CRISPR/Cas12a-Based Biosensing: Advances in Mechanisms and Applications for Nucleic Acid Detection
Abstract
1. Introduction
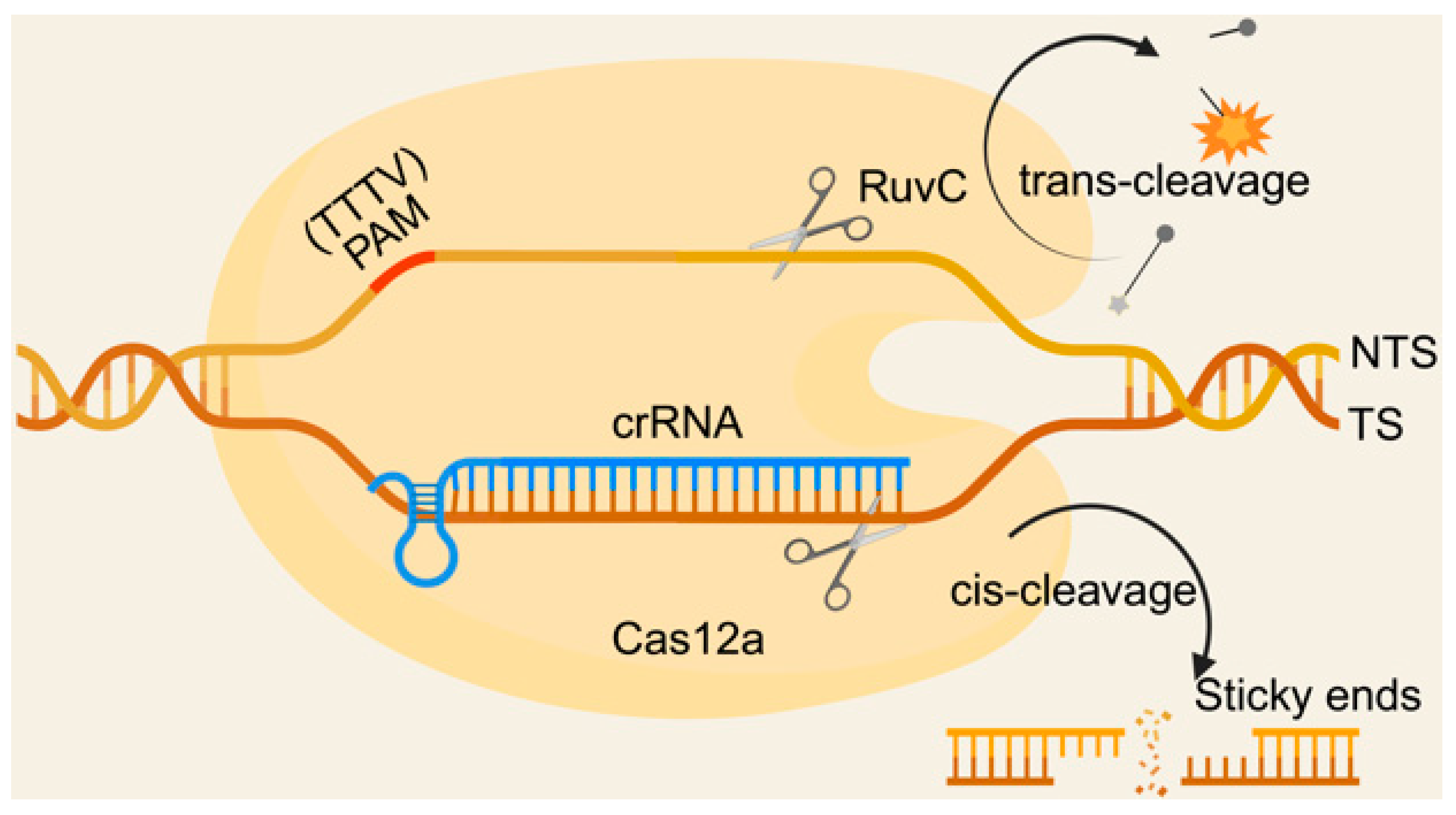
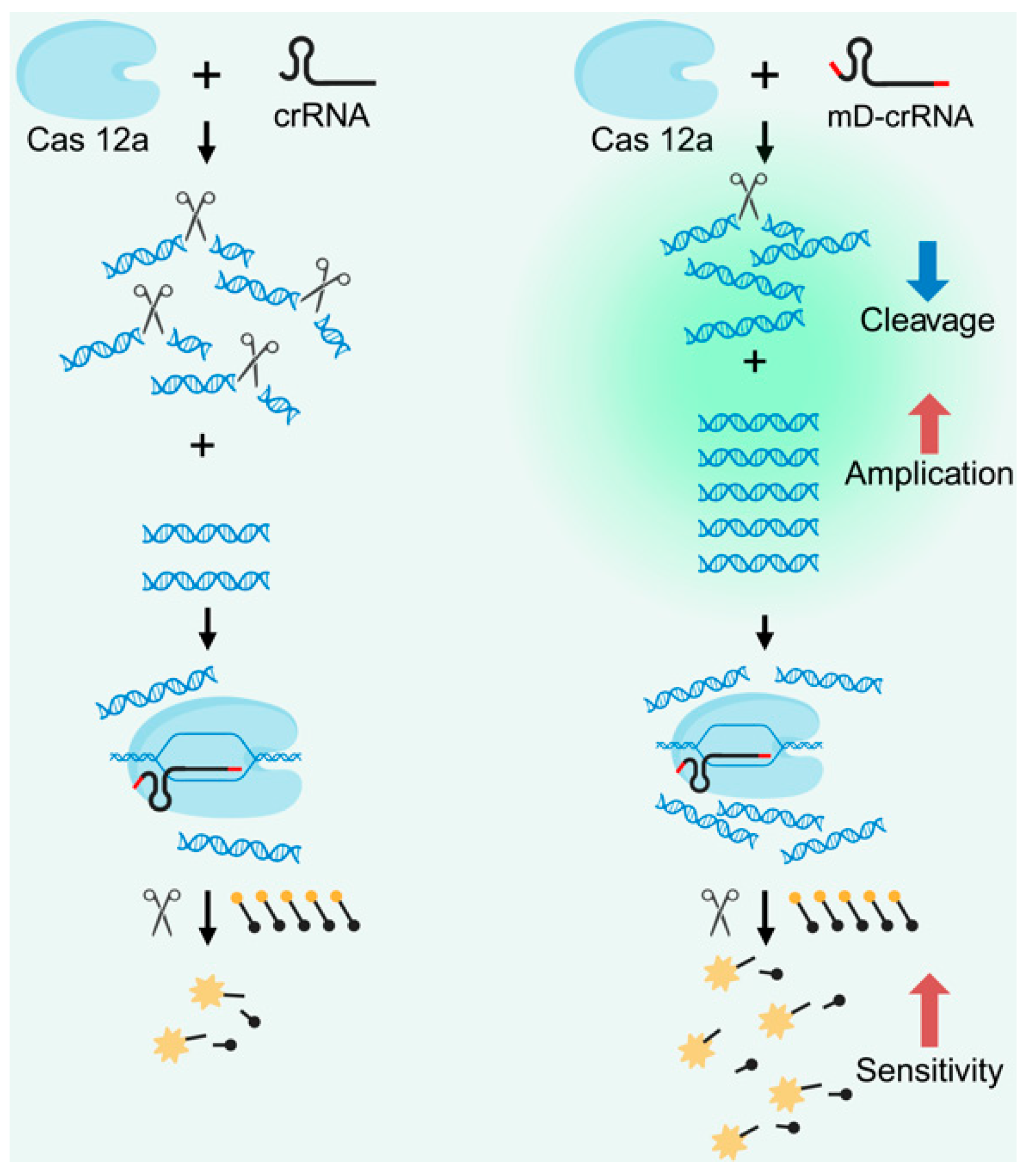
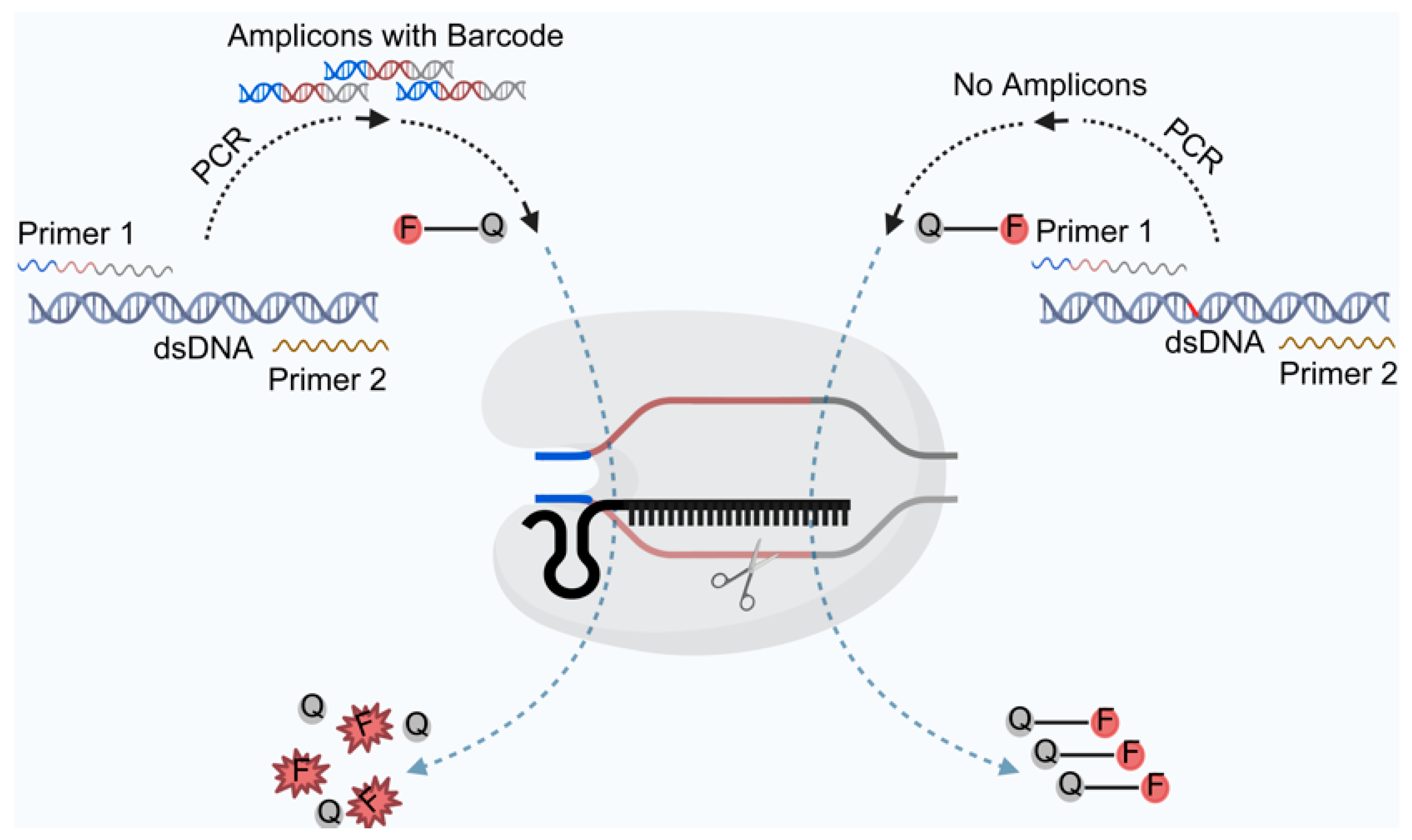
2. Principles of Cas12a Nucleic Acid Detection Technology for Biosensors
3. Advances in CRISPR/Cas12a-Based Biosensors for Nucleic Acid Detection in Disease Diagnosis
3.1. CRISPR/Cas12a-Based Biosensors for Viral Detection
3.2. CRISPR/Cas12a-Based Biosensors for Detection of Mycoplasma and Bacteria
3.3. CRISPR/Cas12a-Based Biosensors for Parasite Detection
3.4. CRISPR/Cas12a-Based Biosensors for Tumor Detection
3.5. CRISPR/Cas12a-Based Biosensors for Genetic Disease Detection
3.6. CRISPR/Cas12a-Based Biosensors for Multisystem Disease Detection
4. Innovations and Challenges of CRISPR/Cas12a-Based Biosensors in Clinical Research
5. Summary
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PCR–RFLP | Polymerase chain reaction–restriction fragment length polymorphism |
| MLPA | Multiplex ligation-dependent probe amplification |
| DHPLC | Denaturing high-performance liquid chromatography |
| qPCR | Quantitative real-time polymerase chain reaction |
| dPCR | Digital polymerase chain reaction |
| NGS | Next-generation sequencing |
| CRISPR | Clustered regularly interspaced short palindromic repeats |
| Cas | CRISPR-associated |
| PAM | Protospacer adjacent motif |
| crRNA | CRISPR RNA |
| TS | Target strand |
| NTS | Non-target strand |
| ssDNA | Single-stranded DNA |
| RPA | Recombinase polymerase amplification |
| HPV | Human papillomavirus |
| SARS-CoV-2 | Severe acute respiratory syndrome coronavirus |
| DETECTR | DNA endonuclease-targeted CRISPR trans reporter |
| RT-LAMP | Reverse transcription loop-mediated isothermal amplification |
| ASFV | African swine fever virus |
| MPXV | Mpox Virus |
| RhV | Rhinovirus |
| HAdV | Human adenovirus |
| GBS | Streptococcus agalactiae |
| IA | Influenza A |
| IB | Influenza B |
| RSV | Respiratory syncytial virus |
| RT–qPCR | Reverse transcription–polymerase chain reaction |
| HOLMES | one-HOur Low-cost Multipurpose highly Efficient System |
| ECF | East Coast fever |
| RAA | Recombinase-assisted isothermal amplification |
| NSCLC | Non-small cell lung cancer |
| MPNs | Myeloproliferative neoplasms |
| CACBA | Aptamer chemiluminescence assay |
| RCT | Rolling circle transcription |
| hOGG1 | Human 8-hydroxyguanine DNA glycosidase |
| FEN1 | Flap endonuclease 1 |
| CagA | Cytotoxin-associated gene A |
| VacA | Vacuolar cytotoxin A |
| LAMP | Loop-mediated isothermal amplification |
| LHON | Leber’s hereditary optic neuropathy |
| SMA | Spinal muscular atrophy |
| DMD | Duchenne muscular dystrophy |
| NAD+ | Nicotinamide adenine dinucleotide |
| microRNAs | miRNAs |
| LFA | Lateral flow assay |
| AcrVA5 | V-A anti-CRISPR protein 5 |
| MS | Mass spectrometry |
| PCSDA | PLA-induced Cascade strand displacement and amplification |
| MCDA | Multiple cross-displacement amplification |
| J2 | Single second-stage juvenile |
| TTSD | Toehold-triggered strand displacement |
References
- Huo, B.; Hu, Y.; Gao, Z.; Li, G. Recent advances on functional nucleic acid-based biosensors for detection of food contaminants. Talanta 2021, 222, 121565. [Google Scholar] [CrossRef]
- Smith, S.J.; Nemr, C.R.; Kelley, S.O. Chemistry-Driven Approaches for Ultrasensitive Nucleic Acid Detection. J. Am. Chem. Soc. 2017, 139, 1020–1028. [Google Scholar] [CrossRef] [PubMed]
- Nanoplasmonic acceleration of nucleic acid amplification for pathogen detection. Nat. Nanotechnol. 2023, 18, 846–847. [CrossRef]
- Yang, N.; Ji, Y.; Wang, A.; Tang, J.; Liu, S.; Zhang, X.; Xu, L.; He, Y. An integrated nucleic acid detection method based on a microfluidic chip for collection and culture of rice false smut spores. Lab. Chip. 2022, 22, 4894–4904. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Pan, L.; Ma, X.; Li, T.; Wang, F.; Yang, D.; Li, M.; Wang, P. Detection of SARS-CoV-2 RNA with a plasmonic chiral biosensor. Biosens. Bioelectron. 2023, 237, 115526. [Google Scholar] [CrossRef]
- Trujillo-González, A.; Edmunds, R.C.; Becker, J.A.; Hutson, K.S. Parasite detection in the ornamental fish trade using environmental DNA. Sci. Rep. 2019, 9, 5173. [Google Scholar] [CrossRef]
- Das, S.; Bano, S.; Kapse, P.; Kundu, G.C. CRISPR based therapeutics: A new paradigm in cancer precision medicine. Mol. Cancer 2022, 21, 85. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Lee, S.R. Rapid Detection of Red Rot Disease Pathogens (Pythium chondricola and P. porphyrae) in Pyropia yezoensis (Rhodophyta) with PCR-RFLP. Plant. Dis. 2022, 106, 30–33. [Google Scholar] [CrossRef]
- Lindstedt, K.; Buczek, D.; Pedersen, T.; Hjerde, E.; Raffelsberger, N.; Suzuki, Y.; Brisse, S.; Holt, K.; Samuelsen, Ø.; Sundsfjord, A. Detection of Klebsiella pneumoniae human gut carriage: A comparison of culture, qPCR, and whole metagenomic sequencing methods. Gut Microbes 2022, 14, 2118500. [Google Scholar] [CrossRef]
- Ahmed, W.; Bivins, A.; Metcalfe, S.; Smith, W.J.M.; Ziels, R.; Korajkic, A.; McMinn, B.; Graber, T.E.; Simpson, S.L. RT-qPCR and ATOPlex sequencing for the sensitive detection of SARS-CoV-2 RNA for wastewater surveillance. Water Res. 2022, 220, 118621. [Google Scholar] [CrossRef]
- Pomari, E.; Piubelli, C.; Perandin, F.; Bisoffi, Z. Digital PCR: A new technology for diagnosis of parasitic infections. Clin. Microbiol. Infect. 2019, 25, 1510–1516. [Google Scholar] [CrossRef]
- Coakley, M.; Villacampa, G.; Sritharan, P.; Swift, C.; Dunne, K.; Kilburn, L.; Goddard, K.; Pipinikas, C.; Rojas, P.; Emmett, W.; et al. Comparison of Circulating Tumor DNA Assays for Molecular Residual Disease Detection in Early-Stage Triple-Negative Breast Cancer. Clin. Cancer Res. 2024, 30, 895–903. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, A.; Miranda, S.; Azevedo, N.F.; Cerqueira, L.; Azevedo, A.S. Imaging biofilms using fluorescence in situ hybridization: Seeing is believing. Front. Cell. Infect. Microbiol. 2023, 13, 1195803. [Google Scholar] [CrossRef] [PubMed]
- Sesboue, C.; Galtier, J.; Jeanneau, M.; Chauvel, A.; Laharanne, E.; Amintas, S.; Merlio, J.P.; Bouabdallah, K.; Gros, F.X.; de Leval, L.; et al. Combined Reverse-Transcriptase Multiplex Ligation-Dependent Probe Amplification and Next-Generation Sequencing Analyses to Assign Unclassified BCL2(-)/BCL6(-) Nonrearranged Small B-Cell Lymphoid Neoplasms as Follicular or Nodal Marginal Zone Lymphoma. Mod. Pathol. 2023, 36, 100043. [Google Scholar] [CrossRef] [PubMed]
- Gauthier, N.P.G.; Chorlton, S.D.; Krajden, M.; Manges, A.R. Agnostic Sequencing for Detection of Viral Pathogens. Clin. Microbiol. Rev. 2023, 36, e0011922. [Google Scholar] [CrossRef]
- Barrangou, R.; Marraffini, L.A. CRISPR-Cas systems: Prokaryotes upgrade to adaptive immunity. Mol. Cell. 2014, 54, 234–244. [Google Scholar] [CrossRef] [PubMed]
- Horvath, P.; Barrangou, R. CRISPR/Cas, the immune system of bacteria and archaea. Science 2010, 327, 167–170. [Google Scholar] [CrossRef]
- Makarova, K.S.; Haft, D.H.; Barrangou, R.; Brouns, S.J.; Charpentier, E.; Horvath, P.; Moineau, S.; Mojica, F.J.; Wolf, Y.I.; Yakunin, A.F.; et al. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 2011, 9, 467–477. [Google Scholar] [CrossRef]
- Makarova, K.S.; Wolf, Y.I.; Alkhnbashi, O.S.; Costa, F.; Shah, S.A.; Saunders, S.J.; Barrangou, R.; Brouns, S.J.; Charpentier, E.; Haft, D.H.; et al. An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 2015, 13, 722–736. [Google Scholar] [CrossRef]
- Zhou, J.; Liu, G.; Zhao, Y.; Zhang, R.; Tang, X.; Li, L.; Jia, X.; Guo, Y.; Wu, Y.; Han, Y.; et al. An efficient CRISPR-Cas12a promoter editing system for crop improvement. Nat. Plants 2023, 9, 588–604. [Google Scholar] [CrossRef]
- Oppel, F.; Schürmann, M.; Goon, P.; Albers, A.E.; Sudhoff, H. Specific Targeting of Oncogenes Using CRISPR Technology. Cancer Res. 2018, 78, 5506–5512. [Google Scholar] [CrossRef] [PubMed]
- Enghiad, B.; Huang, C.; Guo, F.; Jiang, G.; Wang, B.; Tabatabaei, S.K.; Martin, T.A.; Zhao, H. Cas12a-assisted precise targeted cloning using in vivo Cre-lox recombination. Nat. Commun. 2021, 12, 1171. [Google Scholar] [CrossRef]
- Talwar, C.S.; Park, K.H.; Ahn, W.C.; Kim, Y.S.; Kwon, O.S.; Yong, D.; Kang, T.; Woo, E. Detection of Infectious Viruses Using CRISPR-Cas12-Based Assay. Biosensors 2021, 11, 301. [Google Scholar] [CrossRef]
- Kostyusheva, A.; Brezgin, S.; Babin, Y.; Vasilyeva, I.; Glebe, D.; Kostyushev, D.; Chulanov, V. CRISPR-Cas systems for diagnosing infectious diseases. Methods 2022, 203, 431–446. [Google Scholar] [CrossRef]
- Li, X.; Chen, X.; Mao, M.; Peng, C.; Wang, Z. Accelerated CRISPR/Cas12a-based small molecule detection using bivalent aptamer. Biosens. Bioelectron. 2022, 217, 114725. [Google Scholar] [CrossRef]
- Hu, M.; Cheng, X.; Wu, T. Modular CRISPR/Cas12a synergistic activation platform for detection and logic operations. Nucleic Acids Res. 2024, 52, 7384–7396. [Google Scholar] [CrossRef]
- Yue, Y.; Wang, S.; Jin, Q.; An, N.; Wu, L.; Huang, H. A triple amplification strategy using GR-5 DNAzyme as a signal medium for ultrasensitive detection of trace Pb(2+) based on CRISPR/Cas12a empowered electrochemical biosensor. Anal. Chim. Acta 2023, 1263, 341241. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zong, N.; Ye, F.; Mei, Y.; Qu, J.; Jiang, X. Dual-CRISPR/Cas12a-Assisted RT-RAA for Ultrasensitive SARS-CoV-2 Detection on Automated Centrifugal Microfluidics. Anal. Chem. 2022, 94, 9603–9609. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Liu, T.; Guan, L.; Xu, Z.; Xiong, T.; Zhang, Y.; Song, J.; Liu, X.; Yang, Y.; Hao, X. A highly sensitive Lock-Cas12a biosensor for detection and imaging of miRNA-21 in breast cancer cells. Talanta 2024, 273, 125938. [Google Scholar] [CrossRef]
- Zhang, M.; Liu, C.; Shi, Y.; Wu, J.; Wu, J.; Chen, H. Selective endpoint visualized detection of Vibrio parahaemolyticus with CRISPR/Cas12a assisted PCR using thermal cycler for on-site application. Talanta 2020, 214, 120818. [Google Scholar] [CrossRef]
- Wu, H.; He, J.S.; Zhang, F.; Ping, J.; Wu, J. Contamination-free visual detection of CaMV35S promoter amplicon using CRISPR/Cas12a coupled with a designed reaction vessel: Rapid, specific and sensitive. Anal. Chim. Acta 2020, 1096, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Fang, R.; Li, Y.; Deng, F.; Liu, X.; Yang, D. CRISPR/Cas12a-powered CLASA towards OTA ultrasensitive detection in cereal samples. Microchem. J. 2024, 196, 109691. [Google Scholar] [CrossRef]
- Yu, S.; Lei, X.; Qu, C. MicroRNA Sensors Based on CRISPR/Cas12a Technologies: Evolution From Indirect to Direct Detection. Crit. Rev. Anal. Chem. 2024, 15, 1–17. [Google Scholar] [CrossRef]
- Li, X.; Dang, Z.; Tang, W.; Zhang, H.; Shao, J.; Jiang, R.; Zhang, X.; Huang, F. Detection of Parasites in the Field: The Ever-Innovating CRISPR/Cas12a. Biosensors 2024, 14, 145. [Google Scholar] [CrossRef] [PubMed]
- Lei, X.; Cao, S.; Liu, T.; Wu, Y.; Yu, S. Non-canonical CRISPR/Cas12a-based technology: A novel horizon for biosensing in nucleic acid detection. Talanta 2024, 271, 125663. [Google Scholar] [CrossRef]
- Swarts, D.C.; van der Oost, J.; Jinek, M. Structural Basis for Guide RNA Processing and Seed-Dependent DNA Targeting by CRISPR-Cas12a. Mol. Cell. 2017, 66, 221–233.e224. [Google Scholar] [CrossRef]
- Swarts, D.C.; Jinek, M. Mechanistic Insights into the cis- and trans-Acting DNase Activities of Cas12a. Mol. Cell. 2019, 73, 589–600.e584. [Google Scholar] [CrossRef] [PubMed]
- Yamano, T.; Nishimasu, H.; Zetsche, B.; Hirano, H.; Slaymaker, I.M.; Li, Y.; Fedorova, I.; Nakane, T.; Makarova, K.S.; Koonin, E.V.; et al. Crystal Structure of Cpf1 in Complex with Guide RNA and Target DNA. Cell 2016, 165, 949–962. [Google Scholar] [CrossRef]
- Dai, Y.; Somoza, R.A.; Wang, L.; Welter, J.F.; Li, Y.; Caplan, A.I.; Liu, C.C. Exploring the Trans-Cleavage Activity of CRISPR-Cas12a (cpf1) for the Development of a Universal Electrochemical Biosensor. Angew. Chem. Int. Ed. Engl. 2019, 58, 17399–17405. [Google Scholar] [CrossRef]
- Liu, N.; Liu, R.; Zhang, J. CRISPR-Cas12a-mediated label-free electrochemical aptamer-based sensor for SARS-CoV-2 antigen detection. Bioelectrochemistry 2022, 146, 108105. [Google Scholar] [CrossRef]
- Zhou, M.; Zhang, C.; Chen, M.; Hu, Z.; Li, M.; Li, Z.; Wu, L.; Liang, D. A protospacer adjacent motif-free, multiplexed, and quantitative nucleic acid detection platform with barcode-based Cas12a activity. MedComm 2023, 4, e310. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.S.; Ma, E.; Harrington, L.B.; Da Costa, M.; Tian, X.; Palefsky, J.M.; Doudna, J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 2018, 360, 436–439. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Tian, D.; Liu, Y.; Lin, Z.; Lyon, C.J.; Lai, W.; Fusco, D.; Drouin, A.; Yin, X.; Hu, T.; et al. Ultra-sensitive and high-throughput CRISPR-p owered COVID-19 diagnosis. Biosens. Bioelectron. 2020, 164, 112316. [Google Scholar] [CrossRef]
- Chen, Y.; Shi, Y.; Chen, Y.; Yang, Z.; Wu, H.; Zhou, Z.; Li, J.; Ping, J.; He, L.; Shen, H.; et al. Contamination-free visual detection of SARS-CoV-2 with CRISPR/Cas12a: A promising method in the point-of-care detection. Biosens. Bioelectron. 2020, 169, 112642. [Google Scholar] [CrossRef]
- Wang, R.; Qian, C.; Pang, Y.; Li, M.; Yang, Y.; Ma, H.; Zhao, M.; Qian, F.; Yu, H.; Liu, Z.; et al. opvCRISPR: One-pot visual RT-LAMP-CRISPR platform for SARS-cov-2 detection. Biosens. Bioelectron. 2021, 172, 112766. [Google Scholar] [CrossRef]
- Pang, B.; Xu, J.; Liu, Y.; Peng, H.; Feng, W.; Cao, Y.; Wu, J.; Xiao, H.; Pabbaraju, K.; Tipples, G.; et al. Isothermal Amplification and Ambient Visualization in a Single Tube for the Detection of SARS-CoV-2 Using Loop-Mediated Amplification and CRISPR Technology. Anal. Chem. 2020, 92, 16204–16212. [Google Scholar] [CrossRef]
- Hu, M.; Qiu, Z.; Bi, Z.; Tian, T.; Jiang, Y.; Zhou, X. Photocontrolled crRNA activation enables robust CRISPR-Cas12a diagnostics. Proc. Natl. Acad. Sci. USA 2022, 119, e2202034119. [Google Scholar] [CrossRef]
- Sun, Q.; Lin, H.; Li, Y.; Yuan, L.; Li, B.; Ma, Y.; Wang, H.; Deng, X.; Chen, H.; Tang, S. A photocontrolled one-pot isothermal amplification and CRISPR-Cas12a assay for rapid detection of SARS-CoV-2 Omicron variants. Microbiol. Spectr. 2024, 12, e0364523. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Q.; Zhou, M.; Deng, W.; Gao, Q.; Li, Z.; Wu, L.; Liang, D. Sensitive and visual detection of SARS-CoV-2 using RPA-Cas12a one-step assay with ssDNA-modified crRNA. Anal. Chim. Acta 2024, 1309, 342693. [Google Scholar] [CrossRef]
- Luan, H.; Wang, S.; Ju, L.; Liu, T.; Shi, H.; Ge, S.; Jiang, S.; Wu, J.; Peng, J. KP177R-based visual assay integrating RPA and CRISPR/Cas12a for the detection of African swine fever virus. Front. Immunol. 2024, 15, 1358960. [Google Scholar] [CrossRef]
- Wang, H.; Su, A.; Bao, C.; Liang, C.; Xu, W.; Chang, J.; Xu, S. A CRISPR/Cas12a-SERS platform for amplification-free detection of African swine fever virus genes. Talanta 2024, 267, 125225. [Google Scholar] [CrossRef] [PubMed]
- Qian, S.; Chen, Y.; Peng, C.; Wang, X.; Wu, H.; Che, Y.; Wang, H.; Xu, J.; Wu, J. Dipstick-based rapid nucleic acids purification and CRISPR/Cas12a-mediated isothermal amplification for visual detection of African swine fever virus. Talanta 2022, 242, 123294. [Google Scholar] [CrossRef]
- Han, C.; Liu, Q.; Luo, X.; Zhao, J.; Zhang, Z.; He, J.; Ge, F.; Ding, W.; Luo, Z.; Jia, C.; et al. Development of a CRISPR/Cas12a-mediated aptasensor for Mpox virus antigen detection. Biosens. Bioelectron. 2024, 257, 116313. [Google Scholar] [CrossRef] [PubMed]
- Low, S.J.; O’Neill, M.T.; Kerry, W.J.; Krysiak, M.; Papadakis, G.; Whitehead, L.W.; Savic, I.; Prestedge, J.; Williams, L.; Cooney, J.P.; et al. Rapid detection of monkeypox virus using a CRISPR-Cas12a mediated assay: A laboratory validation and evaluation study. Lancet Microbe 2023, 4, e800–e810. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Xu, L.; Liu, Q.; Cao, Y.; Yang, K.; Song, X.; Shao, Y.; Tu, J.; Qi, K. Enzymatic recombinase amplification coupled with CRISPR-Cas12a for ultrasensitive, rapid, and specific Porcine circovirus 3 detection. Mol. Cell. Probes 2021, 59, 101763. [Google Scholar] [CrossRef]
- Dou, B.; Zhang, Y.; Gao, H.; Zhang, S.; Zheng, J.; Lu, X.; Liu, S.; Zhou, H.; Hun, X. CRISPR/Cas12a-based MUSCA-PEC strategy for HSV-1 assay. Anal. Chim. Acta 2023, 1250, 340955. [Google Scholar] [CrossRef]
- Wang, H.; Li, H.; Tang, B.; Ye, C.; Han, M.; Teng, L.; Yue, M.; Li, Y. Fast and sensitive differential diagnosis of pseudorabies virus-infected versus pseudorabies virus-vaccinated swine using CRISPR-Cas12a. Microbiol. Spectr. 2024, 12, e0261723. [Google Scholar] [CrossRef]
- Tan, Q.; Shi, Y.; Duan, C.; Li, Q.; Gong, T.; Li, S.; Duan, X.; Xie, H.; Li, Y.; Chen, L. Simple, sensitive, and visual detection of 12 respiratory pathogens with one-pot-RPA-CRISPR/Cas12a assay. J. Med. Virol. 2024, 96, e29624. [Google Scholar] [CrossRef]
- Zeng, Q.; Zhou, M.; Hu, Z.; Deng, W.; Li, Z.; Wu, L.; Liang, D. Rapid and sensitive Cas12a-based one-step nucleic acid detection with ssDNA-modified crRNA. Anal. Chim. Acta 2023, 1276, 341622. [Google Scholar] [CrossRef]
- Shen, J.; Chen, Z.; Xie, R.; Li, J.; Liu, C.; He, Y.; Ma, X.; Yang, H.; Xie, Z. CRISPR/Cas12a-Assisted isothermal amplification for rapid and specific diagnosis of respiratory virus on an microfluidic platform. Biosens. Bioelectron. 2023, 237, 115523. [Google Scholar] [CrossRef]
