MCR Expression Conferring Varied Fitness Costs on Host Bacteria and Affecting Bacteria Virulence
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, Cell Lines, and Culture Conditions
2.2. Antimicrobial Susceptibility Testing
2.3. Bacteria Growth Kinetics Detection
2.4. Confocal Microscopy Imaging Using LIVE/DEAD Staining
2.5. Membrane Permeability Test Assay
2.6. Cell Infection Experiment
2.7. G. mellonella Infection Model
2.8. Statistical Analysis
3. Results
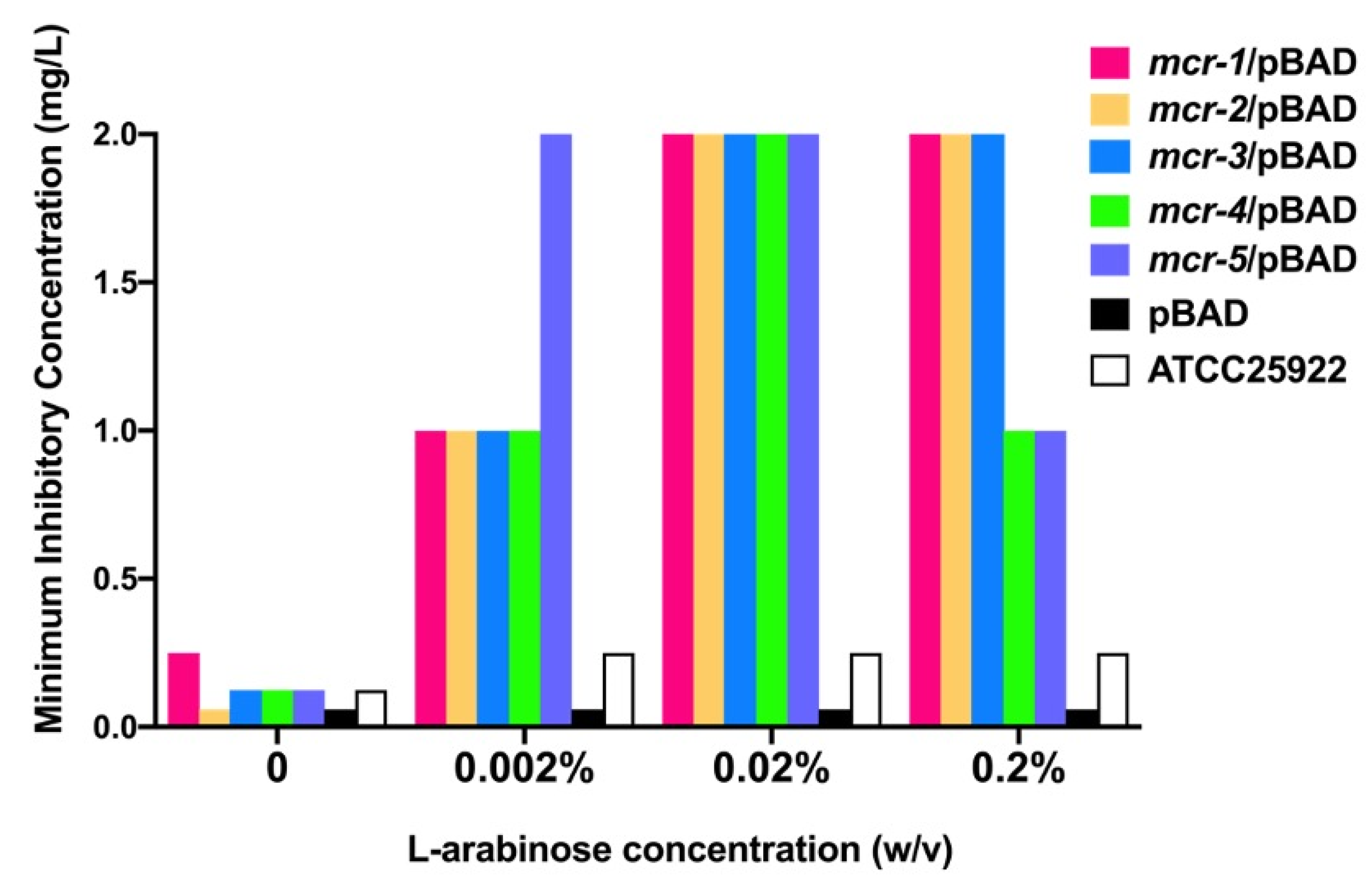
3.1. Mcr-1~5 Mediated Colistin Resistance Levels
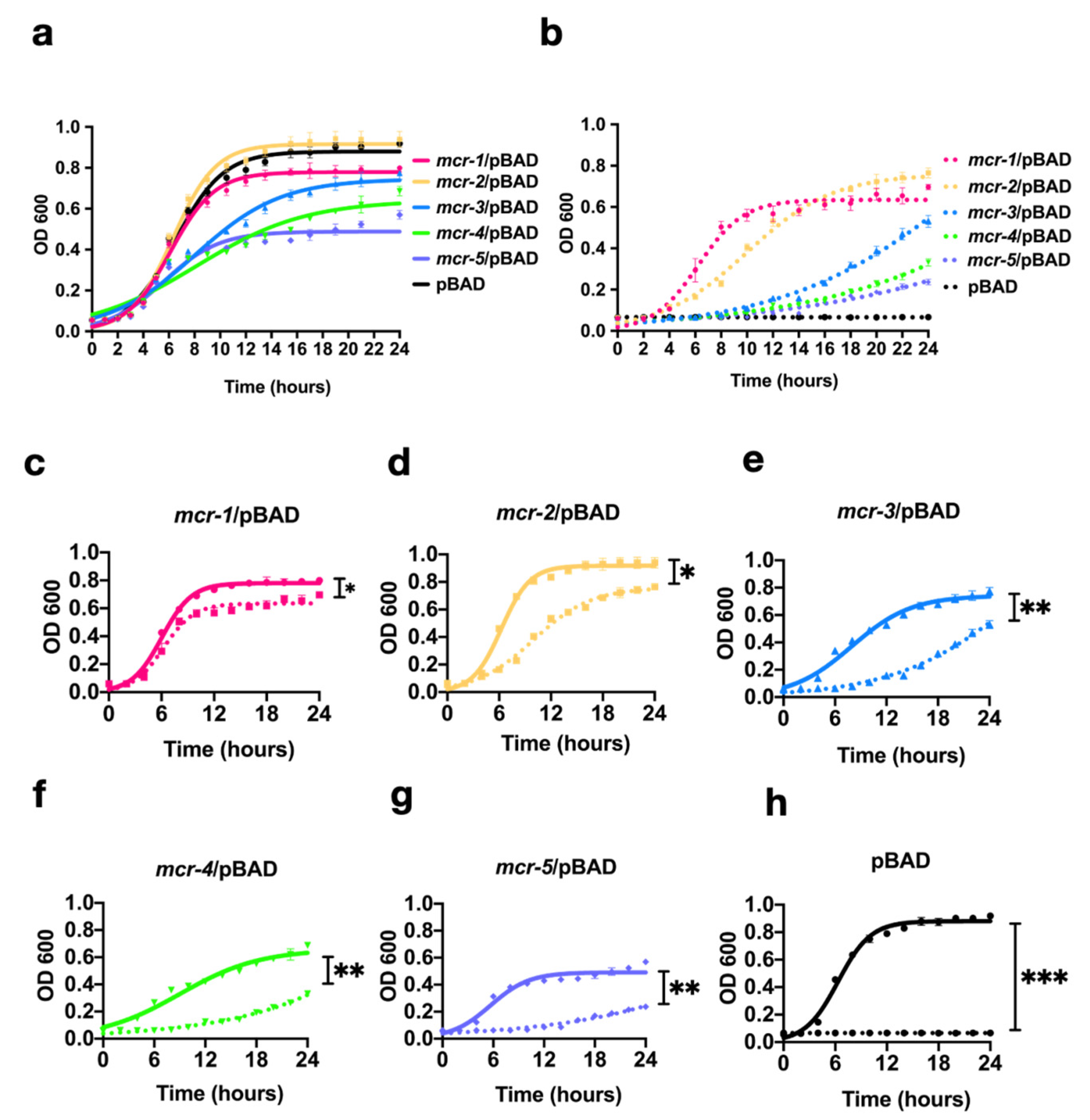
3.2. MCR-1~5 Expression Conferred Varied Fitness Costs on the Host Strain
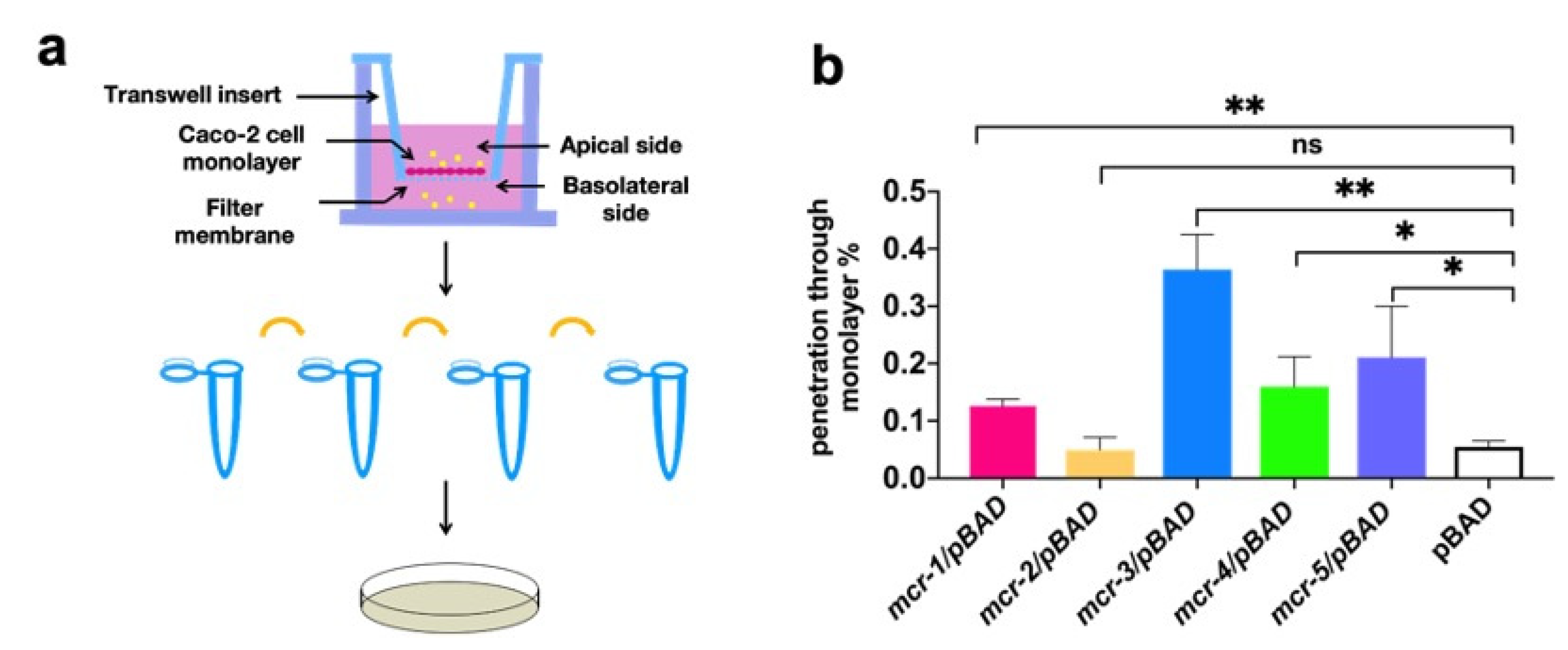
3.3. MCR Expression Increased Bacterial Virulence In Vitro and In Vivo
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Primers | Sequences | Source |
|---|---|---|
| mcr-1-F | ggctaacaggaggaattaaccatggATGATGCAGCATACTTCTGTGTG | This study |
| mcr-1-R | gccaaaacagccaagcttcgaattcTCAGCGGATGAATGCGGTG | This study |
| mcr-2-F | ggctaacaggaggaattaaccatggATGACATCACATCACTCTTG | This study |
| mcr-2-R | gccaaaacagccaagcttcgaattcTTACTGGATAAATGCCGC | This study |
| mcr-3-F | ggctaacaggaggaattaaccatggATGCCTTCCCTTATAAAAATAAAAATTG | This study |
| mcr-3-R | gccaaaacagccaagcttcgaattcTTATTGAACATTACGACATTGAC | This study |
| mcr-4-F | ggctaacaggaggaattaaccatggGTGATTTCTAGATTTAAGACGTTATC | This study |
| mcr-4-R | gccaaaacagccaagcttcgaattcCTAATACCTGCAAGGTGC | This study |
| mcr-5-F | ggctaacaggaggaattaaccatggATGCGGTTGTCTGCATTTATC | This study |
| mcr-5-R | gccaaaacagccaagcttcgaattcTCATTGTGGTTGTCCTTTTC | This study |
References
- Yang, Q.; Li, M.; Spiller, O.B.; Andrey, D.O.; Hinchliffe, P.; Li, H.; MacLean, C.; Niumsup, P.; Powell, L.; Pritchard, M.; et al. Balancing mcr-1 expression and bacterial survival is a delicate equilibrium between essential cellular defence mechanisms. Nat. Commun. 