Bacterial Natural Product Drug Discovery for New Antibiotics: Strategies for Tackling the Problem of Antibiotic Resistance by Efficient Bioprospecting
Abstract
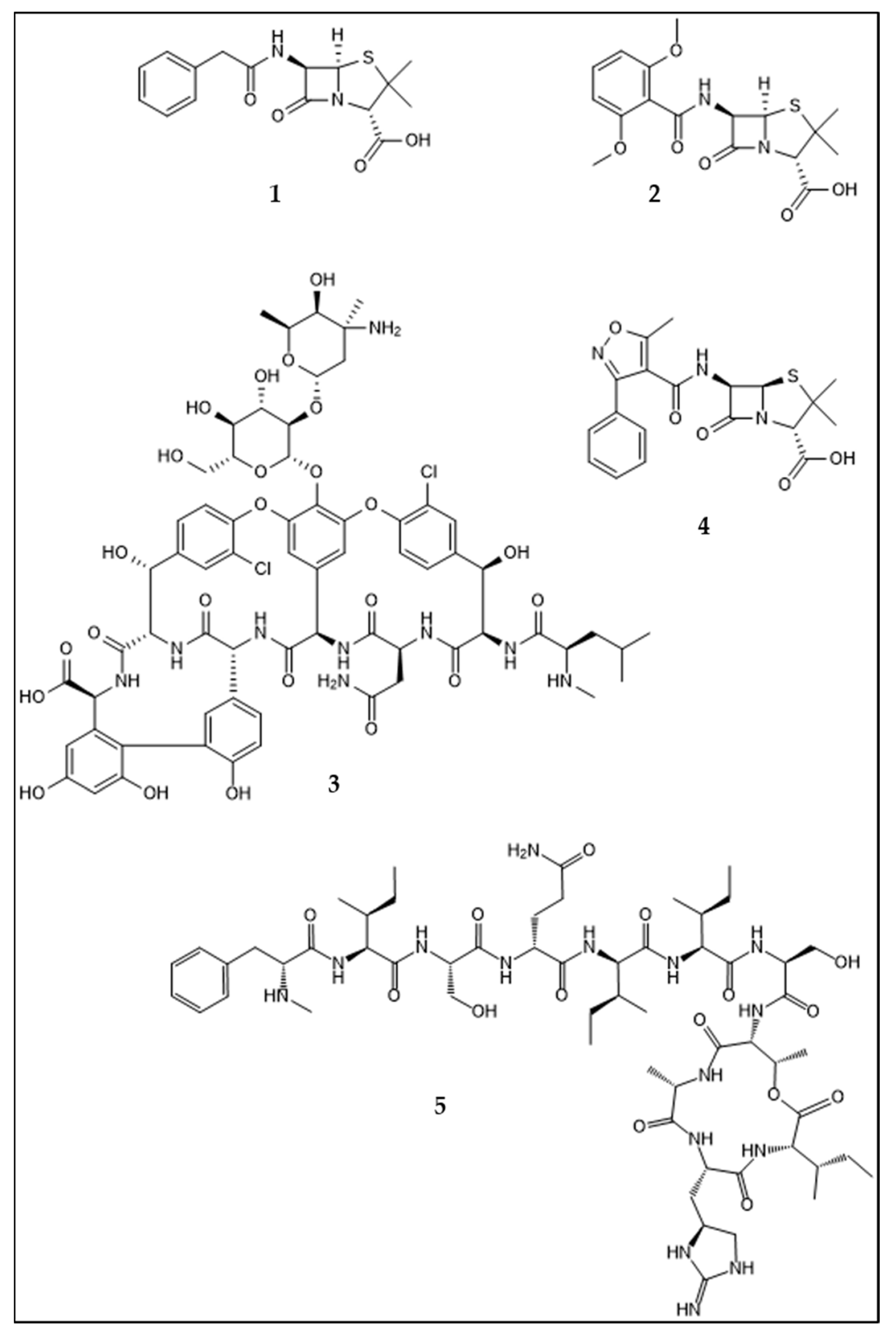
:1. Introduction: Development of Antibiotic Resistance in Staphylococcus aureus. An Example for Acquisition of Resistance to Antibiotics in Clinical Use
2. Mechanisms and Acquisition of Antibiotic Resistance
3. The Ancient Origin of Antibiotic Resistance
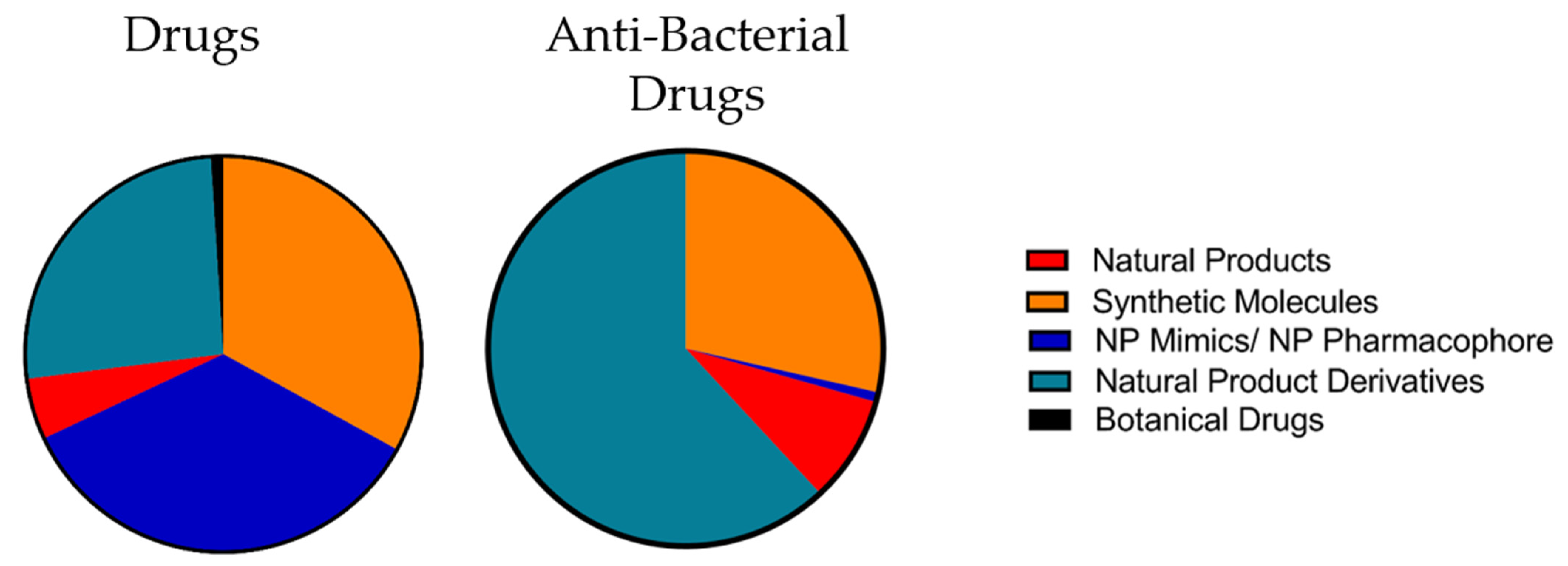
4. Natural Products in Drug Discovery
4.1. Chemical Characteristics of Natural Products
4.2. Suitability of Natural Products for Drug Discovery
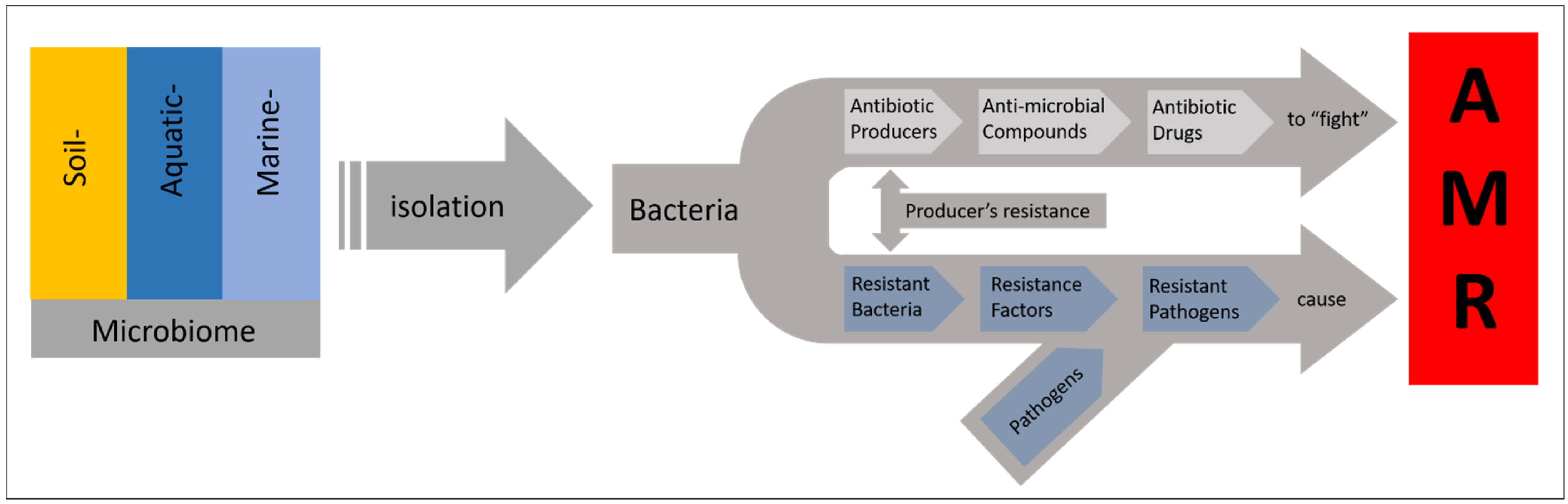
5. Microorganisms as Producer of Natural Products and Hurdles in Bioprospecting
6. Reaching out in Less Investigated Environments to Find Novel Isolates and Compounds
7. Selecting the Proven Prolific Producers of Natural Products–Characteristics and Indicators for the Biosynthetic Potential of Bacteria
8. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Coates, A.; Hu, Y.; Bax, R.; Page, C. The future challenges facing the development of new antimicrobial drugs. Nat. Rev. Drug Discov. 2002, 1, 895. [Google Scholar] [CrossRef]
- Fleming, A. Penicillin’s finder assays its future. N. Y. Times 1945, 26, 21. [Google Scholar]
- Bax, R.P.; Anderson, R.; Crew, J.; Fletcher, P.; Johnson, T.; Kaplan, E.; Knaus, B.; Kristinsson, K.; Malek, M.; Strandberg, L. Antibiotic resistance—What can we do? Nat. Med. 1998, 4, 545. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. The Evolving Threat of Antimicrobial Resistance: Options for Actions; World Health Organization: Geneva, Switzerland, 2012. [Google Scholar]
- Rice, L.B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: No ESKAPE. J. Infect. Dis. 2008, 197, 1079. [Google Scholar] [CrossRef] [PubMed]
- Chambers, H.F. The changing epidemiology of Staphylococcus aureus? Emerg. Infect. Dis. 2001, 7, 178–182. [Google Scholar] [CrossRef] [PubMed]
- Murray, B.E.; Moellering, R.C. Patterns and mechanisms of antibiotic resistance. Med. Clin. N. Am. 1978, 62, 899–923. [Google Scholar] [CrossRef]
- Tsubakishita, S.; Kuwahara-Arai, K.; Sasaki, T.; Hiramatsu, K. Origin and Molecular Evolution of the Determinant of Methicillin Resistance in Staphylococci. Antimicrob. Agents Chemother. 2010, 54, 4352. [Google Scholar] [CrossRef] [Green Version]
- Jevons, M.P.; Rolinson, G.N.; Knox, R. “Celbenin”-Resistant Staphylococci. Br. Med. J. 1961, 1, 124–126. [Google Scholar] [CrossRef]
- Leclercq, R.; Derlot, E.; Duval, J.; Courvalin, P. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N. Engl. J. Med. 1988, 319, 157. [Google Scholar] [CrossRef]
- Hiramatsu, K.; Hanaki, H.; Ino, T.; Yabuta, K.; Oguri, T.; Tenover, F.C. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J. Antimicrob. Chemother. 1997, 40, 135–136. [Google Scholar] [CrossRef]
- Sievert, D.M.; Boulton, M.L.; Stoltman, G.; Johnson, D.; Stobierski, M.G.; Downes, F.P.; Somsel, P.A.; Rudrik, J.T.; Brown, W.; Hafeez, W.; et al. Staphylococcus aureus resistant to vancomycin-United States. JAMA J. Am. Med. Assoc. 2002, 288, 824. [Google Scholar]
- Kapoor, G.; Saigal, S.; Elongavan, A. Action and resistance mechanisms of antibiotics: A guide for clinicians. J. Anaesthesiol. Clin. Pharm. 2017, 33, 300–305. [Google Scholar] [CrossRef] [PubMed]
- Aslam, B.; Wang, W.; Arshad, M.I.; Khurshid, M.; Muzammil, S.; Rasool, M.H.; Nisar, M.A.; Alvi, R.F.; Aslam, M.A.; Qamar, M.U.; et al. Antibiotic resistance: A rundown of a global crisis. Infect. Drug Resist. 2018, 11, 1645–1658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hiramatsu, K.; Cui, L.; Kuroda, M.; Ito, T. The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends Microbiol. 2001, 9, 486–493. [Google Scholar] [CrossRef]
- Gardete, S.; Tomasz, A. Mechanisms of vancomycin resistance in Staphylococcus aureus. J. Clin. Investig. 2014, 124, 2836–2840. [Google Scholar] [CrossRef] [PubMed]
- Brandi, M.; Limbago, A.J.; Kallen, W.Z.; Eggers, P.; McDougal, L.K.; Albrechta, V.S. Report of the 13th Vancomycin-Resistant Staphylococcus aureus Isolate from the United States. J. Clin. Microbiol. 2014, 52, 998. [Google Scholar] [CrossRef] [Green Version]
- Arthur, M.; Molinas, C.; Depardieu, F.; Courvalin, P. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J. Bacteriol. 1993, 175, 117. [Google Scholar] [CrossRef] [Green Version]
- Arthur, M.; Courvalin, P. Genetics and mechanisms of glycopeptide resistance in enterococci. Antimicrob. Agents Chemother. 1993, 37, 1563. [Google Scholar] [CrossRef] [Green Version]
- Olsen, J.E.; Christensen, H.; Aarestrup, F.M. Diversity and evolution of blaZ from Staphylococcus aureus and coagulase-negative staphylococci. J. Antimicrob. Chemother. 2006, 57, 450–460. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Agidi, S.; Lejeune, J.T. Diversity of staphylococcal cassette chromosome in coagulase-negative staphylococci from animal sources. J. Appl. Microbiol. 2009, 107, 1375–1383. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Wang, Y.; Walsh, T.R.; Yi, L.X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Zhao, F.; Feng, Y.; Lü, X.; McNally, A.; Zong, Z. IncP Plasmid Carrying Colistin Resistance Gene mcr-1 in Klebsiella pneumoniae from Hospital Sewage. Antimicrob. Agents Chemother. 2017, 61, e02229-16. [Google Scholar] [CrossRef] [Green Version]
- Carroll, L.M.; Gaballa, A.; Guldimann, C.; Sullivan, G.; Henderson, L.O.; Wiedmann, M. Identification of Novel Mobilized Colistin Resistance Gene mcr-9 in a Multidrug-Resistant, Colistin-Susceptible Salmonella enterica Serotype Typhimurium Isolate. mBio 2019, 10, e00853-19. [Google Scholar] [CrossRef] [Green Version]
- Kozhevin, P.; Vinogradova, K.; Bulgakova, V. The soil antibiotic resistome. Mosc. Univ. Soil Sci. Bull. 2013, 68, 53–59. [Google Scholar] [CrossRef]
- Mindlin, S.; Soina, V.; Petrova, M.; Gorlenko, Z. Isolation of antibiotic resistance bacterial strains from Eastern Siberia permafrost sediments. Russ. J. Genet. 2008, 44, 27–34. [Google Scholar] [CrossRef]
- Pawlowski, A.C.; Wang, W.; Koteva, K.; Barton, H.A.; McArthur, A.G.; Wright, G.D. A diverse intrinsic antibiotic resistome from a cave bacterium. Nat. Commun. 2016, 7, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Costa, V.M.; King, C.E.; Kalan, L.; Morar, M.; Sung, W.W.L.; Schwarz, C.; Froese, D.; Zazula, G.; Calmels, F.; Debruyne, R.; et al. Antibiotic resistance is ancient. Nature 2011, 477, 457. [Google Scholar] [CrossRef] [PubMed]
- Garau, G.; Di Guilmi, A.M.; Hall, B.G. Structure-Based Phylogeny of the Metallo-β-Lactamases. Antimicrob. Agents Chemother. 2005, 49, 2778. [Google Scholar] [CrossRef] [Green Version]
- Hall, B.G.; Barlow, M. Evolution of the serine β-lactamases: Past, present and future. Drug Resist. Updates 2004, 7, 111–123. [Google Scholar] [CrossRef]
- Aminov, R.I. The role of antibiotics and antibiotic resistance in nature. Environ. Microbiol. 2009, 11, 2970–2988. [Google Scholar] [CrossRef]
- Macia, M.D.; Blanquer, D.; Togores, B.; Sauleda, J.; Perez, J.L.; Oliver, A. Hypermutation Is a Key Factor in Development of Multiple-Antimicrobial Resistance in Pseudomonas aeruginosa Strains Causing Chronic Lung Infections. Antimicrob. Agents Chemother. 2005, 49, 3382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sköld, O. Sulfonamide resistance: Mechanisms and trends. Drug Resist. Updates 2000, 3, 155–160. [Google Scholar] [CrossRef]
- Lupo, A.; Coyne, S.; Berendonk, T. Origin and evolution of antibiotic resistance: The common mechanisms of emergence and spread in water bodies. Front. Microbiol. 2012, 3, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goossens, H.; Ferech, M.; Vander Stichele, R.; Elseviers, M. Outpatient antibiotic use in Europe and association with resistance: A cross-national database study. Lancet 2005, 365, 579–587. [Google Scholar] [CrossRef]
- Bartlett, J.G.; Gilbert, D.N.; Spellberg, B. Seven ways to preserve the miracle of antibiotics. Clin. Infect. Dis. 2013, 56, 1445. [Google Scholar] [CrossRef]
- Knapp, C.W.; Dolfing, J.; Ehlert, P.A.I.; Graham, D.W. Evidence of Increasing Antibiotic Resistance Gene Abundances in Archived Soils since 1940. Environ. Sci. Technol. 2010, 44, 580–587. [Google Scholar] [CrossRef]
- Newman, D.; Cragg, G. Natural Products as Sources of New Drugs over the Last 25 Years. J. Nat. Prod. 2007, 70, 461–477. [Google Scholar] [CrossRef] [Green Version]
- Hatosy, S.M.; Martiny, A.C. The ocean as a global reservoir of antibiotic resistance genes. Appl. Environ. Microbiol. 2015, 81, 7593–7599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Neill, J. The Review on Antimicrobial Resistance. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations; H.M. Government & Wellcome Trust: London, UK, 2016. Available online: https://wellcomecollection.org/works/thvwsuba (accessed on 31 May 2016).
- Kasanah, N.; Hamann, M.T. Development of antibiotics and the future of marine microorganisms to stem the tide of antibiotic resistance. Curr. Opin. Investig. Drugs 2004, 5, 827–837. [Google Scholar] [PubMed]
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. Pharm. Ther. 2015, 40, 277. [Google Scholar]
- Holmes, A.H.; Moore, L.S.P.; Sundsfjord, A.; Steinbakk, M.; Regmi, S.; Karkey, A.; Guerin, P.J.; Piddock, L.J.V. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 2016, 387, 176–187. [Google Scholar] [CrossRef]
- Waglechner, N.; Wright, G.D. Antibiotic resistance: It’s bad, but why isn’t it worse? BMC Biol. 2017, 15, 84. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef]
- Newman, D.; Cragg, G. Natural Products as Sources of New Drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef] [Green Version]
- Henkel, T.; Brunne, R.M.; Müller, H.; Reichel, F. Statistical Investigation into the Structural Complementarity of Natural Products and Synthetic Compounds. Angew. Chem. Int. Ed. 1999, 38, 643–647. [Google Scholar] [CrossRef]
- Feher, M.; Schmidt, J.M. Property distributions: Differences between drugs, natural products, and molecules from combinatorial chemistry. J. Chem. Inf. Comput. Sci. 2003, 43, 218–227. [Google Scholar] [CrossRef]
- Evans, B.E.; Rittle, K.E.; Bock, M.G.; Dipardo, R.M.; Freidinger, R.M.; Whitter, W.L.; Lundell, G.F.; Veber, D.F.; Anderson, P.S.; Chang, R.S. Methods for drug discovery: Development of potent, selective, orally effective cholecystokinin antagonists. J. Med. Chem. 1988, 31, 2235. [Google Scholar] [CrossRef] [PubMed]
- Welsch, M.E.; Snyder, S.A.; Stockwell, B.R. Privileged scaffolds for library design and drug discovery. Curr. Opin. Chem. Biol. 2010, 14, 347–361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breinbauer, R.; Vetter, I.R.; Waldmann, H. From protein domains to drug candidates—Natural products as guiding principles in the design and synthesis of compound libraries. Angew. Chem. Int. Ed. 2002, 41, 2878–2890. [Google Scholar] [CrossRef]
- Baltz, R.H. Renaissance in antibacterial discovery from actinomycetes. Curr. Opin. Pharmacol. 2008, 8, 557–563. [Google Scholar] [CrossRef] [PubMed]
- Ganesan, A. The impact of natural products upon modern drug discovery. Curr. Opin. Chem. Biol. 2008, 12, 306–317. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Koehn, F.; Carter, G. The evolving role of natural products in drug discovery. Nat. Rev. Drug Discov. 2005, 4, 206–220. [Google Scholar] [CrossRef] [PubMed]
- Baker, D.D.; Chu, M.; Oza, U.; Rajgarhia, V. The value of natural products to future pharmaceutical discovery. Nat. Prod. Rep. 2007, 24, 1225–1244. [Google Scholar] [CrossRef]
- János, B. Bioactive Microbial Metabolites. J. Antibiot. 2005, 58, 1. [Google Scholar] [CrossRef] [Green Version]
- Payne, D.J.; Gwynn, M.N.; Holmes, D.J.; Pompliano, D.L. Drugs for bad bugs: Confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 2006, 6, 29. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M.; Battershill, C.N. Therapeutic agents from the sea: Biodiversity, chemo-evolutionary insight and advances to the end of Darwin’s 200th year. Diving Hyperb. Med. 2009, 39, 216–225. [Google Scholar] [PubMed]
- Schinke, C.; Martins, T.; Queiroz, S.C.N.; Melo, I.S.; Reyes, F.G.R. Antibacterial Compounds from Marine Bacteria, 2010–2015. J. Nat. Prod. 2017, 80, 1215–1228. [Google Scholar] [CrossRef] [PubMed]
- Kong, D.X.; Jiang, Y.Y.; Zhang, H.Y. Marine natural products as sources of novel scaffolds: Achievement and concern. Drug Discov. Today 2010, 15, 884–886. [Google Scholar] [CrossRef] [PubMed]
- Jose, P.A.; Jha, B. New Dimensions of Research on Actinomycetes: Quest for Next Generation Antibiotics. Front. Microbiol. 2016, 7, 1295. [Google Scholar] [CrossRef]
- Zhang, G.; Li, J.; Zhu, T.; Gu, Q.; Li, D. Advanced tools in marine natural drug discovery. Curr. Opin. Biotechnol. 2016, 42, 13–23. [Google Scholar] [CrossRef]
- Covington, B.C.; Xu, F.; Seyedsayamdost, M.R. A Natural Product Chemist’s Guide to Unlocking Silent Biosynthetic Gene Clusters. Annu. Rev. Biochem. 2021, 90, 32. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Cobb, R.E.; Zhao, H. Recent advances in natural product discovery. Curr. Opin. Biotechnol. 2014, 30, 230–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewis, W.H.; Tahon, G.; Geesink, P.; Sousa, D.Z.; Ettema, T.J.G. Innovations to culturing the uncultured microbial majority. Nat. Rev. Microbiol. 2021, 19, 225–240. [Google Scholar] [CrossRef]
- Zhang, L.; An, R.; Wang, J.; Sun, N.; Zhang, S.; Hu, J.; Kuai, J. Exploring novel bioactive compounds from marine microbes. Curr. Opin. Microbiol. 2005, 8, 276–281. [Google Scholar] [CrossRef]
- Dührkop, K.; Fleischauer, M.; Ludwig, M.; Aksenov, A.A.; Melnik, A.V.; Meusel, M.; Dorrestein, P.C.; Rousu, J.; Böcker, S. SIRIUS 4: A rapid tool for turning tandem mass spectra into metabolite structure information. Nat. Methods 2019, 16, 299–302. [Google Scholar] [CrossRef] [Green Version]
- Guijas, C.; Montenegro-Burke, J.R.; Domingo-Almenara, X.; Palermo, A.; Warth, B.; Hermann, G.; Koellensperger, G.; Huan, T.; Uritboonthai, W.; Aisporna, A.E.; et al. METLIN: A Technology Platform for Identifying Knowns and Unknowns. Anal. Chem. 2018, 90, 3156–3164. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medema, M.H.; Blin, K.; Cimermancic, P.; de Jager, V.; Zakrzewski, P.; Fischbach, M.A.; Weber, T.; Takano, E.; Breitling, R. antiSMASH: Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011, 39, 339–346. [Google Scholar] [CrossRef]
- Ling, L.L.; Schneider, T.; Peoples, A.J.; Spoering, A.L.; Engels, I.; Conlon, B.P.; Mueller, A.; Schäberle, T.F.; Hughes, D.E.; Epstein, S.; et al. A new antibiotic kills pathogens without detectable resistance. Nature 2015, 517, 455. [Google Scholar] [CrossRef]
- Wright, G. Antibiotics: An irresistible newcomer. Nature 2015, 517, 442–444. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, M.W. Teixobactin: A novel anti-infective agent. Expert Rev. Anti-Infect. Ther. 2019, 17, 1–3. [Google Scholar] [CrossRef]
- Lebar, M.D.; Heimbegner, J.L.; Baker, B.J. Cold-water marine natural products. Nat. Prod. Rep. 2007, 24, 774–797. [Google Scholar] [CrossRef] [PubMed]
- Skropeta, D. Deep-sea natural products. Nat. Prod. Rep. 2008, 25, 1131–1166. [Google Scholar] [CrossRef] [Green Version]
- Challinor, V.L.; Bode, H.B. Bioactive natural products from novel microbial sources. Ann. N. Y. Acad. Sci. 2015, 13541, 82–97. [Google Scholar] [CrossRef]
- Barka, E.A.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Klenk, H.P.; Clement, C.; Ouhdouch, Y.; Wezel, G.P.V. Taxonomy, Physiology, and Natural Products of Actinobacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 1–43. [Google Scholar] [CrossRef] [Green Version]
- Watve, M.; Tickoo, R.; Jog, M.; Bhole, B. How many antibiotics are produced by the genus Streptomyces? Arch. Microbiol. 2001, 176, 386–390. [Google Scholar] [CrossRef] [PubMed]
- Nett, M.; Ikeda, H.; Moore, B.S. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat. Prod. Rep. 2009, 26, 1362–1384. [Google Scholar] [CrossRef]
- William, F.; Paul, R.J. Developing a new resource for drug discovery: Marine actinomycete bacteria. Nat. Chem. Biol. 2006, 2, 666. [Google Scholar] [CrossRef]
- Gaspari, F.; Paitan, Y.; Mainini, M.; Losi, D.; Ron, E.Z.; Marinelli, F. Myxobacteria isolated in Israel as potential source of new anti-infectives. J. Appl. Microbiol. 2005, 98, 429–439. [Google Scholar] [CrossRef] [PubMed]
- Davila-Cespedes, A.; Hufendiek, P.; Crusemann, M.; Schaberle, T.; Konig, G. Marine-derived myxobacteria of the suborder Nannocystineae: An underexplored source of structurally intriguing and biologically active metabolites. Beilstein J. Org. Chem. 2016, 12, 969–984. [Google Scholar] [CrossRef]
- Gemperlein, K.; Zaburannyi, N.; Garcia, R.; La Clair, J.; Müller, R. Metabolic and Biosynthetic Diversity in Marine Myxobacteria. Mar. Drugs 2018, 16, 314. [Google Scholar] [CrossRef] [Green Version]
- Schäberle, T.F.; Goralski, E.; Neu, E.; Erol, O.; Hölzl, G.; Dörmann, P.; Bierbaum, G.; König, G.M. Marine myxobacteria as a source of antibiotics--comparison of physiology, polyketide-type genes and antibiotic production of three new isolates of Enhygromyxa salina. Mar. Drugs 2010, 8, 2466. [Google Scholar] [CrossRef] [Green Version]
- Schäberle, T.F.; Lohr, F.; Schmitz, A.; König, G.M. Antibiotics from myxobacteria. Nat. Prod. Rep. 2014, 31, 953. [Google Scholar] [CrossRef]
- Nunnery, J.K.; Mevers, E.; Gerwick, W.H. Biologically active secondary metabolites from marine cyanobacteria. Curr. Opin. Biotechnol. 2010, 21, 787–793. [Google Scholar] [CrossRef] [Green Version]
- Pham, J.V.; Yilma, M.A.; Feliz, A.; Majid, M.T.; Maffetone, N.; Walker, J.R.; Kim, E.; Cho, H.J.; Reynolds, J.M.; Song, M.C.; et al. A Review of the Microbial Production of Bioactive Natural Products and Biologics. Front. Microbiol. 2019, 10, 1404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Y.; Chen, J.; Hu, G.; Yu, J.; Zhu, X.; Lin, Y.; Chen, S.; Yuan, J.; Taglialatela-Scafati, O. Statistical Research on the Bioactivity of New Marine Natural Products Discovered during the 28 Years from 1985 to 2012. Mar. Drugs 2015, 13, 202. [Google Scholar] [CrossRef] [PubMed]
- Donadio, S.; Monciardini, P.; Sosio, M. Polyketide synthases and nonribosomal peptide synthetases: The emerging view from bacterial genomics. Nat. Prod. Rep. 2007, 24, 1073–1109. [Google Scholar] [CrossRef] [PubMed]
- Belknap, K.C.; Park, C.J.; Barth, B.M.; Andam, C.P. Genome mining of biosynthetic and chemotherapeutic gene clusters in Streptomyces bacteria. Sci. Rep. 2020, 10, 2003. [Google Scholar] [CrossRef] [PubMed]
- Rath, C.M.; Janto, B.; Earl, J.; Ahmed, A.; Hu, F.Z.; Hiller, L.; Dahlgren, M.; Kreft, R.; Yu, F.; Wolff, J.J.; et al. Meta-omic characterization of the marine invertebrate microbial consortium that produces the chemotherapeutic natural product ET-743. ACS Chem. Biol. 2011, 6, 1244. [Google Scholar] [CrossRef] [Green Version]
- Lago, J.; Rodríguez, L.P.; Blanco, L.; Vieites, J.M.; Cabado, A.G. Tetrodotoxin, an Extremely Potent Marine Neurotoxin: Distribution, Toxicity, Origin and Therapeutical Uses. Mar. Drugs 2015, 13, 6384. [Google Scholar] [CrossRef] [PubMed]
- Kristoffersen, V.; Rämä, T.; Isaksson, J.; Andersen, J.H.; Gerwick, W.H.; Hansen, E. Characterization of Rhamnolipids Produced by an Arctic Marine Bacterium from the Pseudomonas fluorescence Group. Mar. Drugs 2018, 16, 163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schneider, Y.K. Bacterial Natural Product Drug Discovery for New Antibiotics: Strategies for Tackling the Problem of Antibiotic Resistance by Efficient Bioprospecting. Antibiotics 2021, 10, 842. https://doi.org/10.3390/antibiotics10070842
Schneider YK. Bacterial Natural Product Drug Discovery for New Antibiotics: Strategies for Tackling the Problem of Antibiotic Resistance by Efficient Bioprospecting. Antibiotics. 2021; 10(7):842. https://doi.org/10.3390/antibiotics10070842
Chicago/Turabian StyleSchneider, Yannik K. 2021. "Bacterial Natural Product Drug Discovery for New Antibiotics: Strategies for Tackling the Problem of Antibiotic Resistance by Efficient Bioprospecting" Antibiotics 10, no. 7: 842. https://doi.org/10.3390/antibiotics10070842
APA StyleSchneider, Y. K. (2021). Bacterial Natural Product Drug Discovery for New Antibiotics: Strategies for Tackling the Problem of Antibiotic Resistance by Efficient Bioprospecting. Antibiotics, 10(7), 842. https://doi.org/10.3390/antibiotics10070842





