific roles need to be conducted in the future. The function of Wbl family proteins has been reported in a number of studies. Some of them had the capacity to form a protein-protein complex and change the structural conformation to regulate its targets, while some Wbl proteins can also modulate sigma factors [,,,], suggesting that they may play broad roles in multiple pathways. In conclusion, mining of the RNA-seq data provided a useful alternative approach for searching target genes, which may be related to secondary metabolisms. It would be more effective to activate silent gene clusters if combined with other strategies.
3. Materials and Methods
3.1. Strains and Growth Condition
Streptomyces ansochromogenes 7100, a nikkomycin producer, was used as the wild-type strain. S. ansochromogenes and its derivatives were grown on MS (2% soya flour, 2% mannitol) agar at 28 °C for harvesting spores. For nikkomycin or tylosin analogue compounds (1, 2, 3) production, the strains were firstly grown in SP (3% mannitol, 1% potato starch, 0.8% yeast extract, 0.5% neutral soy peptone) as a seed medium and then inoculated 5% (v/v = 5/100) seed culture into SP (3% mannitol, 1% potato starch, 0.8% yeast extract, 0.5% neutral soy peptone) for further fermentation at 28 °C for 5–8 days. For the production of oviedomycin, S. ansochromogenes 7100 and its derivatives were grown in the same seed medium and then 1% (v/v = 1/100) of seed culture was inoculated into a GYM medium (0.4% glucose, 0.4% malt extract, 1% yeast extract) for further fermentation at 28 °C for 6 days. Disruption mutants of san 7324 (ctg1_705), san7324L (ctg1_3665), the double gene disruption mutant (Δsan7324–7324L) and complementary strain were previously constructed []. The wblA disruption mutant (ΔwblA) and its complementary strain were previously constructed [].
3.2. HPLC Analysis of Oviedomycin
The cultures were filtered through gauze, extracted with chloroform, dried in vacuo and then re-dissolved in 1 mL of methanol. The detection of oviedomycin was performed by HPLC with a ZORBAX SB-C18 reverse phase column (4.6 mm × 250 mm, 5 μm; Agilent) as described previously [].
3.3. PacBio Sequencing, Assembly and Gene Annotation
The 64 μg of high-quality genomic DNA was used to prepare the SMRTbell library, and then sequenced on a PacBio RS II platform [].
Prior to assembly, raw reads containing low-quality were filtered. After pre-process, a total of 2.09 Gb of the usable data were retained for the following assembly (Table S1). We used Canu software to assemble the subreads into scaffolds. The final assembly size is 9.56 Mb with scaffold N50 of 9.21M (Table S2). Repeat sequences were identified by Repeat Masker software. The estimated repeat sequences account for 2.37% of the genome assemblies (Table S3). Prodigal was performed to predict genes. In total, 8325 protein-encoding genes were generated for S. ansochromogenes 7100 (Table S4). The tRNA-coding genes were predicted by tRNAscan-SE (v1.3.1) software. rRNA and other ncRNA-coding genes were identified by Infernal 1.1 against Rfam database (http://rfam.xfam.org/, access on 9 September 2021) (Table S5). Annotation of predicted genes was conducted by BLASTP against NCBI non-redundant protein sequence database (NR), Swissprot and TrEMBL protein databases with E-value < 10−5. Gene Ontology (GO) terms were retrieved from NR outputs by Blast2GO software []. Pathway analysis was performed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) annotation service KAAS (Table S6).
3.4. RNA Isolation, Library Construction and Sequencing
The mycelia of S. ansochromogenes 7100 and ΔwblA were harvested at 24 h and 48 h from a 50-mL culture by centrifugation at 12,000 rpm for 15 min at 4 °C. The cell pellets were quickly frozen in liquid N2 and then stored at −80 °C. Total RNAs were isolated using the TRIzol reagent, and DNase I was used to digest the total DNA according to the manufacturer’s protocol. RNA degradation or contamination was monitored with 1% agarose gel electrophoresis. RNA purity was checked by using the NanoPhotometer® spectrophotometer (IMPLEN, Westlake Village, CA, USA). Strand-specific RNA-Seq libraries were constructed and sequenced by the Illumina HiSeq 2000 system, 100 bp paired-end reads were generated and the RNA-seq data were analyzed by Majorbio company.