2017, 8, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Tian, G.; Zhang, R.; Shen, Y.; Tyrrell, M.J.; Huang, X.; Zhou, H.; Lei, L.; Li, H.; Doi, Y.; et al. Prevalence, risk factors, outcomes, and molecular epidemiology of mcr-1-positive Enterobacteriaceae in patients and healthy adults from China: An epidemiological and clinical study. Lancet Infect. Dis. 2017, 17, 390–399. [Google Scholar] [CrossRef] [Green Version]
- Voigt, A.M.; Ciorba, P.; Döhla, M.; Exner, M.; Felder, C.; Lenz-Plet, F.; Sib, E.; Skutlarek, D.; Schmithausen, R.M.; Faerber, H.A. The investigation of antibiotic residues, antibiotic resistance genes and antibiotic-resistant organisms in a drinking water reservoir system in Germany. Int. J. Hyg. Environ. Health 2020, 224, 113449. [Google Scholar] [CrossRef]
- Wang, X.; Wang, Y.; Ying, Z.; Li, J.; Yin, W.; Wang, S.; Shen, J.; Shen, Z.; Wang, Y. Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg. Microbes Infect. 2018, 7, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, W.; Li, H.; Shen, Y.; Liu, Z.; Wang, S.; Shen, Z.; Zhang, R.; Walsh, T.R.; Shen, J.; Wang, Y. Novel Plasmid-Mediated Colistin Resistance Gene mcr-3 in Escherichia coli. mBio 2017, 8, e00543-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carattoli, A.; Villa, L.; Feudi, C.; Curcio, L.; Orsini, S.; Luppi, A.; Pezzotti, G.; Magistrali, F.C. Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Eurosurveillance 2017, 22, 30589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borowiak, M.; Fischer, J.; Hammerl, J.A.; Hendriksen, R.S.; Szabo, I.; Malorny, B. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J. Antimicrob. Chemother. 2017, 72, 3317–3324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- AbuOun, M.; Stubberfield, J.E.; Duggett, A.N.; Kirchner, M.; Dormer, L.; Nunez-Garcia, J.; Randall, P.L.; Lemma, F.; Crook, W.D.; Teale, C.; et al. mcr-1 and mcr-2 (mcr-6.1) variant genes identified in Moraxella species isolated from pigs in Great Britain from 2014 to 2015. J. Antimicrob. Chemother. 2017, 72, 2745–2749. [Google Scholar] [CrossRef] [Green Version]
