Surface Plasmon Resonance Aptasensors: Emerging Design and Deployment Landscape
Abstract
1. Introduction
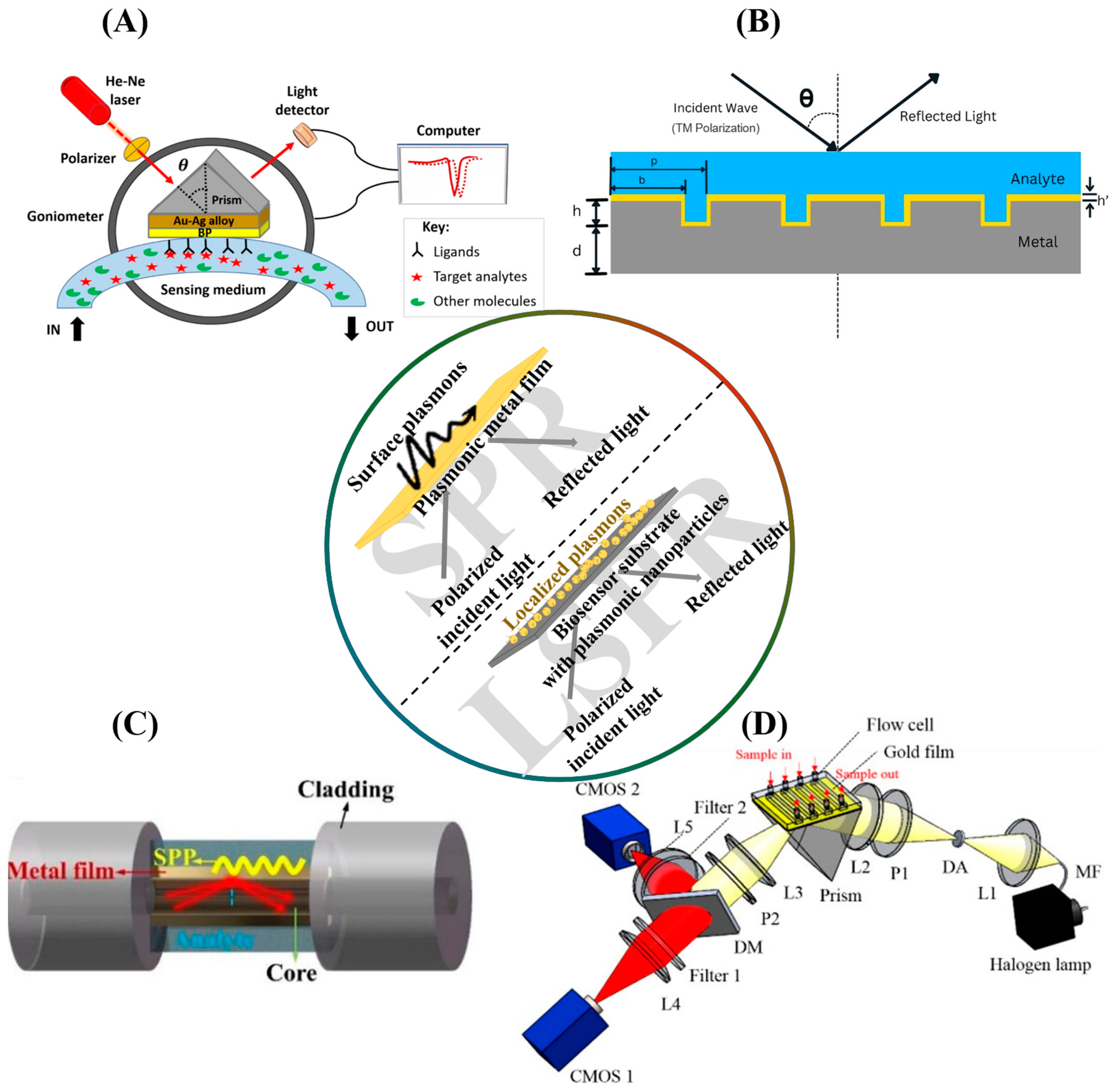
2. SPR Biosensors: Theoretical Basis and Applications
2.1. The Principle of Surface Plasmon Resonance
2.2. SPR Configurations
2.3. SPR Applications
3. Design Considerations in SPR Aptasensors
3.1. Aptamer Selection and Properties
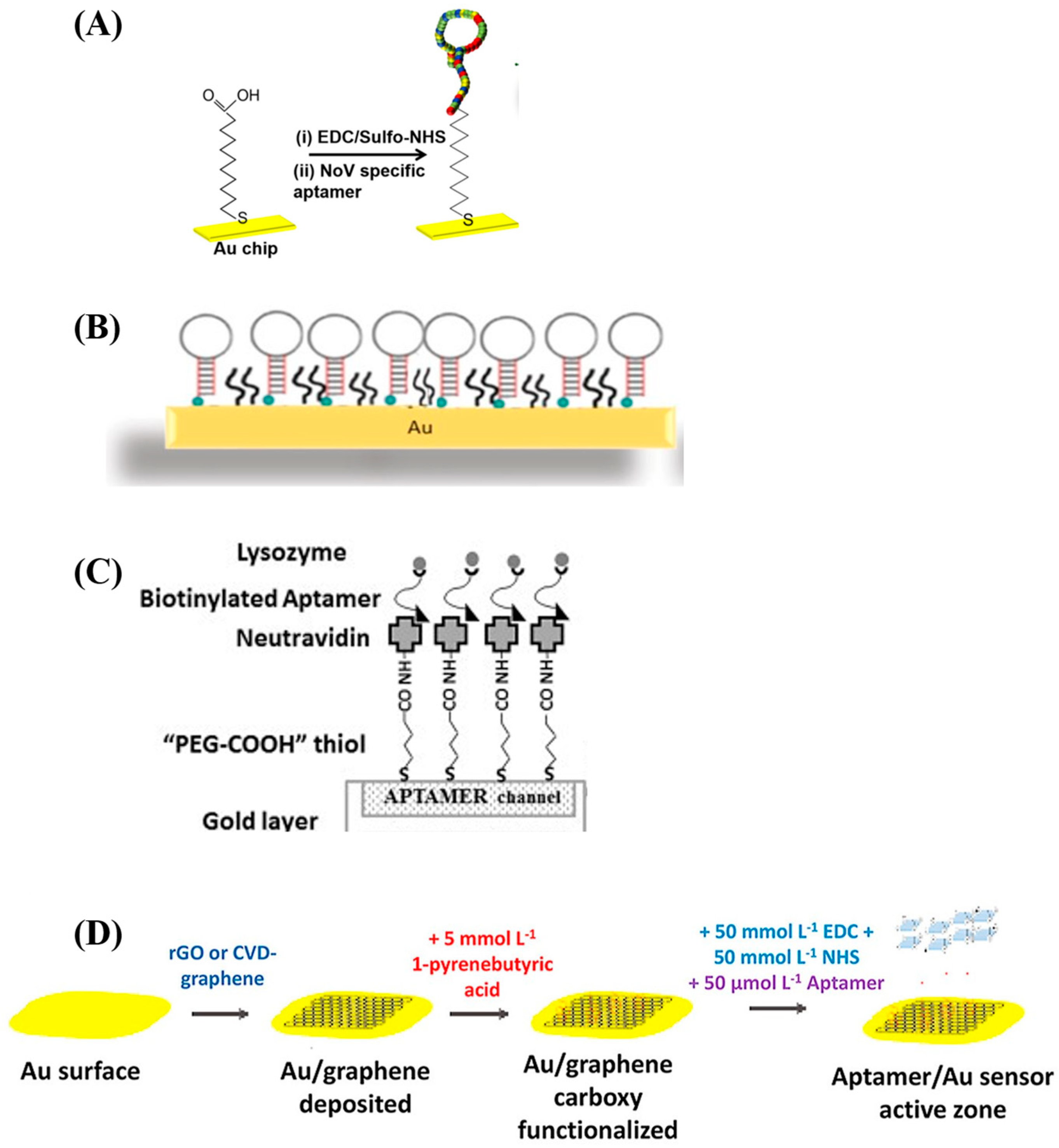
3.2. Aptamer Surface Immobilization Strategies
| Analyte of Interest | 5′-Aptamer Variable Sequence-3′ | Aptamer Preparation Method | Sensor Surface Preparation | Ref. |
|---|---|---|---|---|
| Whole Avian Influenza Virus—H5Nx | -CCC AGG TCG TGG TGG GTA CTG CGT ATG TGC- -TAG CCC CAG GCG GTG CGA GCT ACT GCC ATT GCAC- -TAA CGG TGT GGC CCG GGG GTA CAG CGC ACT- -TAC AAG TTG GAG GGG TTA AAT GTC TGC CGC- -GGC ATC GTT GGT TAA CCT CAT CAC GCG GGC- -TAA ATG GGC GTG GGA ATG ACT CTA CGG GGC- | Graphene oxide-assisted SELEX (GO-SELEX) and negative SELEX | Streptavidin captures biotinylated aptamers. Di-thiopropoinic acid (DTP) self-assembled monolayers are formed on gold film surface and carboxylic acid groups are activated via EDC/NHS reaction to covalently bind streptavidin which is used to capture biotinylated aptamers. | [67] |
| Visceral Adipose Tissue-Derived Serpin (vaspin) | -TGA TGG TGT GGC GGG GGG GGC CTG GGG GGG GCC GCC GAT G- | GO-SELEX | DTP self-assembly, streptavidin amine coupling, and capture of 5’T10-biotinylated aptamer. | [68] |
| Norovirus Capsid Protein | - -GCT AGC GAA TTC CGT ACG AAG GGC GAA TTC CAC ATT GGG CTG CAG CCC GGG GGA TCC- -GTC TGT AGT AGG GAG GAT GGT CCG GGG CCC CGA GAC GAC GTT ATC AGG C- -CGT ACG GAA TTC GCT AGC ACG GGG CTT AAG GAA TAC AGA TGT ACT ACC GAG CTC ATG AGG ATC CGA GCT CCA CGT G- -CGT ACG GAA TTC GCT AGC CGA CGG TCA ATG CTC GTG AGC CAG TAC ACA CAA TAT ATG TGG ATC CGA GCT CCA CGT G- | Ordinary SELEX with next-generation sequencing | Mercaptoundecanoic acid (MUA) self-assembly and onward amine coupling of 5’-amine-modified aptamers. | [65] |
| Salmonella typhimurium Outer Membrane Proteins | -TAT GGC GGC GTC ACC CGA CGG GGA CTT GAC ATT ATG ACA G- -GAG GAA AGT CTA TAG CAG AGG AGA TGT GTG AAC CGA GTA A- | Magnetic bead SELEX | Cysteamine self-assembly, carboxymethylated dextran coating, and amine-coupling of amine-modified aptamers. | [69] |
| Salmonella typhimurium | -TAT GGC GGC GTC ACC CGA CGG GGA CTT GAC ATT ATG ACA G- | - | Self-assembly of thiolated aptamer molecules on gold nanoparticles. | [24] |
| Ochratoxin, Aflatoxin, Adenosine Triphosphate, and Potassium Ions. | -TTT TTG TGG GTA GGG GGG GTT GGA CCA CAC CAA CC- -TTT TTA ACC TGG GGG AGT ATT GCG GAG GAA GGT- -TTT TTG TTG GGC ACG TGT TGT CTC TCT GTG TCT CGT GCC CTT CGC TAG GCC CAC A- -TTT TTG ATC GGG TGT GGG TGG CGT AAA GGG AGC ATC GGA CA- | - | Self-assembly of thiolated aptamers on gold nanorods. | [25] |
| 25-Hydroxyvitamin D3 | -AGC AGC ACA GAG GTC ATG GGG GGT GTG ACT TTG GTG TGC CTA TGC GTG CTA CGG AA- | GO-SELEX | Self-assembly of thiolated aptamers on gold nanorods. | [26] |
| Whole Staphylococcus aureus Cells | -TCC CAC GAT CTC ATT AGT CTG TGG ATA AGC GTG GGA CGT CTA TGA- | Whole-cell SELEX | Self-assembly of thiolated aptamers, following reduction of disulfide bonds in oligonucleotide molecules. | [23] |
| Ochratoxin | -TTT TTG ATC GGG TGT GGG TGG CGT AAA GGG AGC ATC GGA CA- | - | Self-assembly of thiolated aptamers on gold nanorods. | [70] |
| Whole Shigella Cells | -TTT TTT TTT TTT AGT CTT TCG CTG TTG CTG CTG ATG CC- -Cy5.5-GGC ATC AGC AGC AAC AGC GAA AGA CT- | Whole-cell SELEX | Streptavidin capture of biotinylated aptamer fragment. | [71] |
| Lysozyme Allergen | -GGG AAT GGA TCC ACA TCT ACG AAT TCA TCA GGG CTA AAG AG- | Robotic SELEX | Neutravidin capture of biotinylated aptamer. | [66] |
| HER2 | -TCT AAA AGG ATT CTT CCC AAG GGG ATC CAA TTC AAA CAG- | Adherent whole-cell SELEX with next-generation sequencing | Self-assembly of thiol-modified aptamers on gold-coated optical fiber surface. | [72] |
| MCF-7 Breast Cancer Cells | -GCA GTT GAT CCT TTG GAT ACC CTG G- | - | Thiol coupling of aptamer cysteine group to gold surface. | [73] |
| Tyrosine Kinase-7 | -ATC TAA CTG CTG CGC CGC CGG GAA AAT ACT GTA CGG TTA GAT TTT TTT TTT- | - | Thiol coupling of aptamers to gold nanostar surface. | [74] |
4. Translational Perspective of SPR Aptasensors
4.1. Drug Discovery Applications
4.2. Diagnostics and Disease Monitoring
4.3. Regulatory Considerations and Broader Impact
5. Challenges and Limitations of SPR Aptasensors
6. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CTC | Circulating tumor cell |
| ctDNA | Circulating tumor DNA |
| DNA | Deoxyribonucleic acid |
| DTP | Di-thiopropionate |
| EDC | Ethyl di-methyl aminopropyl carbodiimide |
| FDA | Food and Drug Agency |
| GO-SELEX | Graphene oxide-based SELEX |
| HER2 | Receptor tyrosine protein kinase |
| HIV | Human Immunodeficiency Virus |
| IVDMDD | In Vitro Diagnostic Medical Device Directive |