- Yang, J.; Barua, N.; Rahman, M.N.; Li, C.; Lo, N.; Yeong, K.Y.; Tsang, T.F.; Yang, X.; Cheung, Y.Y.; Tsang, A.K.L.; et al. Rapid SARS-CoV-2 Variants Enzymatic Detection (SAVED) by CRISPR-Cas12a. Microbiol. Spectr. 2022, 10, e0326022. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Qiu, Z.; Jiang, Y.; Zhu, D.; Zhou, X. Exploiting the orthogonal CRISPR-Cas12a/Cas13a trans-cleavage for dual-gene virus detection using a handheld device. Biosens. Bioelectron. 2022, 196, 113701. [Google Scholar] [CrossRef]
- Hu, T.; Ke, X.; Li, W.; Lin, Y.; Liang, A.; Ou, Y.; Chen, C. CRISPR/Cas12a-Enabled Multiplex Biosensing Strategy Via an Affordable and Visual Nylon Membrane Readout. Adv. Sci. 2023, 10, e2204689. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, N.; Zhang, L.; Lu, Y.; Shen, M.; Zhang, Y.; Feng, L.; Jing, J.; Cheng, J.; Xu, Y. A Wax Interface-Enabled One-Pot Multiplexed Nucleic Acid Testing Platform for Rapid and Sensitive Detection of Viruses and Variants. Small Methods 2024, 8, e2400030. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.N.; Ma, A.X.; Wang, D.X.; Dai, Z.Q.; Wu, S.L.; Lu, S.; Zhu, L.N.; Jiang, H.X.; Pang, D.W.; Kong, D.M. Allosteric Activator-Regulated CRISPR/Cas12a System Enables Biosensing and Imaging of Intracellular Endogenous and Exogenous Targets. Anal. Chem. 2024, 96, 6426–6435. [Google Scholar] [CrossRef]
- Yu, C.; Chen, H.-J.; Liu, H.-Y.; Ning, D.; Wang, L.; Shi, X.-H.; Wang, Z.-G.; Pang, D.-W.; Liu, S.-L. Dual-lock-and-key virus-mimicking nanoprobes for ultra-high accurate and sensitive imaging of viral infections in vivo. Nano Today 2024, 59, 102527. [Google Scholar] [CrossRef]
- Li, S.Y.; Cheng, Q.X.; Wang, J.M.; Li, X.Y.; Zhang, Z.L.; Gao, S.; Cao, R.B.; Zhao, G.P.; Wang, J. CRISPR-Cas12a-assisted nucleic acid detection. Cell. Discov. 2018, 4, 20. [Google Scholar] [CrossRef]
- Wang, B.; Wang, R.; Wang, D.; Wu, J.; Li, J.; Wang, J.; Liu, H.; Wang, Y. Cas12aVDet: A CRISPR/Cas12a-Based Platform for Rapid and Visual Nucleic Acid Detection. Anal. Chem. 2019, 91, 12156–12161. [Google Scholar] [CrossRef]
- Deng, Z.; Hu, H.; Tang, D.; Liang, J.; Su, X.; Jiang, T.; Hu, X.; Ying, W.; Zhen, D.; Xiao, X.; et al. Ultrasensitive, Specific, and Rapid Detection of Mycoplasma pneumoniae Using the ERA/CRISPR-Cas12a Dual System. Front. Microbiol. 2022, 13, 811768. [Google Scholar] [CrossRef]
- Jia, N.; Zhou, J.; Xiao, F.; Zheng, B.; Huang, X.; Sun, C.; Fu, J.; Xu, