Electrochemical Detection of Cancer Biomarkers: From Molecular Sensing to Clinical Translation
Abstract
1. Introduction
Scope and Methodology
2. Principles of Electrochemical Biosensors for Cancer Detection
2.1. Electrochemical Transduction Methods
2.2. Biorecognition Elements
2.3. Signal Amplification Strategies
2.4. Comparison with Conventional Diagnostic Methods
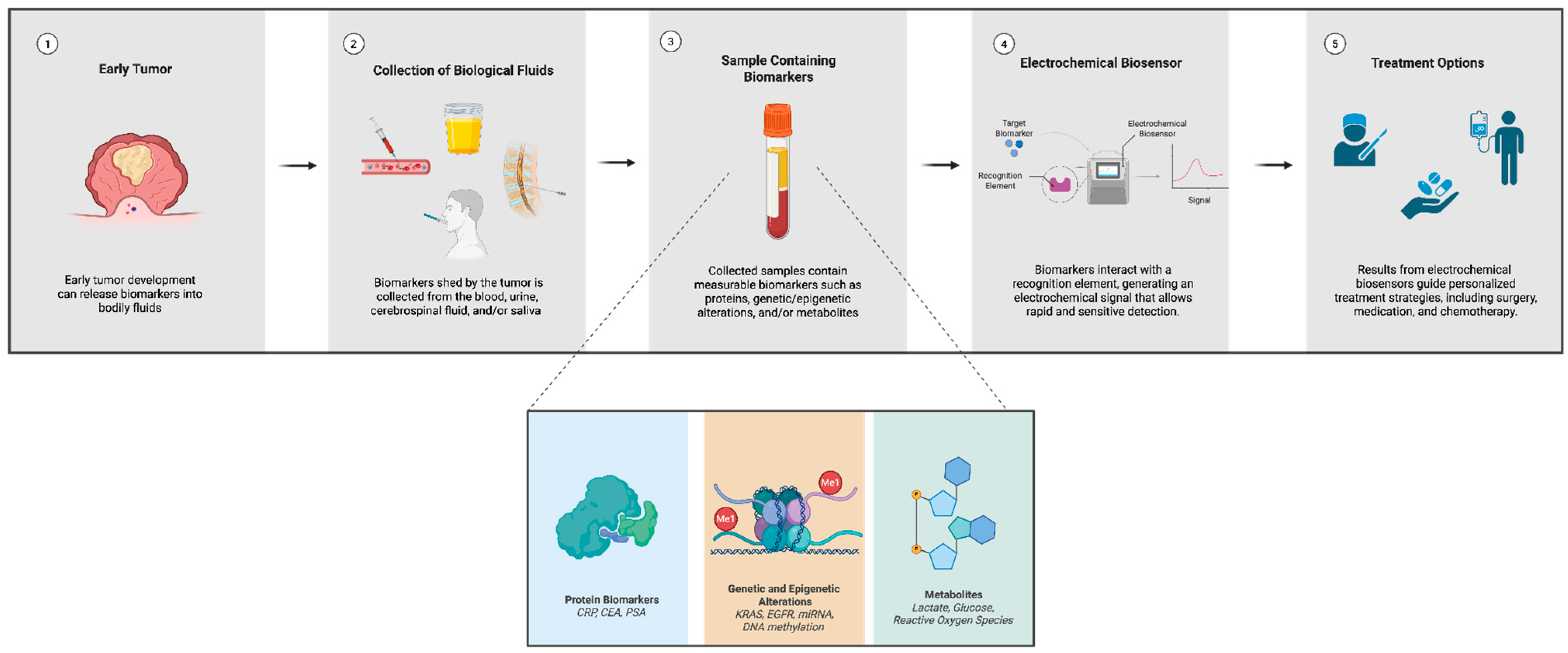
3. Cancer Biomarkers and Their Electrochemical Detection
3.1. Protein Biomarkers
3.1.1. CRP
3.1.2. CEA
3.1.3. PSA
3.2. Genetic and Epigenetic Biomarkers
3.3. Metabolite-Based Detection
4. Nanomaterials in Electrochemical Cancer Biosensors
4.1. Carbon-Based Nanomaterials
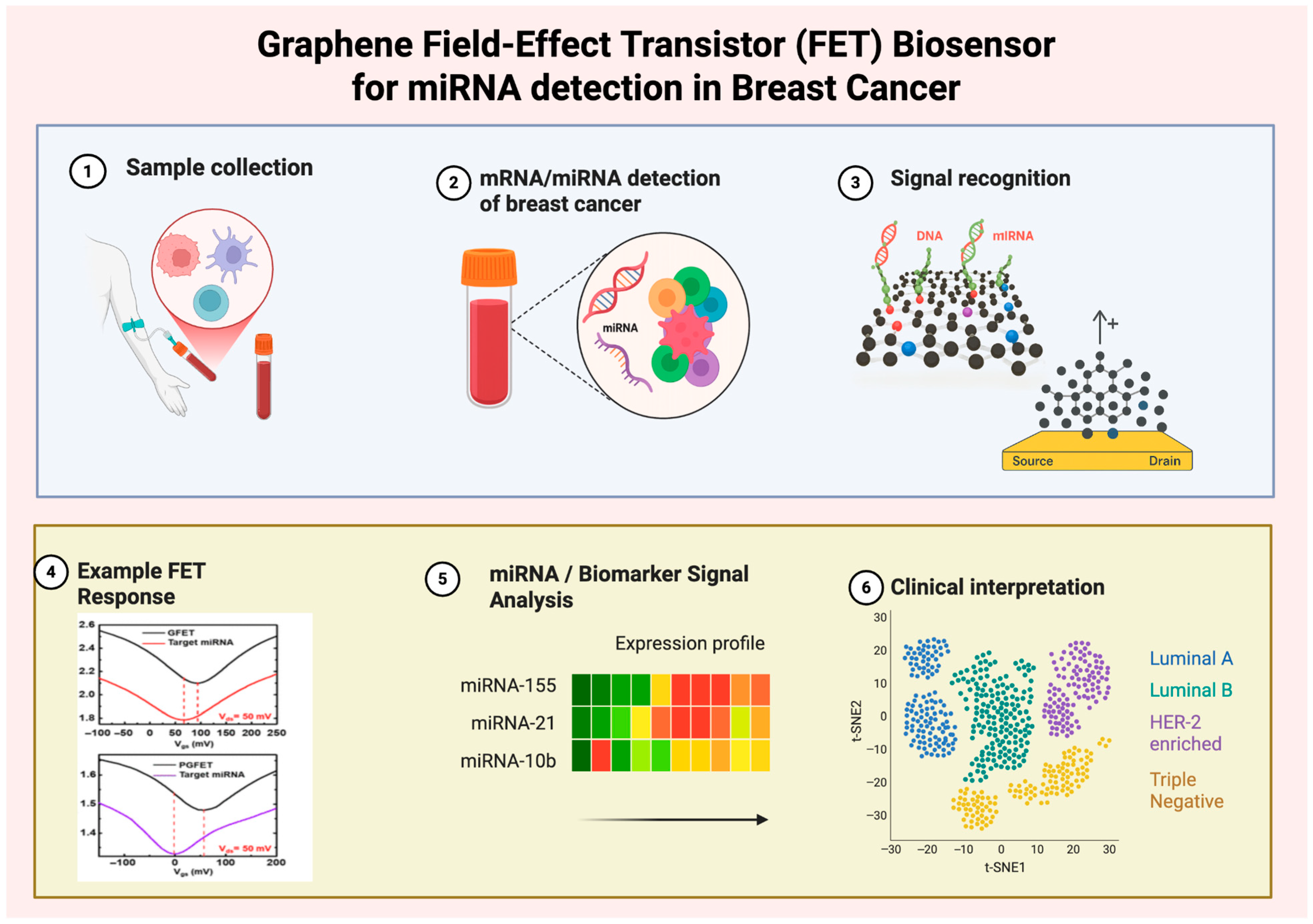
Graphene
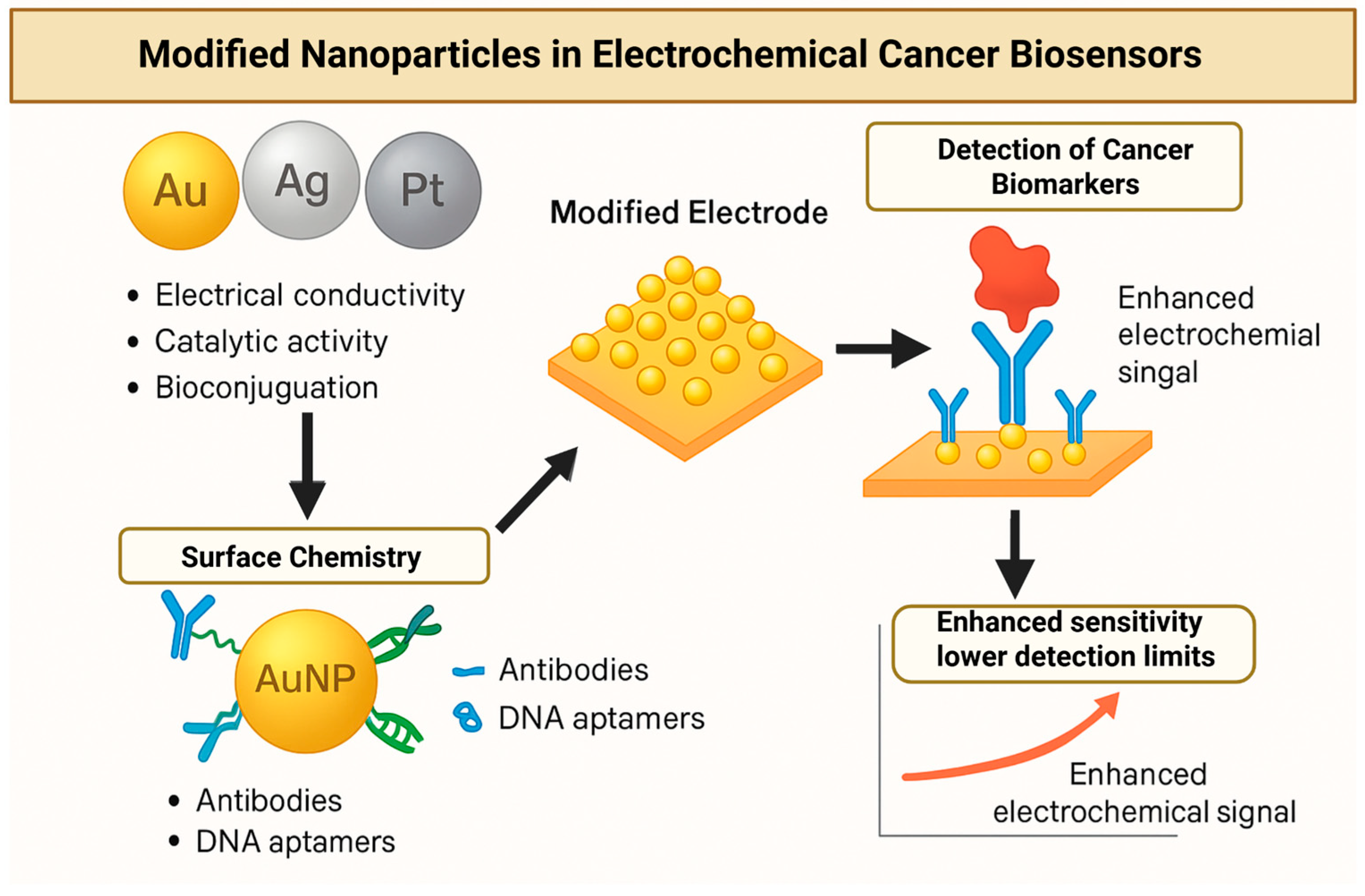
4.2. Metal and Metal Oxide Nanoparticles
4.2.1. Gold Nanoparticles (AuNPs)
4.2.2. Silver and Silver Oxide Nanoparticles
| References | Biomarker/Target | Sensor Type/ Modification | Detection Limit (LOD) | Linear Range | Testing Medium | Interference Study | Real-Sample Validation |
|---|---|---|---|---|---|---|---|
| Huang et al. [101] | Carcinoembryonic antigen (CEA) | Sandwich immunosensor with Ag-Au NPs on graphene | 8 pg/mL | 10–1.2 × 105 pg/mL | Human serum (spiked) | Not reported | Yes; spiked serum (92–105% recovery) |
| Ortega et al. [100] | EpCAM (circulating tumor cells) | Microfluidic immunosensor; AgNP chitosan film with HRP tag | 2.7 pg/mL | 2.7–2000 pg/mL | Spiked human serum/blood | Not reported | Yes; blood serum |
| Lee et al. [97] | Glucose | GOx/AuNP (AuCL4)/polypyrrole/graphite | Not reported | 0.1 pg/mL–0.7 mM | PBS, Human serum | Not reported | PBS, Human serum |
4.3. Conducting Polymers and Hybrid Nanocomposites
4.4. Nanomaterial-Based Electrochemical Cancer Biosensors for Point-of-Care in Low-Resource Settings
5. Advanced Strategies for Electrochemical Cancer Biosensors
5.1. CRISPR-Based Electrochemical Biosensors
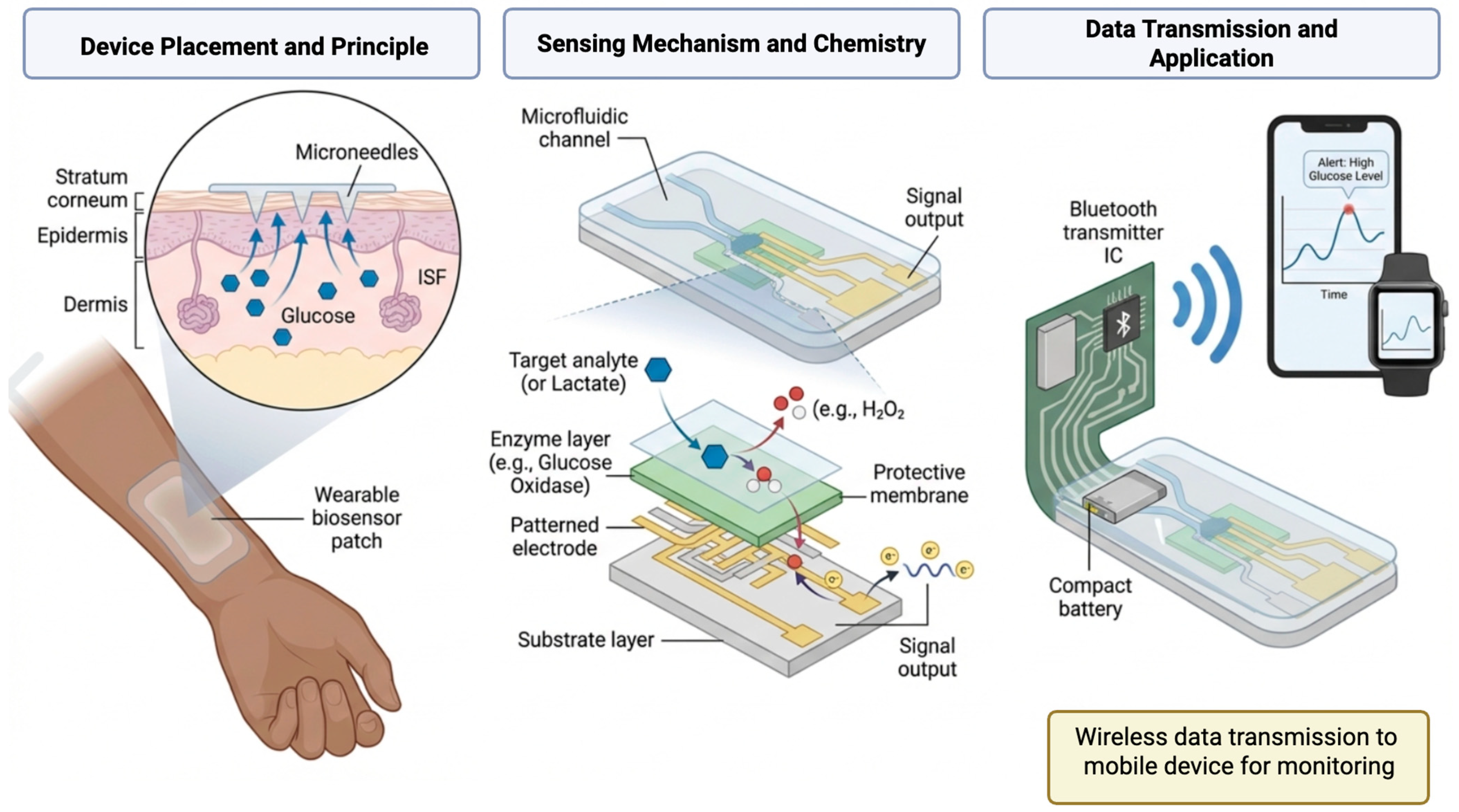
5.2. Wearable and Implantable Electrochemical Sensors
5.3. AI-Integrated Electrochemical Biosensors
5.4. Microfluidics and Lab-on-a-Chip Platforms
6. Challenges and Future Perspectives
7. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, X.; Gole, J.; Gore, A.; He, Q.; Lu, M.; Min, J.; Yuan, Z.; Yang, X.; Jiang, Y.; Zhang, T.; et al. Non-invasive early detection of cancer four years before conventional diagnosis using a blood test. Nat. Commun. 2020, 11, 3475. [Google Scholar] [CrossRef] [PubMed]
- Goddard, K.A.B.; Feuer, E.J.; Mandelblatt, J.S.; Meza, R.; Holford, T.R.; Jeon, J.; Lansdorp-Vogelaar, I.; Gulati, R.; Stout, N.K.; Howlader, N.; et al. Estimation of Cancer Deaths Averted From Prevention, Screening, and Treatment Efforts, 1975–2020. JAMA Oncol. 2025, 11, 162–167. [Google Scholar] [CrossRef] [PubMed]
- Sanko, V.; Kuralay, F. Label-Free Electrochemical Biosensor Platforms for Cancer Diagnosis: Recent Achievements and Challenges. Biosensors 2023, 13, 333. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Xia, J.; Li, L.; Zeng, Y.; Zhao, J.; Li, G. Electrochemical Biosensors for Cancer Diagnosis: Multitarget Analysis to Present Molecular Characteristics of Tumor Heterogeneity. JACS Au 2024, 4, 4655–4672. [Google Scholar] [CrossRef]
- Fu, L.; Karimi-Maleh, H. Leveraging electrochemical sensors to improve efficiency of cancer detection. World J. Clin. Oncol. 2024, 15, 360–366. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Y.; Bo, L.; Zhou, E.; Chen, Y.; Naranmandakh, S.; Xie, W.; Ru, Q.; Chen, L.; Zhu, Z.; et al. Progress of linking gut microbiota and musculoskeletal health: Casualty, mechanisms, and translational values. Gut Microbes 2023, 15, 2263207. [Google Scholar] [CrossRef]
- Topkaya, S.N.; Azimzadeh, M.; Ozsoz, M. Electrochemical Biosensors for Cancer Biomarkers Detection: Recent Advances and Challenges. Electroanalysis 2016, 28, 1402–1419. [Google Scholar] [CrossRef]
- Ben Moussa, F.; Kutner, W.; Beduk, T.; Sena-Torralba, A.; Mostafavi, E. Electrochemical bio- and chemosensors for cancer biomarkers: Natural (with antibodies) versus biomimicking artificial (with aptamers and molecularly imprinted polymers) recognition. Talanta 2024, 267, 125259. [Google Scholar] [CrossRef]
- Pulumati, A.; Pulumati, A.; Dwarakanath, B.S.; Verma, A.; Papineni, R.V.L. Technological advancements in cancer diagnostics: Improvements and limitations. Cancer Rep. 2023, 6, e1764. [Google Scholar] [CrossRef]
- Tseng, L.-J.; Matsuyama, A.; MacDonald-Dickinson, V. Histology: The gold standard for diagnosis? Can. Vet. J. 2023, 64, 389–391. [Google Scholar]
- Sohrabi, N.; Valizadeh, A.; Farkhani, S.M.; Akbarzadeh, A. Basics of DNA biosensors and cancer diagnosis. Artif. Cells Nanomed. Biotechnol. 2016, 44, 654–663. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.; Ahommed, M.; Daizy, M.; Bacchu, M.; Ali, M.; Al-Mamun, M.; Aly, M.S.; Khan, M.; Hossain, S. Recent development in electrochemical biosensors for cancer biomarkers detection. Biosens. Bioelectron. X 2021, 8, 100075. [Google Scholar] [CrossRef]