| LSPR | Localized surface plasmon resonance |
| MUA | Mercapto-undecanoic acid |
| NHS | N-hydroxy succinimide |
| RNA | Ribonucleic acid |
| SAMR | Chinese State Administration for Market Regulation |
| SARS-CoV2 | Severe acute respiratory syndrome coronavirus 2 |
| SELEX | Systematic Evolution of Ligands by Exponential Enrichment |
| SPR | Surface plasmon resonance |
| SPRi | Surface plasmon resonance imaging |
References
- Stuart, D.D.; Van Zant, W.; Valiulis, S.; Malinick, A.S.; Hanson, V.; Cheng, Q. Trends in Surface Plasmon Resonance Biosensing: Materials, Methods, and Machine Learning. Anal. Bioanal. Chem. 2024, 416, 5221–5232. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, X.; Wu, L.; Ding, L.; Effah, C.Y.; Wu, Y.; Xiong, Y.; He, L. Construction and Bioapplications of Aptamer-Based Dual Recognition Strategy. Biosens. Bioelectron. 2022, 195, 113661. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.D.; Szostak, J.W. In Vitro Selection of RNA Molecules That Bind Specific Ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Yüce, M.; Ullah, N.; Budak, H. Trends in Aptamer Selection Methods and Applications. Analyst 2015, 140, 5379–5399. [Google Scholar] [CrossRef]
- Kulabhusan, P.K.; Hussain, B.; Yüce, M. Current Perspectives on Aptamers as Diagnostic Tools and Therapeutic Agents. Pharmaceutics 2020, 12, 646. [Google Scholar] [CrossRef]
- Cai, S.; Yan, J.; Xiong, H.; Liu, Y.; Peng, D.; Liu, Z. Investigations on the Interface of Nucleic Acid Aptamers and Binding Targets. Analyst 2018, 143, 5317–5338. [Google Scholar] [CrossRef]
- Yüce, M.; Kurt, H.; Hussain, B.; Ow-Yang, C.W.; Budak, H. Exploiting Stokes and Anti-Stokes Type Emission Profiles of Aptamer-Functionalized Luminescent Nanoprobes for Multiplex Sensing Applications. ChemistrySelect 2018, 3, 5814–5823. [Google Scholar] [CrossRef]
- Yüce, M.; Kurt, H.; Hussain, B.; Budak, H. Systematic Evolution of Ligands by Exponential Enrichment for Aptamer Selection. In Biomedical Applications of Functionalized Nanomaterials: Concepts, Development and Clinical Translation; Elsevier: Amsterdam, The Netherlands, 2018; pp. 211–243. [Google Scholar] [CrossRef]
- Fano, U. The Theory of Anomalous Diffraction Gratings and of Quasi-Stationary Waves on Metallic Surfaces (Sommerfeld’s Waves). JOSA 1941, 31, 213–222. [Google Scholar] [CrossRef]
- Burstein, E.; Chen, W.P.; Chen, Y.J.; Hartstein, A. Surface Polaritons—Propagating Electromagnetic Modes at Interfaces. J. Vac. Sci. Technol. 1974, 11, 1004–1019. [Google Scholar] [CrossRef]
- Liedberg, B.; Nylander, C.; Lunström, I. Surface Plasmon Resonance for Gas Detection and Biosensing. Sens. Actuators 1983, 4, 299–304. [Google Scholar] [CrossRef]
- Kurihara, K.; Nakamura, K.; Suzuki, K. Asymmetric SPR Sensor Response Curve-Fitting Equation for the Accurate Determination of SPR Resonance Angle. Sens. Actuators B Chem. 2002, 86, 49–57. [Google Scholar] [CrossRef]
- Butt, M.A. Surface Plasmon Resonance-Based Biodetection Systems: Principles, Progress and Applications—A Comprehensive Review. Biosensors 2025, 15, 35. [Google Scholar] [CrossRef] [PubMed]
- Günaydın, B.N.; Çetinkaya, A.O.; Torabfam, M.; Tütüncüoğlu, A.; Kayalan, C.I.; Bayazıt, M.K.; Yüce, M.; Kurt, H. Plasmonic Group IVB Transition Metal Nitrides: Fabrication Methods and Applications in Biosensing, Photovoltaics and Photocatalysis. Adv. Colloid. Interface Sci. 2024, 333, 103298. [Google Scholar] [CrossRef] [PubMed]
- Andalibi Miandoab, S.; Talebzadeh, R. Ultra-Sensitive and Selective 2D Hybrid Highly Doped Semiconductor-Graphene Biosensor Based on SPR and SEIRA Effects in the Wide Range of Infrared Spectral. Opt. Mater. 2022, 129, 112572. [Google Scholar] [CrossRef]
- Khan, Q.; Sohrab, A.; Khan, M.A.; Khesro, A. Surface Plasmon Polariton Resonance Sensing at Metal-Dielectric Interface Based on Wavelength Interrogation. Plasmonics 2024, 20, 943–950. [Google Scholar] [CrossRef]
- Halpern, A.R.; Chen, Y.; Corn, R.M.; Kim, D. Surface Plasmon Resonance Phase Imaging Measurements of Patterned Monolayers and DNA Adsorption onto Microarrays. Anal. Chem. 2011, 83, 2801–2806. [Google Scholar] [CrossRef]
- Bonyár, A. Label-Free Nucleic Acid Biosensing Using Nanomaterial-Based Localized Surface Plasmon Resonance Imaging: A Review. ACS Appl. Nano Mater. 2020, 3, 8506–8521. [Google Scholar] [CrossRef]
- Saxena, S.; Rastogi, Y.; Sharma, N.K. Theoretical Analysis of Diffraction Grating-Based SPR Sensor Using the Rigorous Coupled Wave Analysis Method. Plasmonics 2025. [Google Scholar] [CrossRef]
- Zhang, S.; Han, B.; Zhang, Y.-N.; Liu, Y.; Zheng, W.; Zhao, Y.; Zhang, S.; Han, B.; Zhang, Y.-N.; Liu, Y.; et al. Multichannel Fiber Optic SPR Sensors: Realization Methods, Application Status, and Future Prospects. Laser Photon. Rev. 2022, 16, 2200009. [Google Scholar] [CrossRef]