3.5. Quality Control and Reads Mapping to the Reference Genome
The raw data were filtered using a Perl program by removing low-quality sequences, reads with more than 5% of N bases (unknown bases) and reads containing adaptor sequences. Then the clean data were mapped to the genome sequences of S. ansochromogenes 7100 by the Bowtie2 program [].
3.6. Quantification of Gene Expression Level and Differential Expression Analysis
An abundance of transcripts was estimated using RSEM and transformed into formula FPKM (fragments per kilobase per million mapped reads). Differentially expressed genes (DEGs) were calculated by using R package DESeq []. Genes with an adjusted p-value (p-adjust) < 0.001 found by DESeq were assigned as differential expressions, and those with fold change |Log2FC| ≥ 1 were defined to be either up- or down-regulated genes, respectively. The Heatmap was made by R package heatmap.2.
3.7. RT-PCR Analysis
For RT-PCR, 1 μg of RNA was reverse-transcribed to cDNA using a SuperRT cDNA Synthesis Kit (CWBIO, Beijing, China) []. To perform transcriptional analysis of san7324 and san7324L, primer pair (san7324-RT-F: ACGCTGCGGCTGGTGGTCAT, san7324-RT-R: CCGTTCCCTCCCACAGCTTGAT) and another primer pair (san7324L-RT-F: CCCGTCATCAAGCTGTGGGAGG, san7324L-RT-R: TCAGGTGCTGGGCCACGAAC) were respectively used for PCR amplification with cDNAs of WT and ΔwblA as templates. The transcription of hrdB using primers (hrdB-RT-F: GCTGGCCAAGGAACTCGACAT, hrdB-RT-R: CGAAGCGCATGGAGACGACG) was used as an internal control.
3.8. Data Availability
The genomic DNA sequence of S. ansochromogenes 7100 is deposited in the China National Microbiology Data Center (NMDC) with accession numbers NMDC60029072. RNA-seq raw data were deposited in the China National Microbiology Data Center (NMDC) with accession numbers NMDC40009909 (WT_24 h), NMDC40009910 (WT_48 h), NMDC40009911 (ΔwblA_24 h), NMDC40009912 (ΔwblA_48 h).
4. Conclusions
RNA-seq analyses demonstrated that high-level transcription of most genes was associated with the production of nikkomycin in WT and TACs in ΔwblA appeared at 24 h, but decreased significantly at 48 h. Interestingly, double mutation of san7324 and san7324L (Δsan7324/7324L) led to the production of oviedomycin, which was not produced in wild-type S. ansochromogenes under laboratory conditions. It was suggested that RsbR homologs (San7324/7324L) might negatively regulate oviedomycin biosynthesis. Moreover, the findings would provide an effective approach to dissect the production and biosynthetic pathways of natural secondary metabolites, and also provide insights into the activation of cryptic natural products.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/antibiotics10091097/s1, Figure S1: Saturation curves of sequencing data in RNA-seq, Figure S2: Differentially expressed genes involved in nikkomycin biosynthesis in both WT and ΔwblA, Figure S3: HPLC analysis of nikkomycin production in WT and Δsan7324, Table S1: Statistics of filtered sequencing data, Table S2: Assembly of genome sequencing data, Table S3: Summary of repeat elements identified, Table S4: Statistics of prediction of the proteins, Table S5: Non-coding RNAs, Table S6: Annotation of genes, Table S7: Genome mapping statistics of RNA-seq data, Table S8: Significantly expressed genes in Venn diagram, Table S9: FPKM value of part of differentially expressed genes.
Author Contributions
Y.L. performed experiments, bioinformatics analysis and wrote the original manuscript; H.Y. performed bioinformatics analysis and wrote part of the manuscript; H.G. participated in fermentation and confirmed oviedomycin structure; J.L. (Jingjing Li) performed the RT-PCR experiment; J.Z., H.X. and J.L. (Jine Li) revised the manuscript; H.T. conceived and supervised the whole research work, and also revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Key Research and Development Program of China (2020YFA0907800) and the National Natural Science Foundation of China (31771378 and 31800029).
Data Availability Statement
The data presented in this study are openly available in China National Microbiology Data Center (NMDC), reference number of the BioProject is [NMDC10017856]. Data of genomic DNA of S. ansochromogenes 7100 can be found in NMDC60029072, RNA-seq data of S. ansochromogenes 7100 can be found in NMDC40009909 (WT_24 h), NMDC40009910 (WT_48 h), NMDC40009911 (ΔwblA_24 h), NMDC40009912 (ΔwblA_48 h).
Acknowledgments
We thank Cheng Lu for RNA isolation and Jingjing Yang for her kind guidance when we upload the data in China National Microbiology Data Center (NMDC).
Conflicts of Interest
All authors agree and declare no conflict of interest.
References
- Li, Y.; Li, J.; Tian, Z.; Xu, Y.; Zhang, J.; Liu, W.; Tan, H. Coordinative Modulation of Chlorothricin Biosynthesis by Binding of the Glycosylated Intermediates and End Product to a Responsive Regulator ChlF1. J. Biol. Chem. 2016, 291, 5406–5417. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Zhang, J.; Zheng, J.; Guan, H.; Liu, W.; Tan, H. Co-expression of a SARP Family Activator ChlF2 and a Type II Thioesterase ChlK Led to High Production of Chlorothricin in Streptomyces antibioticus DSM 40725. Front. Bioeng. Biotechnol. 2020, 8, 1013. [Google Scholar] [CrossRef]
- Liu, G.; Chater, K.F.; Chandra, G.; Niu, G.; Tan, H. Molecular Regulation of Antibiotic Biosynthesis in Streptomyces. Microbiol. Mol. Biol. Rev. 2013, 77, 112–143. [Google Scholar] [CrossRef] [Green Version]
- Rutledge, P.; Challis, G. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat. Rev. Genet. 2015, 13, 509–523. [Google Scholar] [CrossRef]
- Li, Y.; Tan, H. Biosynthesis and molecular regulation of secondary metabolites in microorganisms. Sci. China Life Sci. 2017, 60, 935–938. [Google Scholar] [CrossRef]
- Schroeckh, V.; Scherlach, K.; Nützmann, H.-W.; Shelest, E.; Schmidt-Heck, W.; Schuemann, J.; Martin, K.; Hertweck, C.; Brakhage, A.A. Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA 2009, 106, 14558–14563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Zhang, J.; Tian, Y.; Tan, H. Enhancement of salinomycin production by ribosome engineering in Streptomyces albus. Sci. China Life Sci. 2019, 62, 276–279. [Google Scholar] [CrossRef] [PubMed]
- Van Santen, J.A.; Kautsar, S.A.; Medema, M.H.; Linington, R.G. Microbial natural product databases: Moving forward in the multi-omics era. Nat. Prod. Rep. 2021, 38, 264–278. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Medema, M.H.; Kottmann, R.; Lee, S.Y.; Weber, T. The antiSMASH database, a comprehensive database of microbial secondary metabolite biosynthetic gene clusters. Nucleic Acids Res. 2017, 45, D555–D559. [Google Scholar] [CrossRef] [Green Version]
- Blin, K.; Kim, H.U.; Medema, M.H.; Weber, T. Recent development of antiSMASH and other computational approaches to mine secondary metabolite biosynthetic gene clusters. Brief. Bioinform. 2019, 20, 1103–1113. [Google Scholar] [CrossRef] [PubMed]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptivek-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.-Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Guo, Z.; Tang, W.; Chen, Y. Activation of three natural product biosynthetic gene clusters from Streptomyces lavendulae CGMCC 4.1386 by a reporter-guided strategy. Synth. Syst. Biotechnol. 2018, 3, 254–260. [Google Scholar] [CrossRef]
- Ji, Z.-Y.; Nie, Q.-Y.; Yin, Y.; Zhang, M.; Pan, H.-X.; Hou, X.-F.; Tang, G.-L. Activation and Characterization of Cryptic Gene Cluster: Two Series of Aromatic Polyketides Biosynthesized by Divergent Pathways. Angew. Chem. Int. Ed. 2019, 58, 18046–18054. [Google Scholar] [CrossRef]
- Liu, N.; Guan, H.; Niu, G.; Jiang, L.; Li, Y.; Zhang, J.; Li, J.; Tan, H. Molecular mechanism of mureidomycin biosynthesis activated by introduction of an exogenous regulatory gene ssaA into Streptomyces roseosporus. Sci. China Life Sci. 2021, 1–15. [Google Scholar] [CrossRef]
- Bush, M.J. The actinobacterial WhiB-like (Wbl) family of transcription factors. Mol. Microbiol. 2018, 110, 663–676. [Google Scholar] [CrossRef]
- Kim, J.S.; Lee, H.N.; Kim, P.; Lee, H.S.; Kim, E.S. Negative Role of wblA in Response to Oxidative Stress in Streptomyces coelicolor. J. Microbiol. Biotechnol. 2012, 22, 736–741. [Google Scholar] [CrossRef]
- Nah, H.-J.; Park, J.; Choi, S.; Kim, E.-S. WblA, a global regulator of antibiotic biosynthesis in Streptomyces. J. Ind. Microbiol. Biotechnol. 2021, 48. [Google Scholar] [CrossRef]
- Huang, X.; Ma, T.; Tian, J.; Shen, L.; Zuo, H.; Hu, C.; Liao, G. wblA, a pleiotropic regulatory gene modulating morphogenesis and daptomycin production in Streptomyces roseosporus. J. Appl. Microbiol. 2017, 123, 669–677. [Google Scholar] [CrossRef]
- Singh, A.; Guidry, L.; Narasimhulu, K.V.; Mai, D.; Trombley, J.; Redding, K.E.; Giles, G.; Lancaster, J.R.; Steyn, A.J.C. Mycobacterium tuberculosis WhiB3 responds to O2 and nitric oxide via its [4Fe–4S] cluster and is essential for nutrient starvation survival. Proc. Natl. Acad. Sci. USA 2007, 104, 11562–11567. [Google Scholar] [CrossRef] [Green Version]
- Burian, J.; Yim, G.; Hsing, M.; Axerio-Cilies, P.; Cherkasov, A.; Spiegelman, G.B.; Thompson, C.J. The mycobacterial antibiotic resistance determinant WhiB7 acts as a transcriptional activator by binding the primary sigma factor SigA (RpoV). Nucleic Acids Res. 2013, 41, 10062–10076. [Google Scholar] [CrossRef] [PubMed]
- Garg, S.; Alam, S.; Bajpai, R.; Kishan, K.R.; Agrawal, P. Redox biology of Mycobacterium tuberculosis H37Rv: Protein-protein interaction between GlgB and WhiB1 involves exchange of thiol-disulfide. BMC Biochem. 2009, 10, 1. [Google Scholar] [CrossRef] [Green Version]
- Tan, H.; Wu, W.; Tian, Y.; Yang, H.; Song, Y.; Dong, K.; Huang, X.; Song, D. Studies on the differentiation and properties of stretptomyces ansochromogens. Acta Microbiol. Sin. 1994, 34, 398–402. [Google Scholar]
- Pan, Y.; Wang, L.; He, X.; Tian, Y.; Liu, G.; Tan, H. SabR enhances nikkomycin production via regulating the transcriptional level of sanG, a pathway-specific regulatory gene in Streptomyces ansochromogenes. BMC Microbiol. 2011, 11, 164. [Google Scholar] [CrossRef] [Green Version]
- Lu, C.; Liao, G.; Zhang, J.; Tan, H. Identification of novel tylosin analogues generated by a wblA disruption mutant of Streptomyces ansochromogenes. Microb. Cell Factories 2015, 14, 173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palaniappan, K.; Chen, I.-M.A.; Chu, K.; Ratner, A.; Seshadri, R.; Kyrpides, N.; Ivanova, N.N.; Mouncey, N.J. IMG-ABC v.5.0: An update to the IMG/Atlas of Biosynthetic Gene Clusters Knowledgebase. Nucleic Acids Res. 2019, 48, D422–D430. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niu, G.; Tan, H. Nucleoside antibiotics: Biosynthesis, regulation, and biotechnology. Trends Microbiol. 2015, 23, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Moy, B.E.; Seshu, J. STAS Domain Only Proteins in Bacterial Gene Regulation. Front. Cell Infect. Microbiol. 2021, 11, 679982. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Xin, Y.-P.; Shan, Y.; Zhang, X.; Song, H.-H.; Fang, W.-H. Phosphorylation residue T175 in RsbR protein is required for efficient induction of sigma B factor and survival of Listeria monocytogenes under acidic stress. J. Zhejiang Univ. Sci. B 2019, 20, 660–669. [Google Scholar] [CrossRef] [PubMed]
- Van der Steen, J.B.; Ávila-Pérez, M.; Knippert, D.; Vreugdenhil, A.; van Alphen, P.; Hellingwerf, K.J. Differentiation of Function among the RsbR Paralogs in the General Stress Response of Bacillus subtilis with Regard to Light Perception. J. Bacteriol. 2012, 194, 1708–1716. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.; Li, J.; Li, Y.; Cai, Y.; Tan, H. Effects of san7324 and san7324L disruption on morphological differentiation and nikkomycin production of Streptomyces ansochromogenes. Acta Microbiol. Sin. 2019, 59, 235–246. [Google Scholar]
- Xu, J.; Zhang, J.; Zhuo, J.; Li, Y.; Tian, Y.; Tan, H. Activation and mechanism of a cryptic oviedomycin gene cluster via the disruption of a global regulatory gene, adpA, in Streptomyces ansochromogenes. J. Biol. Chem. 2017, 292, 19708–19720. [Google Scholar] [CrossRef] [Green Version]
- Fowler-Goldsworthy, K.; Gust, B.; Mouz, S.; Chandra, G.; Findlay, K.C.; Chater, K.F. The actinobacteria-specific gene wblA controls major developmental transitions in Streptomyces coelicolor A3(2). Microbiology 2011, 157, 1312–1328. [Google Scholar] [CrossRef] [Green Version]
- Stewart, M.Y.Y.; Bush, M.; Crack, J.C.; Buttner, M.; Le Brun, N.E. Interaction of the Streptomyces Wbl protein WhiD with the principal sigma factor sigma(σHrdB) depends on the WhiD [4Fe–4S] cluster. J. Biol. Chem. 2020, 295, 9752–9765. [Google Scholar] [CrossRef] [Green Version]
- Zhiwu, Q.; Gui, S.; Dequan, L.; Pan, L.; Wang, S.; Ke, W.; Liang, D.; Ding, Y. A precise chloroplast genome of Nelumbo nucifera (Nelumbonaceae) evaluated with Sanger, Illumina MiSeq, and PacBio RS II sequencing platforms: Insight into the plastid evolution of basal eudicots. BMC Plant Biol. 2014, 14, 289. [Google Scholar] [CrossRef] [Green Version]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, J.; Li, Y.; Guan, H.; Zhang, J.; Tan, H. Enhancement of neomycin production by engineering the entire biosynthetic gene cluster and feeding key precursors in Streptomyces fradiae CGMCC 4.576. Appl. Microbiol. Biotechnol. 2019, 103, 2263–2275. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).