- Ahmed, M.U.; Saaem, I.; Wu, P.C.; Brown, A.S. Personalized diagnostics and biosensors: A review of the biology and technology needed for personalized medicine. Crit. Rev. Biotechnol. 2014, 34, 180–196. [Google Scholar] [CrossRef] [PubMed]
- Ahirwar, R. Recent advances in nanomaterials-based electrochemical immunosensors and aptasensors for HER2 assessment in breast cancer. Microchim. Acta 2021, 188, 317. [Google Scholar] [CrossRef]
- Diaconu, I.; Cristea, C.; Hârceagă, V.; Marrazza, G.; Berindan-Neagoe, I.; Săndulescu, R. Electrochemical immunosensors in breast and ovarian cancer. Clin. Chim. Acta 2013, 425, 128–138. [Google Scholar] [CrossRef]
- Rajarathinam, T.; Jayaraman, S.; Kim, C.-S.; Lee, J.; Chang, S.-C. Portable Amperometric Biosensor Enhanced with Enzyme-Ternary Nanocomposites for Prostate Cancer Biomarker Detection. Biosensors 2024, 14, 623. [Google Scholar] [CrossRef]
- Sadighbayan, D.; Hasanzadeh, M.; Ghafar-Zadeh, E. Biosensing based on field-effect transistors (FET): Recent progress and challenges. TrAC Trends Anal. Chem. 2020, 133, 116067. [Google Scholar] [CrossRef]
- Seo, G.; Lee, G.; Kim, M.J.; Baek, S.-H.; Choi, M.; Ku, K.B.; Lee, C.-S.; Jun, S.; Park, D.; Kim, H.G.; et al. Rapid Detection of COVID-19 Causative Virus (SARS-CoV-2) in Human Nasopharyngeal Swab Specimens Using Field-Effect Transistor-Based Biosensor. ACS Nano 2020, 14, 5135–5142. [Google Scholar] [CrossRef]
- Chao, L.; Liang, Y.; Hu, X.; Shi, H.; Xia, T.; Zhang, H.; Xia, H. Recent advances in field effect transistor biosensor technology for cancer detection: A mini review. J. Phys. D Appl. Phys. 2021, 55, 153001. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, L.; Xuan, T.; Wang, J.; Wang, X. Label-free Electrochemical Impedance Spectroscopy Aptasensor for Ultrasensitive Detection of Lung Cancer Biomarker Carcinoembryonic Antigen. Front. Chem. 2021, 9, 721008. [Google Scholar] [CrossRef]
- Grabowska, I.; Hepel, M.; Kurzątkowska-Adaszyńska, K. Advances in Design Strategies of Multiplex Electrochemical Aptasensors. Sensors 2022, 22, 161. [Google Scholar] [CrossRef] [PubMed]
- Nasrollahpour, H.; Khalilzadeh, B.; Naseri, A.; Sillanpää, M.; Chia, C.H. Homogeneous Electrochemiluminescence in the Sensors Game: What Have We Learned from Past Experiments? Anal. Chem. 2022, 94, 349–365. [Google Scholar] [CrossRef] [PubMed]
- Che, Z.-Y.; Wang, X.-Y.; Ma, X.; Ding, S.-N. Bipolar electrochemiluminescence sensors: From signal amplification strategies to sensing formats. Coord. Chem. Rev. 2021, 446, 214116. [Google Scholar] [CrossRef]
- Chikkaveeraiah, B.V.; Bhirde, A.A.; Morgan, N.Y.; Eden, H.S.; Chen, X. Electrochemical Immunosensors for Detection of Cancer Protein Biomarkers. ACS Nano 2012, 6, 6546–6561. [Google Scholar] [CrossRef]
- Wang, Y.; Yao, W.; Sun, W.; Fan, Y.; Xiao, J.; Wang, T.; Huang, K.; Liu, L.; Wang, X.; Jiang, H. Aptamer-based electrochemical analysis platform for tumor cells and biomarkers detection. J. Electroanal. Chem. 2024, 960, 118194. [Google Scholar] [CrossRef]
- Liang, T.; Qin, X.; Zhang, Y.; Yang, Y.; Chen, Y.; Yuan, L.; Liu, F.; Chen, Z.; Li, X.; Yang, F. CRISPR/dCas9-Mediated Specific Molecular Assembly Facilitates Genotyping of Mutant Circulating Tumor DNA. Anal. Chem. 2023, 95, 16305–16314. [Google Scholar] [CrossRef]
- Bruch, R.; Baaske, J.; Chatelle, C.; Meirich, M.; Madlener, S.; Weber, W.; Dincer, C.; Urban, G.A. CRISPR/Cas13a-Powered Electrochemical Microfluidic Biosensor for Nucleic Acid Amplification-Free miRNA Diagnostics. Adv. Mater. 2019, 31, e1905311. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, X.; Pang, L.; Geng, P.; Mi, F.; Hu, C.; Peng, F.; Guan, M. Application progress of magnetic molecularly imprinted polymers chemical sensors in the detection of biomarkers. Analyst 2022, 147, 571–586. [Google Scholar] [CrossRef]
- Liu, R.; Zhao, M.; Zhang, X.; Zhang, C.; Ren, B.; Ma, J. Advances and Challenges in Molecularly Imprinted Electrochemical Sensors for Application in Environmental, Biomedicine, and Food Safety. Crit. Rev. Anal. Chem. 2025, 1–19. [Google Scholar] [CrossRef]
- Xu, L.; Wen, Y.; Pandit, S.; Mokkapati, V.R.S.S.; Mijakovic, I.; Li, Y.; Ding, M.; Ren, S.; Li, W.; Liu, G. Graphene-based biosensors for the detection of prostate cancer protein biomarkers: A review. BMC Chem. 2019, 13, 112. [Google Scholar] [CrossRef]
- Wang, B.; Akiba, U.; Anzai, J.-I. Recent Progress in Nanomaterial-Based Electrochemical Biosensors for Cancer Biomarkers: A Review. Molecules 2017, 22, 1048. [Google Scholar] [CrossRef] [PubMed]
- Rostamabadi, P.F.; Heydari-Bafrooei, E. Impedimetric aptasensing of the breast cancer biomarker HER2 using a glassy carbon electrode modified with gold nanoparticles in a composite consisting of electrochemically reduced graphene oxide and single-walled carbon nanotubes. Microchim. Acta 2019, 186, 495. [Google Scholar] [CrossRef] [PubMed]
- Qian, J.; Zhao, L.; Huang, Y.; Zhao, C.; Liu, H.; Liu, X.; Cheng, Z.; Yu, F. A microdroplet SERS-RCA biosensor with enhanced specificity and reproducibility for profiling dual miRNAs in idiopathic pulmonary fibrosis diagnosis and monitoring. Chem. Eng. J. 2024, 482, 149012. [Google Scholar] [CrossRef]
- Xue, X.; Liu, Y.; Wang, Y.; Meng, M.; Wang, K.; Zang, X.; Zhao, S.; Sun, X.; Cui, L.; Pan, L.; et al. MiR-21 and MiR-155 promote non-small cell lung cancer progression by downregulating SOCS1, SOCS6, and PTEN. Oncotarget 2016, 7, 84508–84519. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.A.; Abdulrahman, Z.F.A.; Faraidun, H.N. Circulatory miRNA-155, miRNA-21 target PTEN expression and activity as a factor in breast cancer development. Cell. Mol. Biol. 2020, 66, 44–50. [Google Scholar] [CrossRef]
- Zhang, X.; Xu, Y.; Zhang, X.; Wu, H.; Shen, J.; Chen, R.; Xiong, Y.; Li, J.; Guo, S. Progress on the layer-by-layer assembly of multilayered polymer composites: Strategy, structural control and applications. Prog. Polym. Sci. 2019, 89, 76–107. [Google Scholar] [CrossRef]
- Karunanidhi, M.; Kanniyappan, H.; Zhan, E.; Mathew, R.; Sun, Y.; Wu, J.; Yan, Y.; Munirathinum, G.; Mathew, M.T. PancreaAlert: Intelligent Nanoengineered Biosensor for Pancreatic Cancer. Cancer Res. 2024, 84, 7289. [Google Scholar] [CrossRef]
- Aronson, J.K.; Ferner, R.E. Biomarkers—A General Review. Curr. Protoc. Pharmacol. 2017, 76, 9.23.1–9.23.17. [Google Scholar] [CrossRef]
- Sarhadi, V.K.; Armengol, G. Molecular Biomarkers in Cancer. Biomolecules 2022, 12, 1021. [Google Scholar] [CrossRef]
- Zhu, M.; Ma, Z.; Zhang, X.; Hang, D.; Yin, R.; Feng, J.; Xu, L.; Shen, H. C-reactive protein and cancer risk: A pan-cancer study of prospective cohort and Mendelian randomization analysis. BMC Med. 2022, 20, 301. [Google Scholar] [CrossRef]
- Yang, H.J.; Kim, M.W.; Raju, C.V.; Cho, C.H.; Park, T.J.; Park, J.P. Highly sensitive and label-free electrochemical detection of C-reactive protein on a peptide receptor−gold nanoparticle−black phosphorous nanocomposite modified electrode. Biosens. Bioelectron. 2023, 234, 115382. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Liu, Z.; Yan, Y.; Chen, J.; Yang, R.; Huang, Q.; Jin, M.; Shui, L. Triple signal-enhancing electrochemical aptasensor based on rhomboid dodecahedra carbonized-ZIF67 for ultrasensitive CRP detection. Biosens. Bioelectron. 2022, 207, 114129. [Google Scholar] [CrossRef] [PubMed]
- Ansar, W.; Ghosh, S. C-reactive protein and the biology of disease. Immunol. Res. 2013, 56, 131–142. [Google Scholar] [CrossRef] [PubMed]
- Hall, C.; Clarke, L.; Pal, A.; Buchwald, P.; Eglinton, T.; Wakeman, C.; Frizelle, F. A Review of the Role of Carcinoembryonic Antigen in Clinical Practice. Ann. Coloproctol. 2019, 35, 294–305. [Google Scholar] [CrossRef]
- Niedzielska, J.; Jastrzębski, T. Carcinoembryonic Antigen (CEA): Origin, Role in Oncology, and Concentrations in Serum and Peritoneal Fluid. J. Clin. Med. 2025, 14, 3189. [Google Scholar] [CrossRef]
- Bruserud, Ø.; Aarstad, H.H.; Tvedt, T.H.A. Combined C-Reactive Protein and Novel Inflammatory Parameters as a Predictor in Cancer—What Can We Learn from the Hematological Experience? Cancers 2020, 12, 1966. [Google Scholar] [CrossRef]
- Rawla, P. Epidemiology of Prostate Cancer. World J. Oncol. 2019, 10, 63–89. [Google Scholar] [CrossRef]
- Özyurt, C.; Uludağ, İ.; Sezgintürk, M.K. An ultrasensitive and disposable electrochemical aptasensor for prostate-specific antigen (PSA) detection in real serum samples. Anal. Bioanal. Chem. 2023, 415, 1123–1136. [Google Scholar] [CrossRef]
- Pan, L.-H.; Kuo, S.-H.; Lin, T.-Y.; Lin, C.-W.; Fang, P.-Y.; Yang, H.-W. An electrochemical biosensor to simultaneously detect VEGF and PSA for early prostate cancer diagnosis based on graphene oxide/ssDNA/PLLA nanoparticles. Biosens. Bioelectron. 2017, 89, 598–605. [Google Scholar] [CrossRef]
- Ilango, S.; Paital, B.; Jayachandran, P.; Padma, P.R.; Nirmaladevi, R. Epigenetic alterations in cancer. Front. Biosci.-Landmark 2020, 25, 1058–1109. [Google Scholar] [CrossRef]
- Sun, Z.; Wang, L.; Wu, S.; Pan, Y.; Dong, Y.; Zhu, S.; Yang, J.; Yin, Y.; Li, G. An Electrochemical Biosensor Designed by Using Zr-Based Metal–Organic Frameworks for the Detection of Glioblastoma-Derived Exosomes with Practical Application. Anal. Chem. 2020, 92, 3819–3826. [Google Scholar] [CrossRef]
- Rafanan, J.; Ghani, N.; Kazemeini, S.; Nadeem-Tariq, A.; Shih, R.; Vida, T.A. Modernizing Neuro-Oncology: The Impact of Imaging, Liquid Biopsies, and AI on Diagnosis and Treatment. Int. J. Mol. Sci. 2025, 26, 917. [Google Scholar] [CrossRef] [PubMed]
- Attoye, B.; Pou, C.; Blair, E.; Rinaldi, C.; Thomson, F.; Baker, M.J.; Corrigan, D.K. Developing a Low-Cost, Simple-to-Use Electrochemical Sensor for the Detection of Circulating Tumour DNA in Human Fluids. Biosensors 2020, 10, 156. [Google Scholar] [CrossRef] [PubMed]
- Attoye, B.; Baker, M.J.; Thomson, F.; Pou, C.; Corrigan, D.K. Optimisation of an Electrochemical DNA Sensor for Measuring KRAS G12D and G13D Point Mutations in Different Tumour Types. Biosensors 2021, 11, 42. [Google Scholar] [CrossRef] [PubMed]
- Beylerli, O.; Gareev, I.; Sufianov, A.; Ilyasova, T.; Guang, Y. Long noncoding RNAs as promising biomarkers in cancer. Noncoding RNA Res. 2022, 7, 66–70. [Google Scholar] [CrossRef]
- Moore, L.D.; Le, T.; Fan, G. DNA Methylation and Its Basic Function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef]
- Soda, N.; Gonzaga, Z.J.; Chen, S.; Koo, K.M.; Nguyen, N.-T.; Shiddiky, M.J.A.; Rehm, B.H.A. Bioengineered Polymer Nanobeads for Isolation and Electrochemical Detection of Cancer Biomarkers. ACS Appl. Mater. Interfaces 2021, 13, 31418–31430. [Google Scholar] [CrossRef]
- Wang, G.L.; Zhou, L.Y.; Luo, H.Q.; Li, N.B. Electrochemical strategy for sensing DNA methylation and DNA methyltransferase activity. Anal. Chim. Acta 2013, 768, 76–81. [Google Scholar] [CrossRef]
- Gao, F.; Fan, T.; Ou, S.; Wu, J.; Zhang, X.; Luo, J.; Li, N.; Yao, Y.; Mou, Y.; Liao, X.; et al. Highly efficient electrochemical sensing platform for sensitive detection DNA methylation, and methyltransferase activity based on Ag NPs decorated carbon nanocubes. Biosens. Bioelectron. 2018, 99, 201–208. [Google Scholar] [CrossRef]
- Jiang, H.; Gu, X.; Zuo, Z.; Tian, G.; Liu, J. Prognostic value of circulating tumor cells in patients with bladder cancer: A meta-analysis. PLoS ONE 2021, 16, e0254433. [Google Scholar] [CrossRef]
- Alharthi, S.D.; Kanniyappan, H.; Prithweeraj, S.; Bijukumar, D.; Mathew, M.T. Proteomic-based electrochemical non-invasive biosensor for early breast cancer diagnosis. Int. J. Biol. Macromol. 2023, 253, 126681. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Zhou, R.; Hao, X.; Zhang, W.; Chen, G.; Zhu, T. Circulating biomarkers in perioperative management of cancer patients. Precis. Clin. Med. 2023, 6, pbad018. [Google Scholar] [CrossRef] [PubMed]
- Sproston, N.R.; Ashworth, J.J. Role of C-Reactive Protein at Sites of Inflammation and Infection. Front. Immunol. 2018, 9, 754. [Google Scholar] [CrossRef] [PubMed]
- Hart, P.C.; Rajab, I.M.; Alebraheem, M.; Potempa, L.A. C-Reactive Protein and Cancer—Diagnostic and Therapeutic Insights. Front. Immunol. 2020, 11, 595835. [Google Scholar] [CrossRef]
- Shrotriya, S.; Walsh, D.; Bennani-Baiti, N.; Thomas, S.; Lorton, C. C-Reactive Protein Is an Important Biomarker for Prognosis Tumor Recurrence and Treatment Response in Adult Solid Tumors: A Systematic Review. PLoS ONE 2015, 10, e0143080. [Google Scholar] [CrossRef]
- Boonkaew, S.; Chaiyo, S.; Jampasa, S.; Rengpipat, S.; Siangproh, W.; Chailapakul, O. An origami paper-based electrochemical immunoassay for the C-reactive protein using a screen-printed carbon electrode modified with graphene and gold nanoparticles. Microchim. Acta 2019, 186, 153. [Google Scholar] [CrossRef]
- Pinyorospathum, C.; Chaiyo, S.; Sae-Ung, P.; Hoven, V.P.; Damsongsang, P.; Siangproh, W.; Chailapakul, O. Disposable paper-based electrochemical sensor using thiol-terminated poly(2-methacryloyloxyethyl phosphorylcholine) for the label-free detection of C-reactive protein. Microchim. Acta 2019, 186, 472. [Google Scholar] [CrossRef]
- Yang, M.; Lin, S.-Q.; Liu, X.-Y.; Tang, M.; Hu, C.-L.; Wang, Z.-W.; Zhang, Q.; Zhang, X.; Song, M.-M.; Ruan, G.-T.; et al. Association between C-reactive protein-albumin-lymphocyte (CALLY) index and overall survival in patients with colorectal cancer: From the investigation on nutrition status and clinical outcome of common cancers study. Front. Immunol. 2023, 14, 1131496. [Google Scholar] [CrossRef]
- Das, S.; Dey, M.K.; Devireddy, R.; Gartia, M.R. Biomarkers in Cancer Detection, Diagnosis, and Prognosis. Sensors 2023, 24, 37. [Google Scholar] [CrossRef]
- Jansen, K.; Kornfeld, L.; Lennartz, M.; Rico, S.D.; Kind, S.; Reiswich, V.; Viehweger, F.; Bawahab, A.A.; Fraune, C.; Gorbokon, N.; et al. Carcinoembryonic Antigen Expression in Human Tumors: A Tissue Microarray Study on 13,725 Tumors. Cancers 2024, 16, 4052. [Google Scholar] [CrossRef]
- Yan, Y.; Liu, Z.; Pang, W.; Huang, S.; Deng, M.; Yao, J.; Huang, Q.; Jin, M.; Shui, L. Integrated biosensor array for multiplex biomarkers cancer diagnosis via in-situ self-assembly carbon nanotubes with an ordered inverse-opal structure. Biosens. Bioelectron. 2024, 262, 116528. [Google Scholar] [CrossRef] [PubMed]
- Ngaosri, P.; Karuwan, C.; Wanram, S.; Jarujamrus, P.; Citterio, D.; Amatatongchai, M. A selective dual-signal electrochemical paper-based device using imprinted sensors for voltammetric and impedance analysis of 4-NQO and carcinoembryonic antigen (CEA). Anal. Chim. Acta 2024, 1330, 343273. [Google Scholar] [CrossRef] [PubMed]
- Tzouvadaki, I.; Jolly, P.; Lu, X.; Ingebrandt, S.; de Micheli, G.; Estrela, P.; Carrara, S. Label-Free Ultrasensitive Memristive Aptasensor. Nano Lett. 2016, 16, 4472–4476. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Melo, L.F.; Morales-Rodríguez, M.; Madrigal-Bujaidar, E.; Madrigal-Santillán, E.O.; Morales-González, J.A.; Cruces, R.N.P.; Ramírez, J.A.C.; Damian-Matsumura, P.; Tellez-Plancarte, A.; Batina, N.; et al. Development of a Nanostructured Electrochemical Genosensor for the Detection of the K-ras Gene. J. Anal. Methods Chem. 2022, 2022, 6575140. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Niu, K.; Du, H.; Li, R.; Chen, J.; Lu, X. Sensitive Electrochemical Biosensor for Rapid Screening of Tumor Biomarker TP53 Gene Mutation Hotspot. Biosensors 2022, 12, 658. [Google Scholar] [CrossRef]
- Chiorcea-Paquim, A.-M. Advances in Electrochemical Biosensor Technologies for the Detection of Nucleic Acid Breast Cancer Biomarkers. Sensors 2023, 23, 4128. [Google Scholar] [CrossRef]
- Ferreira, D.C.; Batistuti Sawazaki, M.R.; Bachour Junior, B.; Mulato, M. Electrochemical capacitance-based aptasensor for HER2 detection. Biomed. Microdevices 2025, 27, 5. [Google Scholar] [CrossRef]
- Wuethrich, A.; Dey, S.; Koo, K.M.; Sina, A.A.I.; Trau, M. Simultaneous BRAFV600E Protein and DNA Aberration Detection in Circulating Melanoma Cells Using an Integrated Multimolecular Sensor. In Melanoma: Methods and Protocols; Hargadon, K.M., Ed.; Springer: New York, NY, USA, 2021; pp. 265–276. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Chu, Y.; Gao, Y.; Tang, W.; Qiang, L.; Han, Y.; Gao, J.; Zhang, Y.; Liu, H.; Han, L. Attomolar-Level Ultrasensitive and Multiplex microRNA Detection Enabled by a Nanomaterial Locally Assembled Microfluidic Biochip for Cancer Diagnosis. Anal. Chem. 2021, 93, 5129–5136. [Google Scholar] [CrossRef]
- Dornhof, J.; Kieninger, J.; Muralidharan, H.; Maurer, J.; Urban, G.A.; Weltin, A. Microfluidic organ-on-chip system for multi-analyte monitoring of metabolites in 3D cell cultures. Lab Chip 2022, 22, 225–239. [Google Scholar] [CrossRef]
- Koo, K.-M.; Kim, C.-D.; Kim, T.-H. Recent Advances in Electrochemical Detection of Cell Energy Metabolism. Biosensors 2024, 14, 46. [Google Scholar] [CrossRef]
- Das, S.; Saha, B.; Tiwari, M.; Tiwari, D.K. Diagnosis of cancer using carbon nanomaterial-based biosensors. Sens. Diagn. 2023, 2, 268–289. [Google Scholar] [CrossRef]
- Arshad, F.; Nabi, F.; Iqbal, S.; Khan, R.H. Applications of graphene-based electrochemical and optical biosensors in early detection of cancer biomarkers. Colloids Surf. B Biointerfaces 2022, 212, 112356. [Google Scholar] [CrossRef] [PubMed]
- Pourmadadi, M.; Soleimani Dinani, H.; Saeidi Tabar, F.; Khassi, K.; Janfaza, S.; Tasnim, N.; Hoorfar, M. Properties and Applications of Graphene and Its Derivatives in Biosensors for Cancer Detection: A Comprehensive Review. Biosensors 2022, 12, 269. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Mukherjee, A.; Bytesnikova, Z.; Ashrafi, A.M.; Richtera, L.; Adam, V. 2D graphene-based advanced nanoarchitectonics for electrochemical biosensors: Applications in cancer biomarker detection. Biosens. Bioelectron. 2024, 250, 116050. [Google Scholar] [CrossRef] [PubMed]
- Jin, Q.; Men, K.; Li, G.; Ou, T.; Lian, Z.; Deng, X.; Zhao, H.; Zhang, Q.; Ming, A.; Wei, Q.; et al. Ultrasensitive Graphene Field-Effect Biosensors Based on Ferroelectric Polarization of Lithium Niobate for Breast Cancer Marker Detection. ACS Appl. Mater. Interfaces 2024, 16, 28896–28904. [Google Scholar] [CrossRef]
- Song, J.; Luo, Y.; Hao, Z.; Qu, M.; Huang, C.; Wang, Z.; Yang, J.; Liang, Q.; Jia, Y.; Song, Q.; et al. Graphene-based wearable biosensors for point-of-care diagnostics: From surface functionalization to biomarker detection. Mater. Today Bio 2025, 32, 101667. [Google Scholar] [CrossRef]
- Sun, M.; Wang, S.; Zhang, Y.; Zhang, Z.; Wang, S.; Wang, Z.; Chen, X.; Liu, H.; Zhang, Y.; Han, L. An ultrasensitive flexible biosensor enabled by high-performance graphene field-effect transistors with defect-free van der Waals contacts for breast cancer miRNA fast detection. Talanta 2025, 287, 127637. [Google Scholar] [CrossRef]
- Zhou, L.; Mao, H.; Wu, C.; Tang, L.; Wu, Z.; Sun, H.; Zhang, H.; Zhou, H.; Jia, C.; Jin, Q.; et al. Label-free graphene biosensor targeting cancer molecules based on non-covalent modification. Biosens. Bioelectron. 2017, 87, 701–707. [Google Scholar] [CrossRef]
- Tao, C.; Rouhi, J. A biosensor based on graphene oxide nanocomposite for determination of carcinoembryonic antigen in colorectal cancer biomarker. Environ. Res. 2023, 238, 117113. [Google Scholar] [CrossRef]