- Feiler, M.; Ziman, M.; Kovac, J.; Kuzma, A.; Uherek, F. Design of Optimal SPR-Based Multimode Waveguide Sensor for a Wide Range of Liquid Analytes. Photonics 2023, 10, 618. [Google Scholar] [CrossRef]
- Petryayeva, E.; Krull, U.J. Localized Surface Plasmon Resonance: Nanostructures, Bioassays and Biosensing—A Review. Anal Chim Acta 2011, 706, 8–24. [Google Scholar] [CrossRef] [PubMed]
- Khateb, H.; Klös, G.; Meyer, R.L.; Sutherland, D.S. Development of a Label-Free LSPR-Apta Sensor for Staphylococcus Aureus Detection. ACS Appl. Bio Mater. 2020, 3, 3066–3077. [Google Scholar] [CrossRef]
- Oh, S.Y.; Heo, N.S.; Shukla, S.; Cho, H.J.; Vilian, A.T.E.; Kim, J.; Lee, S.Y.; Han, Y.K.; Yoo, S.M.; Huh, Y.S. Development of Gold Nanoparticle-Aptamer-Based LSPR Sensing Chips for the Rapid Detection of Salmonella typhimurium in Pork Meat. Sci. Rep. 2017, 7, 10130. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Byun, J.Y.; Jang, H.; Hong, D.; Kim, M.G. A Highly Sensitive and Widely Adaptable Plasmonic Aptasensor Using Berberine for Small-Molecule Detection. Biosens. Bioelectron. 2017, 97, 292–298. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.; Lee, W.; Park, J.; Park, H.; Kim, M.; Kim, W.; Hong, J.; Park, J. Wide-Range Direct Detection of 25-Hydroxyvitamin D3 Using Polyethylene-Glycol-Free Gold Nanorod Based on LSPR Aptasensor. Biosens. Bioelectron. 2021, 181, 113118. [Google Scholar] [CrossRef]
- Wang, L.; Wang, Q.; Wang, T.Q.; Zhao, W.M.; Yin, X.Y.; Jiang, J.X.; Zhang, S.S. Plasmonic Crescent Nanoarray-Based Surface Lattice Resonance Sensor with a High Figure of Merit. Nanoscale 2022, 14, 6144–6151. [Google Scholar] [CrossRef]
- Jiang, J.; Wang, X.; Li, S.; Ding, F.; Li, N.; Meng, S.; Li, R.; Qi, J.; Liu, Q.; Liu, G.L. Plasmonic Nano-Arrays for Ultrasensitive Bio-Sensing. Nanophotonics 2018, 7, 1517–1531. [Google Scholar] [CrossRef]
- Yang, Y.; Murray, J.; Haverstick, J.; Tripp, R.A.; Zhao, Y. Silver Nanotriangle Array Based LSPR Sensor for Rapid Coronavirus Detection. Sens. Actuators B Chem. 2022, 359, 131604. [Google Scholar] [CrossRef]
- Wang, Q.; Ren, Z.H.; Zhao, W.M.; Wang, L.; Yan, X.; Zhu, A.S.; Qiu, F.M.; Zhang, K.K. Research Advances on Surface Plasmon Resonance Biosensors. Nanoscale 2022, 14, 564–591. [Google Scholar] [CrossRef]
- Huo, Z.; Li, Y.; Chen, B.; Zhang, W.; Yang, X.; Yang, X. Recent Advances in Surface Plasmon Resonance Imaging and Biological Applications. Talanta 2023, 255, 124213. [Google Scholar] [CrossRef]
- Xiao, C.; Eriksson, J.; Suska, A.; Filippini, D.; Mak, W.C. Print-and-Stick Unibody Microfluidics Coupled Surface Plasmon Resonance (SPR) Chip for Smartphone Imaging SPR (Smart-ISRP). Anal. Chim. Acta 2022, 1201, 339606. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, Y. Advances in Integrated Digital Microfluidic Platforms for Point-of-Care Diagnosis: A Review. Sens. Diagn. 2022, 1, 648–672. [Google Scholar] [CrossRef]
- Phiri, I.K.; Zekriti, M. Theoretical Investigation of Plasmonic Properties of Gold-Silver Alloys for SPR Biosensing Applications. Results Phys. 2025, 73, 108252. [Google Scholar] [CrossRef]
- Zeng, Y.; Zhou, J.; Sang, W.; Kong, W.; Qu, J.; Ho, H.P.; Zhou, K.; Gao, B.Z.; Chen, J.; Shao, Y. High-Sensitive Surface Plasmon Resonance Imaging Biosensor Based on Dual-Wavelength Differential Method. Front. Chem. 2021, 9, 801355. [Google Scholar] [CrossRef]
- Gürel, B.; Çapkın, E.; Parlar, A.; Özkan, A.; Çorbacıoğlu, M.; Dağlikoca, D.E.; Yüce, M. Optimized Methods for Analytical and Functional Comparison of Biosimilar Mab Drugs: A Case Study for Avastin, Mvasi, and Zirabev. Sci. Pharm. 2022, 90, 36. [Google Scholar] [CrossRef]
- Luz, J.G.G.; Souto, D.E.P.; Machado-Assis, G.F.; De Lana, M.; Kubota, L.T.; Luz, R.C.S.; Damos, F.S.; Martins, H.R. Development and Evaluation of a SPR-Based Immunosensor for Detection of Anti-Trypanosoma Cruzi Antibodies in Human Serum. Sens. Actuators B Chem. 2015, 212, 287–296. [Google Scholar] [CrossRef]
- Yamamichi, J.; Ojima, T.; Yurugi, K.; Iida, M.; Imamura, T.; Ashihara, E.; Kimura, S.; Maekawa, T. Single-Step, Label-Free Quantification of Antibody in Human Serum for Clinical Applications Based on Localized Surface Plasmon Resonance. Nanomedicine 2011, 7, 889–895. [Google Scholar] [CrossRef]
- Fischer, M.B.; Wolfram, W.; Binder, C.J.; Böhmig, G.A.; Wahrmann, M.; Eibl, M.M.; Wolf, H.M. Surface Plasmon Resonance Analysis Shows an IgG-Isotype-Specific Defect in ABO Blood Group Antibody Formation in Patients with Common Variable Immunodeficiency. Front. Immunol. 2015, 6, 136175. [Google Scholar] [CrossRef]
- Kim, J.Y.; Lee, M.H.; Jung, K.I.; Na, H.Y.; Cha, H.S.; Ko, E.M.; Kim, T.J. Detection of Antibodies against Glucose 6-Phosphate Isomerase in Synovial Fluid of Rheumatoid Arthritis Using Surface Plasmon Resonance (BIAcore). Exp. Mol. Med. 2003, 35, 310–316. [Google Scholar] [CrossRef]
- Huang, Y.C.; Chiang, C.Y.; Li, C.H.; Chang, T.C.; Chiang, C.S.; Chau, L.K.; Huang, K.W.; Wu, C.W.; Wang, S.C.; Lyu, S.R. Quantification of Tumor Necrosis Factor-α and Matrix Metalloproteinases-3 in Synovial Fluid by a Fiber-Optic Particle Plasmon Resonance Sensor. Analyst 2013, 138, 4599–4606. [Google Scholar] [CrossRef]
- Gorodkiewicz, E.; Sieńczyk, M.; Regulska, E.; Grzywa, R.; Pietrusewicz, E.; Lesner, A.; Łukaszewski, Z. Surface Plasmon Resonance Imaging Biosensor for Cathepsin G Based on a Potent Inhibitor: Development and Applications. Anal. Biochem. 2012, 423, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Kumari, A.; Yadav, A.; Singh, O.P.; Sharan, P. A Review of Surface Plasmon Resonance (SPR) Technology in Biosensing: Innovations, Applications and Future Trends. J. Opt. 2024. [Google Scholar] [CrossRef]
- Andam, N.; Refki, S.; Ishitobi, H.; Inouye, Y.; Sekkat, Z. Optical Characterization of Ultra-Thin Films of Azo-Dye-Doped Polymers Using Ellipsometry and Surface Plasmon Resonance Spectroscopy. Photonics 2021, 8, 41. [Google Scholar] [CrossRef]
- Joyce, G.F. Amplification, Mutation and Selection of Catalytic RNA. Gene 1989, 82, 83–87. [Google Scholar] [CrossRef]
- Kinzler, K.W.; Vogelstein, B. Whole Genome PCR: Application to the Identification of Sequences Bound by Gene Regulatory Protein. Nucleic Acids Res. 1989, 17, 3645–3653. [Google Scholar] [CrossRef]
- Bock, L.C.; Griffin, L.C.; Latham, J.A.; Vermaas, E.H.; Toole, J.J. Selection of Single-Stranded DNA Molecules that Bind and Inhibit Human Thrombin. Nature 1992, 355, 564–566. [Google Scholar] [CrossRef]
- Tuerk, C.; Gold, L. Systematic Evolution of Ligands by Exponential Enrichment: RNA Ligands to Bacteriophage T4 DNA Polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef]
- Kurt, H.; Eyüpoǧlu, A.E.; Sütlü, T.; Budak, H.; Yüce, M. Plasmonic Selection of SsDNA Aptamers against Fibroblast Growth Factor Receptor. ACS Comb. Sci. 2019, 21, 578–587. [Google Scholar] [CrossRef]
- Zhu, C.; Feng, Z.; Qin, H.; Chen, L.; Yan, M.; Li, L.; Qu, F. Recent Progress of SELEX Methods for Screening Nucleic Acid Aptamers. Talanta 2024, 266, 124998. [Google Scholar] [CrossRef]
- Yılmaz, D.; Muslu, T.; Parlar, A.; Kurt, H.; Yüce, M. SELEX against Whole-Cell Bacteria Resulted in Lipopolysaccharide Binding Aptamers. J. Biotechnol. 2022, 354, 10–20. [Google Scholar] [CrossRef]
- Wu, D.; Gordon, C.K.L.; Shin, J.H.; Eisenstein, M.; Soh, H.T. Directed Evolution of Aptamer Discovery Technologies. Acc. Chem. Res. 2022, 55, 685–695. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Zhu, C.; Zhao, L.; Li, L.; Huang, Y.; Zhang, Y.; Qu, F. Pressure Controllable Aptamers Picking Strategy by Targets Competition. Chin. Chem. Lett. 2021, 32, 218–220. [Google Scholar] [CrossRef]
- Douaki, A.; Garoli, D.; Inam, A.K.M.S.; Angeli, M.A.C.; Cantarella, G.; Rocchia, W.; Wang, J.; Petti, L.; Lugli, P. Smart Approach for the Design of Highly Selective Aptamer-Based Biosensors. Biosensors 2022, 12, 574. [Google Scholar] [CrossRef] [PubMed]
- Ventouri, I.K.; Loeber, S.; Somsen, G.W.; Schoenmakers, P.J.; Astefanei, A. Field-Flow Fractionation for Molecular-Interaction Studies of Labile and Complex Systems: A Critical Review. Anal. Chim. Acta 2022, 1193, 339396. [Google Scholar] [CrossRef]
- Eissa, S.; Siddiqua, A.; Chinnappan, R.; Zourob, M. Electrochemical SELEX Technique for the Selection of DNA Aptamers against the Small Molecule 11-Deoxycortisol. ACS Appl. Bio Mater. 2019, 2, 2624–2632. [Google Scholar] [CrossRef]
- Kulabhusan, P.K.; Pishva, P.; Çapkın, E.; Tambe, P.; Yüce, M. Aptamer-Based Emerging Tools for Viral Biomarker Detection: A Focus on SARS-CoV-2. Curr. Med. Chem. 2022, 30, 910–934. [Google Scholar] [CrossRef]
- Wu, Y.; Belmonte, I.; Sykes, K.S.; Xiao, Y.; White, R.J. Perspective on the Future Role of Aptamers in Analytical Chemistry. Anal. Chem. 2019, 91, 15335–15344. [Google Scholar] [CrossRef]
- Yüce, M.; Kurt, H. How to Make Nanobiosensors: Surface Modification and Characterisation of Nanomaterials for Biosensing Applications. RSC Adv. 2017, 7, 49386–49403. [Google Scholar] [CrossRef]
- Écija-Arenas, Á.; Kirchner, E.M.; Hirsch, T.; Fernández-Romero, J.M. Development of an Aptamer-Based SPR-Biosensor for the Determination of Kanamycin Residues in Foods. Anal. Chim. Acta 2021, 1169, 338631. [Google Scholar] [CrossRef]
- Song, M.S.; Sekhon, S.S.; Shin, W.R.; Kim, H.C.; Min, J.; Ahn, J.Y.; Kim, Y.H. Detecting and Discriminating Shigella Sonnei Using an Aptamer-Based Fluorescent Biosensor Platform. Molecules 2017, 22, 825. [Google Scholar] [CrossRef]
- Macdonald, H.; Bonnet, H.; Van Der Heyden, A.; Defrancq, E.; Spinelli, N.; Coche-Guérente, L.; Dejeu, J. Influence of Aptamer Surface Coverage on Small Target Recognition: A SPR and QCM-D Comparative Study. J. Phys. Chem. C 2019, 123, 13561–13568. [Google Scholar] [CrossRef]
- Dejeu, J.; Bonnet, H.; Spinelli, N.; Defrancq, E.; Coche-Guérente, L.; Van Der Heyden, A.; Labbé, P. Impact of Conformational Transitions on SPR Signals—Theoretical Treatment and Application in Small Analytes/Aptamer Recognition. J. Phys. Chem. C 2018, 122, 21521–21530. [Google Scholar] [CrossRef]
- Martin, J.A.; Chushak, Y.; Chávez, J.L.; Hagen, J.A.; Kelley-Loughnane, N. Microarrays as Model Biosensor Platforms to Investigate the Structure and Affinity of Aptamers. J. Nucleic Acids 2016, 2016, 9718612. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Lee, S.; Lee, H.J. An Aptamer-Aptamer Sandwich Assay with Nanorod-Enhanced Surface Plasmon Resonance for Attomolar Concentration of Norovirus Capsid Protein. Sens. Actuators B Chem. 2018, 273, 1029–1036. [Google Scholar] [CrossRef]
- Mihai, I.; Vezeanu, A.; Polonschii, C.; Albu, C.; Radu, G.L.; Vasilescu, A. Label-Free Detection of Lysozyme in Wines Using an Aptamer Based Biosensor and SPR Detection. Sens. Actuators B Chem. 2015, 206, 198–204. [Google Scholar] [CrossRef]
- Nguyen, V.T.; Seo, H.B.; Kim, B.C.; Kim, S.K.; Song, C.S.; Gu, M.B. Highly Sensitive Sandwich-Type SPR Based Detection of Whole H5Nx Viruses Using a Pair of Aptamers. Biosens. Bioelectron. 2016, 86, 293–300. [Google Scholar] [CrossRef]
- Ahmad Raston, N.H.; Gu, M.B. Highly Amplified Detection of Visceral Adipose Tissue-Derived Serpin (Vaspin) Using a Cognate Aptamer Duo. Biosens. Bioelectron. 2015, 70, 261–267. [Google Scholar] [CrossRef]
- Wang, B.; Park, B.; Xu, B.; Kwon, Y. Label-Free Biosensing of Salmonella enterica Serovars at Single-Cell Level. J. Nanobiotechnol. 2017, 15, 40. [Google Scholar] [CrossRef]
- Lee, B.; Park, J.H.; Byun, J.Y.; Kim, J.H.; Kim, M.G. An Optical Fiber-Based LSPR Aptasensor for Simple and Rapid in-Situ Detection of Ochratoxin A. Biosens. Bioelectron. 2018, 102, 504–509. [Google Scholar] [CrossRef]
- Kumar, S.; Guo, Z.; Singh, R.; Wang, Q.; Zhang, B.; Cheng, S.; Liu, F.Z.; Marques, C.; Kaushik, B.K.; Jha, R. MoS2 Functionalized Multicore Fiber Probes for Selective Detection of Shigella Bacteria Based on Localized Plasmon. J. Light. Technol. 2021, 39, 4069–4081. [Google Scholar] [CrossRef]
- Loyez, M.; Lobry, M.; Hassan, E.M.; DeRosa, M.C.; Caucheteur, C.; Wattiez, R. HER2 Breast Cancer Biomarker Detection Using a Sandwich Optical Fiber Assay. Talanta 2021, 221, 121452. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Hou, Y.; Ye, Z.; Wang, H.; Koh, K.; Shen, Z.; Shu, Y. Label-Free Surface Plasmon Resonance Cytosensor for Breast Cancer Cell Detection Based on Nano-Conjugation of Monodisperse Magnetic Nanoparticle and Folic Acid. Sens. Actuators B Chem. 2014, 201, 433–438. [Google Scholar] [CrossRef]
- Wang, S.S.; Zhao, X.P.; Liu, F.F.; Younis, M.R.; Xia, X.H.; Wang, C. Direct Plasmon-Enhanced Electrochemistry for Enabling Ultrasensitive and Label-Free Detection of Circulating Tumor Cells in Blood. Anal. Chem. 2019, 91, 4413–4420. [Google Scholar] [CrossRef] [PubMed]
- Capkin, E.; Kurt, H.; Gurel, B.; Bicak, D.; Akgun Bas, S.; Daglikoca, D.E.; Yuce, M. Characterization of FcγRIa (CD64) as a Ligand Molecule for Site-Specific IgG1 Capture: A Side-by-Side Comparison with Protein A. Langmuir 2022, 38, 14623–14634. [Google Scholar] [CrossRef] [PubMed]
- Loo, J.F.C.; Yang, C.; Tsang, H.L.; Lau, P.M.; Yong, K.T.; Ho, H.P.; Kong, S.K. An Aptamer Bio-BarCode (ABC) Assay Using SPR, RNase H, and Probes with RNA and Gold-Nanorods for Anti-Cancer Drug Screening. Analyst 2017, 142, 3579–3587. [Google Scholar] [CrossRef]
- Borg, K.N.; Ho, Y.P.; Zeng, S. Recent Developments on Optical Aptasensors for the Detection of Pro-Inflammatory Cytokines with Advanced Nanostructures. Adv. Opt. Mater. 2024, 12, 2400608. [Google Scholar] [CrossRef]
- Chang, C.C. Recent Advancements in Aptamer-Based Surface Plasmon Resonance Biosensing Strategies. Biosensors 2021, 11, 233. [Google Scholar] [CrossRef]
- Hu, J.; Fu, K.; Bohn, P.W. Whole-Cell Pseudomonas Aeruginosa Localized Surface Plasmon Resonance Aptasensor. Anal. Chem. 2018, 90, 2326–2332. [Google Scholar] [CrossRef]
- Kotlarek, D.; Curti, F.; Vorobii, M.; Corradini, R.; Careri, M.; Knoll, W.; Rodriguez-Emmenegger, C.; Dostálek, J. Surface Plasmon Resonance-Based Aptasensor for Direct Monitoring of Thrombin in a Minimally Processed Human Blood. Sens. Actuators B Chem. 2020, 320, 128380. [Google Scholar] [CrossRef]
- Sun, D.; Wu, Y.; Chang, S.J.; Chen, C.J.; Liu, J.T. Investigation of the Recognition Interaction between Glycated Hemoglobin and Its Aptamer by Using Surface Plasmon Resonance. Talanta 2021, 222, 121466. [Google Scholar] [CrossRef]
- Wang, W.W.; Han, X.; Chu, L.Q. Polyadenine-Mediated Immobilization of Aptamers on a Gold Substrate for the Direct Detection of Bacterial Pathogens. Anal. Sci. 2019, 35, 967–972. [Google Scholar] [CrossRef] [PubMed]
- Lou, Z.; Han, H.; Zhou, M.; Wan, J.; Sun, Q.; Zhou, X.; Gu, N. Fabrication of Magnetic Conjugation Clusters via Intermolecular Assembling for Ultrasensitive Surface Plasmon Resonance (SPR) Detection in a Wide Range of Concentrations. Anal. Chem. 2017, 89, 13472–13479. [Google Scholar] [CrossRef] [PubMed]
- Lou, Z.; Wan, J.; Zhang, X.; Zhang, H.; Zhou, X.; Cheng, S.; Gu, N. Quick and Sensitive SPR Detection of Prion Disease-Associated Isoform (PrPSc) Based on Its Self-Assembling Behavior on Bare Gold Film and Specific Interactions with Aptamer-Graphene Oxide (AGO). Colloids Surf. B Biointerfaces 2017, 157, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Mauriz, E. Trends and Challenges of SPR Aptasensors in Viral Diagnostics: A Systematic Review and Meta-Analysis. Biosensors 2025, 15, 245. [Google Scholar] [CrossRef]
- Ravindran, N.; Kumar, S.; Yashini, M.; Rajeshwari, S.; Mamathi, C.A.; Nirmal Thirunavookarasu, S.; Sunil, C.K. Recent Advances in Surface Plasmon Resonance (SPR) Biosensors for Food Analysis: A Review. Crit. Rev. Food Sci. Nutr. 2023, 63, 1055–1077. [Google Scholar] [CrossRef]
- Das, S.; Devireddy, R.; Gartia, M.R. Surface Plasmon Resonance (SPR) Sensor for Cancer Biomarker Detection. Biosensors 2023, 13, 396. [Google Scholar] [CrossRef]
- Zhao, L.; Li, L.; Zhao, Y.; Zhu, C.; Yang, R.; Fang, M.; Luan, Y. Aptamer-Based Point-of-Care-Testing for Small Molecule Targets: From Aptamers to Aptasensors, Devices and Applications. TrAC Trends Anal. Chem. 2023, 169, 117408. [Google Scholar] [CrossRef]
- Chen, X.F.; Zhao, X.; Yang, Z. Aptasensors for the Detection of Infectious Pathogens: Design Strategies and Point-of-Care Testing. Microchim. Acta 2022, 189, 443. [Google Scholar] [CrossRef]
- Lofgren, J.A.; Dhandapani, S.; Pennucci, J.J.; Abbott, C.M.; Mytych, D.T.; Kaliyaperumal, A.; Swanson, S.J.; Mullenix, M.C. Comparing ELISA and Surface Plasmon Resonance for Assessing Clinical Immunogenicity of Panitumumab. J. Immunol. 2007, 178, 7467–7472. [Google Scholar] [CrossRef]
- Loo, J.; Ng, S.P.; Wu, C.M.L.; Kong, S.K. An Aptasensor Using DNA Aptamer and White Light Common-Path SPR Spectral Interferometry to Detect Cytochrome-c for Anti-Cancer Drug Screening. Sens. Actuators B Chem. 2014, 198, 416–423. [Google Scholar] [CrossRef]
- Lee, K.A.; Ahn, J.Y.; Lee, S.H.; Singh Sekhon, S.; Kim, D.G.; Min, J.; Kim, Y.H. Aptamer-Based Sandwich Assay and Its Clinical Outlooks for Detecting Lipocalin-2 in Hepatocellular Carcinoma (HCC). Sci. Rep. 2015, 5, 10897. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Xu, Y.; Jiang, X.; Tan, H.; Ying, B. Translation of Aptamers toward Clinical Diagnosis and Commercialization. Biosens. Bioelectron. 2022, 208, 114168. [Google Scholar] [CrossRef] [PubMed]
- Sergeev, N.V.; Herold, K.E.; Rasooly, A. Regulatory and Validation Issues for Biosensors and Related Bioanalytical Technologies. In Handbook of Biosensors and Biochips; Wiley: Hoboken, NJ, USA, 2007. [Google Scholar]
- Pawnikar, V.; Patel, M. Biosensors in Wearable Medical Devices: Regulatory Framework and Compliance across US, EU, and Indian Markets. Ann. Pharm. Fr. 2025. [Google Scholar] [CrossRef] [PubMed]
- Fischer, W.A.; Wohl, D.A. Inequities in Access to Diagnostics Threatens Global Public Health Security. Lancet Infect. Dis. 2022, 22, 754–756. [Google Scholar] [CrossRef]
- Zhao, Y.; Yavari, K.; Liu, J. Critical Evaluation of Aptamer Binding for Biosensor Designs. TrAC Trends Anal. Chem. 2022, 146, 116480. [Google Scholar] [CrossRef]
- Thakur, M.; Wang, B.; Verma, M.L. Development and Applications of Nanobiosensors for Sustainable Agricultural and Food Industries: Recent Developments, Challenges and Perspectives. Environ. Technol. Innov. 2022, 26, 102371. [Google Scholar] [CrossRef]
- Berktas, I.; Hezarkhani, M.; Haghighi Poudeh, L.; Saner Okan, B. Recent Developments in the Synthesis of Graphene and Graphene-like Structures from Waste Sources by Recycling and Upcycling Technologies: A Review. Graphene Technol. 2020, 5, 59–73. [Google Scholar] [CrossRef]
- Zahra, Q.U.A.; Fang, X.; Luo, Z.; Ullah, S.; Fatima, S.; Batool, S.; Qiu, B.; Shahzad, F. Graphene Based Nanohybrid Aptasensors in Environmental Monitoring: Concepts, Design and Future Outlook. Crit. Rev. Anal. Chem. 2023, 53, 1433–1454. [Google Scholar] [CrossRef]
- Saeb, M.R. Biosensors from Renewable Resources: Decarbonization and Sustainability. Polym. Renew. Resour. 2024, 15, 155–159. [Google Scholar] [CrossRef]
- Chotimah, S.N.; Putra, Y.E.; Ng, J.H.; Tijptaningrum, A.; Sahara, N.S.; Arfianti, E.; Amaliah, R.; Edel, M.J.; Ortiz Rodriguez, M.; Llerga, T.M.; et al. DNA Aptamer Gold Nanoparticle Colorimetric Diagnostic Test Kit of Saliva Samples for SARS_Cov2 Virus Linked to Mobile Phone Application (AptamexTM). medRxiv 2022. [Google Scholar] [CrossRef]
- Jung, Y.J.; Katilius, E.; Ostroff, R.M.; Kim, Y.; Seok, M.; Lee, S.; Jang, S.; Kim, W.S.; Choi, C.M. Development of a Protein Biomarker Panel to Detect Non-Small-Cell Lung Cancer in Korea. Clin. Lung Cancer 2017, 18, e99–e107. [Google Scholar] [CrossRef] [PubMed]
- Apta-Beacons & Apta-Sensors. Available online: https://www.aptagen.com/aptamer-technologies/apta-beacons-and-apta-sensors/ (accessed on 30 April 2025).
- Cryopep-OLIGOBIND® Thrombin Activity Assay. Available online: https://www.cryopep.com/hemostasis-coagulation/catalogue/research/assays-kits/assays-kits-elisa/oligobind-thrombin-activity-assay/ (accessed on 30 April 2025).
- Biogenes Technologies|Home. Available online: https://www.biogenestech.com/index (accessed on 30 April 2025).
- Candia, J.; Cheung, F.; Kotliarov, Y.; Fantoni, G.; Sellers, B.; Griesman, T.; Huang, J.; Stuccio, S.; Zingone, A.; Ryan, B.M.; et al. Assessment of Variability in the SOMAscan Assay. Sci. Rep. 2017, 7, 14248. [Google Scholar] [CrossRef] [PubMed]
- Kaur, H.; Bruno, J.G.; Kumar, A.; Sharma, T.K. Aptamers in the Therapeutics and Diagnostics Pipelines. Theranostics 2018, 8, 4016–4032. [Google Scholar] [CrossRef] [PubMed]
- Meehan, C.; Hamilton, E.L.; Mansour, C.G.; Lecocq, S.; Drake, C.J.; An, Y.; Rodrigues, E.; Penner, G. A New Method for the Reproducible Development of Aptamers (Neomers). PLoS ONE 2025, 20, e0311497. [Google Scholar] [CrossRef]
- Kalra, P.; Dhiman, A.; Cho, W.C.; Bruno, J.G.; Sharma, T.K. Simple Methods and Rational Design for Enhancing Aptamer Sensitivity and Specificity. Front. Mol. Biosci. 2018, 5, 347832. [Google Scholar] [CrossRef]
- A Kind of Method of Terramycin in Enzyme-Linked Aptamers Detection Food of Functionalization Beads Enrichment. Available online: https://patents.google.com/patent/CN104634754B/en (accessed on 30 April 2025).
- Cho, Y.J.; Kim, C.J.; Kim, N.S.; Kim, C.T.; Maeng, J.S.; Kim, T.E.; Lee, M.H. Slide Chip for Detection Sensor of Food-Borne Pathogens and Preparation Method Thereof. U.S. Patent No. 10,036,073, 31 July 2018. [Google Scholar]
- Magnetic Ferric Oxide@aptamer and Its Application in Combination with Fluorescent Test Strips in the Detection of Foodborne Pathogens. Available online: https://patents.google.com/patent/CN113881790A/en (accessed on 30 April 2025).
- Based on the Aptamer Sensor Construction Method of the Two Amplifying Technique of Au NPs and DNA Circulation and the Application in Adenosine Detects Thereof. Available online: https://patents.google.com/patent/CN103439296B/en (accessed on 30 April 2025).
- A Kind of Optical Electro-Chemistry Aptamer Sensor and Its Preparation Method and Application. Available online: https://patents.google.com/patent/CN108845009B/en (accessed on 30 April 2025).
- Thurman, A. Project ADHERE: Clinical Proof-of-Concept of a Tenofovir (TFV) Aptamer-Based Biosensor for Determing Adherence Using Different Dosing Regimens of Disoproxil Fumarate/Emtricitabine. Available online: https://clinicaltrials.gov/study/NCT04870671 (accessed on 28 May 2025).
- Zong, C.; Liu, J. The Arsenic-Binding Aptamer Cannot Bind Arsenic: Critical Evaluation of Aptamer Selection and Binding. Anal. Chem. 2019, 91, 10887–10893. [Google Scholar] [CrossRef]
- Zeng, H.; Zeng, W.; Liang, Y. Application of Multivalent Aptamers in Tumor Diagnosis, Analysis and Therapy (Review). Oncol. Lett. 2025, 30, 325. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, H.; Ding, Y.; Yu, C.; Li, H. Serum Assisted PD-L1 Aptamer Screening for Improving Its Stability. Sci. Rep. 2025, 15, 1848. [Google Scholar] [CrossRef]
- Chung, Y.D.; Tsai, Y.C.; Wang, C.H.; Lee, G.B. Aptamer Selection via Versatile Microfluidic Platforms and Their Diverse Applications. Lab. Chip 2025, 25, 1047–1080. [Google Scholar] [CrossRef]
- Shamshiri, S.; Asghari, A.; Shahdost-Fard, F.; Rajabi, M. Molecular Dynamic Simulation-Empowered Detection of Digoxin by a Green Aptasensing Interface Based on Pseudo-Gold Nanobones@rice Husk-Derived Nanosilica. Talanta 2025, 291, 127885. [Google Scholar] [CrossRef]
- Arshavsky-Graham, S.; Heuer, C.; Jiang, X.; Segal, E. Aptasensors versus Immunosensors—Which Will Prevail? Eng. Life Sci. 2022, 22, 319–333. [Google Scholar] [CrossRef] [PubMed]
- Pasquardini, L.; Cennamo, N.; Arcadio, F.; Zeni, L. A Review of Apta-POF-Sensors: The Successful Coupling between Aptamers and Plastic Optical Fibers for Biosensing Applications. Appl. Sci. 2022, 12, 4584. [Google Scholar] [CrossRef]
- Torun, H.; Bilgin, B.; Ilgu, M.; Batur, N.; Ozturk, M.; Barlas, T.; Guney-Esken, G.; Yanik, C.; Celik, S.; Dogan, O.; et al. Rapid Nanoplasmonic-Enhanced Detection of SARS-CoV-2 and Variants on DNA Aptamer Metasurfaces. Adv. Devices Instrum. 2023, 4, 0008. [Google Scholar] [CrossRef]
- Kavungal, D.; Magalhães, P.; Kumar, S.T.; Kolla, R.; Lashuel, H.A.; Altug, H. Artificial Intelligence-Coupled Plasmonic Infrared Sensor for Detection of Structural Protein Biomarkers in Neurodegenerative Diseases. Sci. Adv. 2023, 9, eadg9644. [Google Scholar] [CrossRef]
- Teixeira, F.M.F.; Portes, A.V.R.; Marques, T.E.M.; Isayama, Y.H.; de Freitas, F.A.N.; Santana, F.C.; da Rocha, A.M.; Moraes, T.F.S.; Andrade, L.M.; Versiani, A.F.; et al. Advanced Computational Techniques for Plasmonic Metasurfaces in the Detection of Neglected Infectious Diseases. Anal. Chem. 2025, 97, 6813–6825. [Google Scholar] [CrossRef]
- Hua, M.Z.; Liu, J.; Roopesh, M.S.; Lu, X. Microfluidic Optical Aptasensor for Small Molecules Based on Analyte-Tuned Growth of Gold Nanoseeds and Machine Learning-Enhanced Spectrum Analysis: Rapid Detection of Mycotoxins. ACS Sens. 2024, 9, 6299–6308. [Google Scholar] [CrossRef]

| Analyte of Interest | 5′-Aptamer Sequence-3′ | Affinity Values | SPR Configuration | Signal Amplification | Limit of Detection | Ref. |
|---|---|---|---|---|---|---|
| Visceral Adipose tissue-Derived Serpin (vaspin) | -TGA TGG TGT GGC GGG GGG GGC CTG GGG GGG GCC GCC GAT G- | KD: 0.17–0.24 µM | Prism-coupled SPR | Sandwich SPR with gold nanoparticle-conjugated secondary aptamer | 3.5–4.7 ng/mL | [68] |
| Norovirus Capsid Protein | -GCT AGC GAA TTC CGT ACG AAG GGC GAA TTC CAC ATT GGG CTG CAG CCC GGG GGA TCC- -GTC TGT AGT AGG GAG GAT GGT CCG GGG CCC CGA GAC GAC GTT ATC AGG C- -CGT ACG GAA TTC GCT AGC ACG GGG CTT AAG GAA TAC AGA TGT ACT ACC GAG CTC ATG AGG ATC CGA GCT CCA CGT G- -CGT ACG GAA TTC GCT AGC CGA CGG TCA ATG CTC GTG AGC CAG TAC ACA CAA TAT ATG TGG ATC CGA GCT CCA CGT G- | KD: 1.73–21.4 nM | Prism-coupled SPR | Sandwich SPR with gold nanorod-conjugated detection aptamers | 70 aM | [65] |
| Salmonella typhimurium Outer Membrane Proteins | -TAT GGC GGC GTC ACC CGA CGG GGA CTT GAC ATT ATG ACA G- -GAG GAA AGT CTA TAG CAG AGG AGA TGT GTG AAC CGA GTA A- | Koff: 5.2 × 10−3–7.4 × 10−3 s−1 | Prism-coupled SPR | - | 3 × 104 CFU mL−1. | [69] |
| Salmonella typhimurium | -TAT GGC GGC GTC ACC CGA CGG GGA CTT GAC ATT ATG ACA G- | Glass-coupled LSPR | - | 104 CFU/mL | [24] | |
| Ochratoxin, Aflatoxin, Adenosine Triphosphate, and Potassium Ions. | -TTT TTG TGG GTA GGG GGG GTT GGA CCA CAC CAA CC- -TTT TTA ACC TGG GGG AGT ATT GCG GAG GAA GGT- -TTT TTG TTG GGC ACG TGT TGT CTC TCT GTG TCT CGT GCC CTT CGC TAG GCC CAC A- -TTT TTG ATC GGG TGT GGG TGG CGT AAA GGG AGC ATC GGA CA- | Glass-coupled LSPR | LSPR peak shift induction by berberine binding to aptamer–target aptamer G-quadruplex | 0.56–1.05 pM | [25] | |
| 25-Hydroxyvitamin D3 | -AGC AGC ACA GAG GTC ATG GGG GGT GTG ACT TTG GTG TGC CTA TGC GTG CTA CGG AA- | KD: 11 nM | Glass-coupled LSPR | - | 0.1 ng/mL | [26] |
| Whole Staphylococcus aureus Cells | -TCC CAC GAT CTC ATT AGT CTG TGG ATA AGC GTG GGA CGT CTA TGA- | KD: 82.8 nM | Glass-coupled LSPR | - | 103 CFU/mL | [23] |
| Ochratoxin | -TTT TTG ATC GGG TGT GGG TGG CGT AAA GGG AGC ATC GGA CA- | Optical Fiber-based LSPR | - | 12 pM | [70] | |
| Whole Shigella Cells | -TTT TTT TTT TTT AGT CTT TCG CTG TTG CTG CTG ATG CC- -Cy5.5—GGC ATC AGC AGC AAC AGC GAA AGA CT- | Multicore Optical Fiber-based LSPR | - | 1.56 CFU/mL | [71] | |
| Lysozyme Allergen | -GGG AAT GGA TCC ACA TCT ACG AAT TCA TCA GGG CTA AAG AG- | KD: 31–65 nM | Prism-coupled SPR | - | 2.4 nM | [66] |
| HER2 | -TCT AAA AGG ATT CTT CCC AAG GGG ATC CAA TTC AAA CAG- | KD: 6.2 nM | Optical Fiber-based SPR | Sandwich SPR with anti-HER2 antibodies as detection probes | 77.4 pM | [72] |
| MCF-7 Breast Cancer Cells | -GCA GTT GAT CCT TTG GAT ACC CTG G- | Prism-coupled SPR | Dual recognition together with folic acid-functionalized magnetic nanoparticles for binding overexpressed FA receptor. | 500 cells/mL | [73] | |
| Tyrosine Kinase 7 Expressed on Circulating Tumor Cells | -ATC TAA CTG CTG CGC CGC CGG GAA AAT ACT GTA CGG TTA GAT TTT TTT TTT- | Electrochemical LSPR | LSPR-generated hot electrons enhance electrochemical current response | 5 cells/mL | [74] | |
| Cytochrome C | -ATC GAT AAG CTT CCA GAG CCG TGT CTG GGG CCG ACC GGC GCA TTG GGT ACG TTG TTG CCG TAG AAT TCC TGC AGC C- | Prism-coupled SPR | Gold nanorods for plasmonic enhancement and RNAse H enzymatic recycling | 80 pM | [76] |
| tRADE Name/Identifier | Biomarker/Target | Biosensor Setup | Developmental Stage | Ref. |
|---|---|---|---|---|
| AptoDetect™-Lung | EGFR1, MMP7, CA6, KIT, CRP, C9, and SERPINA3 | Proteomic profiling | Commercialized | [103] |
| Apta-Beacon™ | Multiple biomarkers | Colorimetric and fluorometric endpoints | Commercialized | [104] |
| OLIGOBIND | Thrombin | Fluorometric ELISA | Commercialized | [105] |
| APTSENS | COVID-19 | Electrochemical biosensor | Commercialized | [106] |
| SOMAscan | Multiple biomarkers | Proteomic profiling | Commercialized | [107] |
| AflaSense | Aflatoxin | Fluorometry | Commercialized | [108] |
| OTA-Sense | Ochratoxin A | Sandwich fluorometry | Commercialized | [109] |
| ApollomerTM | Multiple pathogens | Portable electrochemical biosensor | Commercialized | [110] |
| Patent: CN104634754B | Terramycin | ELISA | Patent active (expected expiration: 2035) | [111] |
| Patent: US10036073B2 | Foodborne pathogens | Fluorometry | Fee-related expired patent (adjusted expiration: 2032) | [112] |
| Patent: CN113881790A | Foodborne pathogens | PCR and fluorometry | Patent active (expected expiration: 2041) | [113] |
| Patent: CN103439296B | Adenosine | SPR | Fee-related expired patent (expected expiration: 2033) | [114] |
| Patent: CN108845009B | Environmental pollutants | Photoelectrochemical sensor | Patent active (expected expiration: 2038) | [115] |
| Project ADHERE (clinicaltrials.gov ID: NCT04870671) | Tenofovir | Electrochemical aptasensor | Early Phase 1 clinical trials completed. | [116] |
| AptameX | COVID-19 | Colorimetric sensor | Commercialized | [102] |
| Identify Proteomic Biomarkers for Outcome Prediction of Lipiodol TACE Treatment (Lipiodol TACE) | Multiple biomarkers | Proteomic profiling | Observational clinical trials ongoing | Clinicaltrials.gov ID: NCT04459468 |
| Electro-Phage and Colorimetric Aptamer Sensors for Clinical Staging and Monitoring of Bladder Cancer | Multiple urinary biomarkers | Colorimetric sensor | Observational clinical trials ongoing | Clinicaltrials.gov ID: NCT02957370 |
| Non-Invasive, Highly Specific Detection of Oxytocin in Biological Fluids | Salivary oxytocin | Aptamer-based electrochemical assay | Observational clinical trials completed | Clinicaltrials.gov ID: NCT03140709 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khalid-Salako, F.; Kurt, H.; Yüce, M. Surface Plasmon Resonance Aptasensors: Emerging Design and Deployment Landscape. Biosensors 2025, 15, 359. https://doi.org/10.3390/bios15060359
Khalid-Salako F, Kurt H, Yüce M. Surface Plasmon Resonance Aptasensors: Emerging Design and Deployment Landscape. Biosensors. 2025; 15(6):359. https://doi.org/10.3390/bios15060359
Chicago/Turabian StyleKhalid-Salako, Fahd, Hasan Kurt, and Meral Yüce. 2025. "Surface Plasmon Resonance Aptasensors: Emerging Design and Deployment Landscape" Biosensors 15, no. 6: 359. https://doi.org/10.3390/bios15060359
APA StyleKhalid-Salako, F., Kurt, H., & Yüce, M. (2025). Surface Plasmon Resonance Aptasensors: Emerging Design and Deployment Landscape. Biosensors, 15(6), 359. https://doi.org/10.3390/bios15060359