- Xu, T.; Zhang, C.; Ji, Y.; Song, J.; Liu, Y.; Guo, Y.; Zhou, K. Identification of mcr-10 carried by self-transmissible plasmids and chromosome in Enterobacter roggenkampii strains isolated from hospital sewage water. Environ. Pollut. 2021, 268, 115706. [Google Scholar] [CrossRef]
- Carroll, L.M.; Gaballa, A.; Guldimann, C.; Sullivan, G.; Henderson, L.O.; Wiedmann, M. Identification of Novel Mobilized Colistin Resistance Gene mcr-9 in a Multidrug-Resistant, Colistin-Susceptible Salmonella enterica Serotype Typhimurium Isolate. mBio 2019, 10, e00853-19. [Google Scholar] [CrossRef] [Green Version]
- Ling, Z.; Yin, W.; Shen, Z.; Wang, Y.; Shen, J.; Walsh, T.R. Epidemiology of mobile colistin resistance genes mcr-1 to mcr-10. J. Antimicrob. Chemother. 2020, 75, 3087–3095. [Google Scholar] [CrossRef]
- Yang, Q.E.; MacLean, C.; Papkou, A.; Pritchard, M.; Powell, L.; Thomas, D.; Andrey, O.D.; Li, M.; Spiller, B.; Yang, W.; et al. Compensatory mutations modulate the competitiveness and dynamics of plasmid-mediated colistin resistance in Escherichia coli clones. ISME J. 2020, 14, 861–865. [Google Scholar] [CrossRef]
- Wu, R.; Yi, L.; Yu, L.; Wang, J.; Liu, Y.; Chen, X.; Lv, L.; Yang, J.; Liu, J. Fitness Advantage of mcr-1–Bearing IncI2 and IncX4 Plasmids in Vitro. Front. Microbiol. 2018, 9, 331. [Google Scholar] [CrossRef]
- Da Silva, G.J.; Domingues, S. Interplay between Colistin Resistance, Virulence and Fitness in Acinetobacter baumannii. Antibiotics 2017, 6, 28. [Google Scholar] [CrossRef] [Green Version]
- Bahador, A.; Farshadzadeh, Z.; Raoofian, R.; Mokhtaran, M.; Pourakbari, B.; Pourhajibagher, M.; Hashemi, B.F. Association of virulence gene expression with colistin-resistance in Acinetobacter baumannii: Analysis of genotype, antimicrobial susceptibility, and biofilm formation. Ann. Clin. Microbiol. Antimicrob. 2018, 17, 24. [Google Scholar] [CrossRef]
- De La Tabla, V.O.; Ortega, A.; Buñuel, F.; Pérez-Vázquez, M.; Marcos, B.; Oteo, J. Detection of the high-risk clone ST131 of Escherichia coli carrying the colistin resistance gene mcr-1 and causing acute peritonitis. Int. J. Antimicrob. Agents 2017, 49, 115–116. [Google Scholar] [CrossRef]
- Simon-Flament, S.; de Toro, M.; Mora, A.; García, V.; Meniño, G.I.; Díaz-Jiménez, D.; Herrera, A.; Blanco, J. Whole Genome Sequencing and Characteristics of mcr-1–Harboring Plasmids of Porcine Escherichia coli Isolates Belonging to the High-Risk Clone O25b:H4-ST131 Clade B. Front. Microbiol. 2020, 11, 387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayashi, W.; Tanaka, H.; Taniguchi, Y.; Iimura, M.; Soga, E.; Kubo, R.; Matsuo, N.; Kawamura, K.; Arakawa, Y.; Nagano, Y.; et al. Acquisition of mcr-1 and Cocarriage of Virulence Genes in Avian Pathogenic Escherichia coli Isolates from Municipal Wastewater Influents in Japan. Appl. Environ. Microbiol. 2019, 85, e01661-19. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, Y.; Walsh, R.T.; Yi, L.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Shi, X.; Yin, W.; Wang, Y.; Shen, Z.; Ding, S.; Wang, S. A Multiplex SYBR Green Real-Time PCR Assay for the Detection of Three Colistin Resistance Genes from Cultured Bacteria, Feces, and Environment Samples. Front. Microbiol. 2017, 8, 2078. [Google Scholar] [CrossRef]
- Wise, M.G.; Estabrook, M.A.; Sahm, D.F.; Stone, G.G.; Kazmierczak, K.M. Prevalence of mcr-type genes among colistin-resistant Enterobacteriaceae collected in 2014–2016 as part of the INFORM global surveillance program. PLoS ONE 2018, 13, e0195281. [Google Scholar] [CrossRef] [Green Version]
- Gelbíčová, T.; Baráková, A.; Florianová, M.; Jamborová, I.; Zelendová, M.; Pospíšilová, L.; Koláčková, I.; Karpíšková, R. Dissemination and Comparison of Genetic Determinants of mcr-Mediated Colistin Resistance in Enterobacteriaceae via Retailed Raw Meat Products. Front. Microbiol. 2019, 10, 2824. [Google Scholar] [CrossRef] [Green Version]
- Alba, P.; Leekitcharoenphon, P.; Franco, A.; Feltrin, F.; Ianzano, A.; Caprioli, A.; Stravino, F.; Hendriksen, S.R.; Bortolaia, V.; Battisti, A. Molecular Epidemiology of mcr-Encoded Colistin Resistance in Enterobacteriaceae From Food-Producing Animals in Italy Revealed Through the EU Harmonized Antimicrobial Resistance Monitoring. Front. Microbiol. 2018, 9, 1217. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Van Dorp, L.; Shaw, L.P.; Bradley, P.; Wang, Q.; Wang, X.; Jin, L.; Zhang, Q.; Liu, Y.; Rieux, A.; et al. The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat. Commun. 2018, 9, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Russell, N.D.; Corvalan, J.R.F.; Gallo, M.L.; Davis, C.G.; Pirofski, L.-A. Production of Protective Human Antipneumococcal Antibodies by Transgenic Mice with Human Immunoglobulin Loci. Infect. Immun. 2000, 68, 1820–1826. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.; Wu, Z.; Wang, Y.; Zhang, R.; Zhou, H.-W.; Wang, S.; Lei, L.; Li, M.; Cai, J.; Tyrrell, J.; et al. Heterogeneous and Flexible Transmission of mcr-1 in Hospital-Associated Escherichia coli. mBio 2018, 9, e00943-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Lorenzo, F.; De Castro, C.; Silipo, A.; Molinaro, A. Lipopolysaccharide structures of Gram-negative populations in the gut microbiota and effects on host interactions. FEMS Microbiol. Rev. 2019, 43, 257–272. [Google Scholar] [CrossRef]
- Wang, X.; Ribeiro, A.A.; Guan, Z.; Abraham, S.N.; Raetz, C.R.H. Attenuated virulence of a Francisella mutant lacking the lipid A 4’-phosphatase. Proc. Natl. Acad. Sci. USA 2007, 104, 4136–4141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hurley, J.C. Antibiotic-Induced Release of Endotoxin: A Reappraisal. Clin. Infect. Dis. 1992, 15, 840–854. [Google Scholar] [CrossRef] [PubMed]
- Hellman, J.; Roberts, J.D.; Tehan, M.M.; Allaire, J.E.; Warren, H.S. Bacterial Peptidoglycan-associated Lipoprotein Is Released into the Bloodstream in Gram-negative Sepsis and Causes Inflammation and Death in Mice. J. Biol. Chem. 2002, 277, 14274–14280. [Google Scholar] [CrossRef] [Green Version]
- Kaito, C.; Yoshikai, H.; Wakamatsu, A.; Miyashita, A.; Matsumoto, Y.; Fujiyuki, T.; Kato, M.; Ogura, Y.; Hayashi, T.; Isogai, T.; et al. Non-pathogenic Escherichia coli acquires virulence by mutating a growth-essential LPS transporter. PLoS Pathog. 2020, 16, e1008469. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Al-Sadi, R.; Said, H.M.; Ma, T.Y. Lipopolysaccharide Causes an Increase in Intestinal Tight Junction Permeability in Vitro and in Vivo by Inducing Enterocyte Membrane Expression and Localization of TLR-4 and CD14. Am. J. Pathol. 2013, 182, 375–387. [Google Scholar] [CrossRef] [Green Version]
- Elhenawy, W.; Bording-Jorgensen, M.; Valguarnera, E.; Haurat, M.F.; Wine, E.; Feldman, M.F. LPS Remodeling Triggers Formation of Outer Membrane Vesicles in Salmonella. mBio 2016, 7, e00940-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Miao, M.; Yan, J.; Wang, M.; Tang, Y.-W.; Kreiswirth, B.N.; Zhang, X.; Chen, L.; Du, H. Expression characteristics of the plasmid-borne mcr-1 colistin resistance gene. Oncotarget 2017, 8, 107596–107602. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yang, L.; Liu, Z.; Yin, W.; Liu, D.; Shen, Y.; Walsh, R.T.; Shao, B.; Wang, Y. Molecular Insights into Functional Differences between mcr-3- and mcr-1-Mediated Colistin Resistance. Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef] [Green Version]


| Name | Description | Source or Reference |
|---|---|---|
| Strains | ||
| SHP45 | WT E. coli strain with mcr-1 of pig origin | [17] |
| DH5α-mcr-2 | Constructed E. coli strain with mcr-2 | [18] |
| WJ1 | WT E. coli strain with mcr-3 of pig origin | [19,20] |
| D32 | WT E. coli strain with mcr-4 of pig origin | Laboratory collection |
| D13 | WT E. coli strain with mcr-5 of pig origin | Laboratory collection |
| mcr-1/pBAD | E. coli TOP10 with mcr-1/pBAD | This study |
| mcr-2/pBAD | E. coli TOP10 with mcr-2/pBAD | This study |
| mcr-3/pBAD | E. coli TOP10 with mcr-3/pBAD | This study |
| mcr-4/pBAD | E. coli TOP10 with mcr-4/pBAD | This study |
| mcr-5/pBAD | E. coli TOP10 with mcr-5/pBAD | This study |
| pBAD | E. coli TOP10 with pBAD | This study |
| ATCC25922 | E. coli, quality control strain | Laboratory collection |
| Cells | ||
| Caco-2 | Human colon carcinoma cell line | Laboratory collection |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.; Liu, Z.; Yin, W.; Yang, L.; Qiao, L.; Song, S.; Ling, Z.; Zheng, R.; Wu, C.; Wang, Y.; et al. MCR Expression Conferring Varied Fitness Costs on Host Bacteria and Affecting Bacteria Virulence. Antibiotics 2021, 10, 872. https://doi.org/10.3390/antibiotics10070872
Li W, Liu Z, Yin W, Yang L, Qiao L, Song S, Ling Z, Zheng R, Wu C, Wang Y, et al. MCR Expression Conferring Varied Fitness Costs on Host Bacteria and Affecting Bacteria Virulence. Antibiotics. 2021; 10(7):872. https://doi.org/10.3390/antibiotics10070872
Chicago/Turabian StyleLi, Wan, Zhihai Liu, Wenjuan Yin, Lu Yang, Lu Qiao, Shikai Song, Zhuoren Ling, Ruicheng Zheng, Congming Wu, Yang Wang, and et al. 2021. "MCR Expression Conferring Varied Fitness Costs on Host Bacteria and Affecting Bacteria Virulence" Antibiotics 10, no. 7: 872. https://doi.org/10.3390/antibiotics10070872
APA StyleLi, W., Liu, Z., Yin, W., Yang, L., Qiao, L., Song, S., Ling, Z., Zheng, R., Wu, C., Wang, Y., & Shen, J. (2021). MCR Expression Conferring Varied Fitness Costs on Host Bacteria and Affecting Bacteria Virulence. Antibiotics, 10(7), 872. https://doi.org/10.3390/antibiotics10070872