- Kalkal, A.; Pradhan, R.; Kadian, S.; Manik, G.; Packirisamy, G. Biofunctionalized Graphene Quantum Dots Based Fluorescent Biosensor toward Efficient Detection of Small Cell Lung Cancer. ACS Appl. Bio Mater. 2020, 3, 4922–4932. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Zhao, Y.; Huang, Q.; Huang, J.; Tao, Y.; Chen, J.; Li, H.-Y.; Liu, H. Electrochemical protein biosensors for disease marker detection: Progress and opportunities. Microsyst. Nanoeng. 2024, 10, 65. [Google Scholar] [CrossRef] [PubMed]
- Siciliano, G.; Alsadig, A.; Chiriacò, M.S.; Turco, A.; Foscarini, A.; Ferrara, F.; Gigli, G.; Primiceri, E. Beyond traditional biosensors: Recent advances in gold nanoparticles modified electrodes for biosensing applications. Talanta 2024, 268, 125280. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Qian, K.; Qi, J.; Liu, Q.; Yao, C.; Song, W.; Wang, Y. Gold nanoparticles superlattices assembly for electrochemical biosensor detection of microRNA-21. Biosens. Bioelectron. 2018, 99, 564–570. [Google Scholar] [CrossRef]
- Lipińska, W.; Grochowska, K.; Siuzdak, K. Enzyme Immobilization on Gold Nanoparticles for Electrochemical Glucose Biosensors. Nanomaterials 2021, 11, 1156. [Google Scholar] [CrossRef]
- Lee, M.-J.; Choi, J.-H.; Shin, J.-H.; Yun, J.; Kim, T.; Kim, Y.-J.; Oh, B.-K. Gold Nanoclusters with Two Sets of Embedded Enzyme Nanoparticles for Applications as Electrochemical Sensors for Glucose. ACS Appl. Nano Mater. 2023, 6, 12567–12577. [Google Scholar] [CrossRef]
- Guo, Q.; Fan, X.; Yan, F.; Wang, Y. Highly sensitive electrochemical immunosensor based on electrodeposited platinum nanostructures confined in silica nanochannels for the detection of the carcinoembryonic antigen. Front. Chem. 2023, 11, 1271556. [Google Scholar] [CrossRef]
- Iriarte-Mesa, C.; López, Y.C.; Matos-Peralta, Y.; de la Vega-Hernández, K.; Antuch, M. Gold, Silver and Iron Oxide Nanoparticles: Synthesis and Bionanoconjugation Strategies Aimed at Electrochemical Applications. Top. Curr. Chem. 2020, 378, 12. [Google Scholar] [CrossRef]
- Ortega, F.G.; Fernández-Baldo, M.A.; Serrano, M.J.; Messina, G.A.; Lorente, J.A.; Raba, J. Epithelial cancer biomarker EpCAM determination in peripheral blood samples using a microfluidic immunosensor based in silver nanoparticles as platform. Sens. Actuators B Chem. 2015, 221, 248–256. [Google Scholar] [CrossRef]
- Huang, J.; Tian, J.; Zhao, Y.; Zhao, S. Ag/Au nanoparticles coated graphene electrochemical sensor for ultrasensitive analysis of carcinoembryonic antigen in clinical immunoassay. Sens. Actuators B Chem. 2015, 206, 570–576. [Google Scholar] [CrossRef]
- Chen, W.; Wang, H.; Tang, H.; Yang, C.; Li, Y. Unique Voltammetry of Silver Nanoparticles: From Single Particle to Aggregates. Anal. Chem. 2019, 91, 14188–14191. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Tan, X.; Wang, P.; Qin, J. Application of Polypyrrole-Based Electrochemical Biosensor for the Early Diagnosis of Colorectal Cancer. Nanomaterials 2023, 13, 674. Available online: https://www.mdpi.com/2079-4991/13/4/674 (accessed on 26 June 2025). [CrossRef] [PubMed]
- Natarajan, B.; Kannan, P.; Maduraiveeran, G.; Alnaser, A.S. Polymer nanocomposite-based biomolecular sensor for healthcare monitoring. Adv. Colloid Interface Sci. 2025, 343, 103557. [Google Scholar] [CrossRef] [PubMed]
- Park, C.S.; Lee, C.; Kwon, O.S. Conducting Polymer Based Nanobiosensors. Polymers 2016, 8, 249. [Google Scholar] [CrossRef]
- Sonika; Verma, S.K.; Samanta, S.; Srivastava, A.K.; Biswas, S.; Alsharabi, R.M.; Rajput, S. Conducting Polymer Nanocomposite for Energy Storage and Energy Harvesting Systems. Adv. Mater. Sci. Eng. 2022, 2022, 2266899. [Google Scholar] [CrossRef]
- El Aamri, M.; Yammouri, G.; Mohammadi, H.; Amine, A.; Korri-Youssoufi, H. Electrochemical Biosensors for Detection of MicroRNA as a Cancer Biomarker: Pros and Cons. Biosensors 2020, 10, 186. [Google Scholar] [CrossRef]
- Mohammadpour-Haratbar, A.; Boraei, S.B.A.; Zare, Y.; Rhee, K.Y.; Park, S.-J. Graphene-Based Electrochemical Biosensors for Breast Cancer Detection. Biosensors 2023, 13, 80. [Google Scholar] [CrossRef]
- Hong, R.; Sun, H.; Li, D.; Yang, W.; Fan, K.; Liu, C.; Dong, L.; Wang, G. A Review of Biosensors for Detecting Tumor Markers in Breast Cancer. Life 2022, 12, 342. [Google Scholar] [CrossRef]
- Lakard, B. Electrochemical Biosensors Based on Conducting Polymers: A Review. Appl. Sci. 2020, 10, 6614. [Google Scholar] [CrossRef]
- Djaalab, E.; Samar, M.E.H.; Zougar, S.; Kherrat, R. Electrochemical Biosensor for the Determination of Amlodipine Besylate Based on Gelatin–Polyaniline Iron Oxide Biocomposite Film. Catalysts 2018, 8, 233. [Google Scholar] [CrossRef]
- Sarti, C.; Falcon, L.; Cincinelli, A.; Martellini, T.; Chianella, I. Development of molecularly imprinted polymer-based electrochemical sensors for the detection of UV filters in aquatic ecosystems. Talanta 2025, 285, 127375. [Google Scholar] [CrossRef] [PubMed]
- Dube, A.; Malode, S.J.; Alodhayb, A.N.; Mondal, K.; Shetti, N.P. Conducting polymer-based electrochemical sensors: Progress, challenges, and future perspectives. Talanta Open 2025, 11, 100395. [Google Scholar] [CrossRef]
- Lin, J.; Chen, Y.; Liu, X.; Jiang, H.; Wang, X. Engineered Intelligent Electrochemical Biosensors for Portable Point-of-Care Diagnostics. Chemosensors 2025, 13, 146. [Google Scholar] [CrossRef]
- Hosseine, M.; Naghib, S.M.; Khodadadi, A. Label-free electrochemical biosensor based on green-synthesized reduced graphene oxide/Fe3O4/nafion/polyaniline for ultrasensitive detection of SKBR3 cell line of HER2 breast cancer biomarker. Sci. Rep. 2024, 14, 11928. [Google Scholar] [CrossRef]
- Junior, D.W.; Kubota, L.T. CRISPR-based electrochemical biosensors: An alternative for point-of-care diagnostics? Talanta 2024, 278, 126467. [Google Scholar] [CrossRef]
- Liu, F.; Peng, J.; Lei, Y.-M.; Liu, R.-S.; Jin, L.; Liang, H.; Liu, H.-F.; Ma, S.-Y.; Zhang, X.-H.; Zhang, Y.-P.; et al. Electrochemical detection of ctDNA mutation in non-small cell lung cancer based on CRISPR/Cas12a system. Sens. Actuators B Chem. 2022, 362, 131807. [Google Scholar] [CrossRef]
- Guan, X.; Zhao, J.; Sha, Z.; Liang, Y.; Huang, J.; Zhang, J.; Sun, S. CRISPR/Cas12a and aptamer-chemiluminescence based analysis for the relative abundance determination of tumor-related protein positive exosomes for breast cancer diagnosis. Biosens. Bioelectron. 2024, 259, 116380. [Google Scholar] [CrossRef]
- Munje, R.D.; Muthukumar, S.; Jagannath, B.; Prasad, S. A New Paradigm in Sweat Based Wearable Diagnostics Biosensors Using Room Temperature Ionic Liquids (RTILs). Sci. Rep. 2017, 7, 1950. Available online: https://www.nature.com/articles/s41598-017-02133-0 (accessed on 26 June 2025). [CrossRef]
- Ciui, B.; Martin, A.; Mishra, R.K.; Brunetti, B.; Nakagawa, T.; Dawkins, T.J.; Lyu, M.; Cristea, C.; Sandulescu, R.; Wang, J. Wearable Wireless Tyrosinase Bandage and Microneedle Sensors: Toward Melanoma Screening. Adv. Healthc. Mater. 2018, 7, e1701264. [Google Scholar] [CrossRef]
- Huang, X.; Chen, L.; Sha, T.; Lin, Y.; Zeng, R.; Xu, J.; Chen, S.; Cai, H.-H.; Zhang, J.; Zhou, H.; et al. In Situ Tyrosinase Monitoring by Wearable Microneedle Patch toward Clinical Melanoma Screening. ACS Nano 2023, 17, 20073–20086. [Google Scholar] [CrossRef]
- Tu, J.; Min, J.; Song, Y.; Xu, C.; Li, J.; Moore, J.; Hanson, J.; Hu, E.; Parimon, T.; Wang, T.-Y.; et al. A wireless patch for the monitoring of C-reactive protein in sweat. Nat. Biomed. Eng. 2023, 7, 1293–1306. [Google Scholar] [CrossRef]
- González-Fernández, E.; Staderini, M.; Marland, J.R.; Gray, M.E.; Uçar, A.; Dunare, C.; Blair, E.O.; Sullivan, P.; Tsiamis, A.; Greenhalgh, S.N.; et al. In vivo application of an implantable tri-anchored methylene blue-based electrochemical pH sensor. Biosens. Bioelectron. 2022, 197, 113728. [Google Scholar] [CrossRef]
- Seo, J.-W.; Fu, K.; Correa, S.; Eisenstein, M.; Appel, E.A.; Soh, H.T. Real-time monitoring of drug pharmacokinetics within tumor tissue in live animals. Sci. Adv. 2022, 8, eabk2901. [Google Scholar] [CrossRef]
- Corsi, M.; Maurina, E.; Surdo, S.; Vandini, E.; Daini, E.; Vilella, A.; Leo, G.; Farshchian, M.; Grisendi, G.; Golinelli, G.; et al. In Vivo and in Situ Monitoring of Doxorubicin Pharmacokinetics with an Implantable Bioresorbable Optical Sensor—PMC. Sci. Adv. 2025, 11, eads0265. Available online: https://pmc.ncbi.nlm.nih.gov/articles/PMC12002126/ (accessed on 26 June 2025). [CrossRef]
- Wang, Y.; Gao, W.; Sun, M.; Feng, B.; Shen, H.; Zhu, J.; Chen, X.; Yu, S. A filter-electrochemical microfluidic chip for multiple surface protein analysis of exosomes to detect and classify breast cancer. Biosens. Bioelectron. 2023, 239, 115590. [Google Scholar] [CrossRef]
- Felici, E.; Regiart, M.D.; Pereira, S.V.; Ortega, F.G.; Angnes, L.; Messina, G.A.; Fernández-Baldo, M.A. Microfluidic Platform Integrated with Carbon Nanofibers-Decorated Gold Nanoporous Sensing Device for Serum PSA Quantification. Biosensors 2023, 13, 390. [Google Scholar] [CrossRef]
| Electrochemical Transduction Method | Advantage(s) | Limitation(s) | Clinical Practicality |
|---|---|---|---|
| Amperometric | High sensitivity; Versatile integration into devices; Compatible with multiple assay formats | Signal depends on biorecognition element stability; prone to background interference | Adaptable for detecting clinically relevant cancer biomarkers in diagnostic laboratories and point-of-care platforms |
| Potentiometric | Expanded range of detectable biomarkers; label-free detection of exosomes, miRNAs, and circulating tumor DNA | Lower sensitivity for macromolecules | Well-suited for tumor microenvironment monitoring and integration into FET formats to broaden detectable biomarker ranges |
| Impedimetric | Label-free, sensitive | Difficulty in miniaturization for point-of-care testing | Capable of detecting clinically relevant cancer biomarkers and has the capacity for integration into point-of-care testing |
| Voltammetric | Low detection limits; Provides detailed redox profiles; Capacity to detect multiple biomarkers | Requires complex and costly instrumentation | Use in multiplexed assays to detect multiple biomarkers in diagnostics |
| Method | Cost | Detection Time | Sensitivity (LOD) | Specificity |
|---|---|---|---|---|
| Electrochemical sensor | Low (few $ per test) | Seconds-minutes | Picomolar-femtomolar (pg-fg/mL) | Very High (specific bioreceptor) |
| ELISA | Moderate ($10–$20/sample) | Hours (2–6 h) | picogram/mL | High (antibody-based) |
| PCR (qPCR) | High (instrument + $5–$20/sample) | Hours (4–8 h) | Attomolar (few DNA copies) | High (primer-specific) |
| Cancer Biomarker | Biomarker Type | Proposed Use | Detection Method | Reference |
|---|---|---|---|---|
| C-reactive protein (CRP) | Protein | -Inflammatory marker associated with cancer prognosis. -Utilized to monitor cancer progression and recurrence. -Aid in diagnosis alongside other markers (e.g., CALLY index). | -CRP-affinity peptide-functionalized label-free electrochemical biosensor -Electrochemical aptasensor -Origami paper-based electrochemical assay -Disposable paper-based electrochemical assay | [39,40,41,42] |
| Carcinoembryonic antigen (CEA) | Protein | -Monitor recurrence of cancers specifically useful with colorectal cancer. | -Integrated multiplex biosensor assay -Electrochemical paper-based detection -Enzyme-free immunosensor | [43,44,45] |
| Prostate-specific antigen (PSA) | Protein | -Screening and monitoring prostate cancer. | -Disposable electrochemical aptasensor -Label-free aptasensor -Dual monitor electrochemical biosensor -Glycosylation analysis | [46,47,48,49,50,51,52] |
| Epidermal growth factor receptor (EGFR) | Genetic | -Identify mutations linked to cancers such as glioblastoma. | -Label free electrochemical biosensor -Capacitive biosensor | [50] |
| KRAS | Genetic | -Detect oncogenic mutation seen in colorectal, lung, and pancreatic cancer. | -DNA biosensor assay -Nanostructured electrochemical genosensor | [52,53,54] |
| miRNA | Genetic | -Detect abnormal miRNA expression. | -Microfluidic biochip | [55] |
| DNA methylation patterns | Genetic | -Detect epigenetic changes such as methylation patterns. | -Polymer Nanobeads and Electrochemical biosensor -Sensing platform based on Ag Nanoparticles | [56,57,58] |
| Lactate | Metabolic | -Monitor tumor metabolism and aggression. | -Microfluidic organ-on-chip electrochemical system -Dual-sensing system | [59] |
| Glucose | Metabolic | -Monitor glycolysis activity in tumor cells. | -Microfluidic organ-on-chip electrochemical system -Dual sensing system | [59] |
| References | Biomarker/Target | Sensor Type/ Modification | Detection Limit (LOD) | Linear Range | Testing Medium | Interference Study | Real-Sample Validation |
|---|---|---|---|---|---|---|---|
| Sun et al. [89] | miRNA-155 (breast cancer) | Flexible GFET, defect-free vdW contacts | 1.92 fM | 10 fM–100 pM | Human serum; human sweat | Not specified | Yes; validated in human serum and sweat samples |
| Zhou et al. [90] | Carcinoembryonic antigen (CEA) | GFET, antibody-modified (non-covariant) | <100 pg/mL | Not specified | Buffer solutions | Not specified | Not reported |
| Tao et al. [91] | CEA (Colorectal cancer) | GO nanocomposite, PPI/GO/GCE | 0.3 pg/mL | 0.1 pg/mL–1000 ng/mL | Buffer/model solutions | Not specified | Not reported |
| Kalkal et al. [92] | Neuron-specific enolase (NSE, lung cancer) | Amine-N-GQDs/AuNPs fluorescent biosensor | 0.09 pg/mL | 0.1 pgm/mL–1000 ng/mL | Buffer/model solutions | Not specified | Not reported |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Nadeem-Tariq, A.; Rafanan, J.R.; Kang, N.; Zhang, S.; Kanniyappan, H.; Merchant, A. Electrochemical Detection of Cancer Biomarkers: From Molecular Sensing to Clinical Translation. Biosensors 2026, 16, 44. https://doi.org/10.3390/bios16010044
Nadeem-Tariq A, Rafanan JR, Kang N, Zhang S, Kanniyappan H, Merchant A. Electrochemical Detection of Cancer Biomarkers: From Molecular Sensing to Clinical Translation. Biosensors. 2026; 16(1):44. https://doi.org/10.3390/bios16010044
Chicago/Turabian StyleNadeem-Tariq, Ahmed, John Russell Rafanan, Nicole Kang, Sunny Zhang, Hemalatha Kanniyappan, and Aftab Merchant. 2026. "Electrochemical Detection of Cancer Biomarkers: From Molecular Sensing to Clinical Translation" Biosensors 16, no. 1: 44. https://doi.org/10.3390/bios16010044
APA StyleNadeem-Tariq, A., Rafanan, J. R., Kang, N., Zhang, S., Kanniyappan, H., & Merchant, A. (2026). Electrochemical Detection of Cancer Biomarkers: From Molecular Sensing to Clinical Translation. Biosensors, 16(1), 44. https://doi.org/10.3390/bios16010044





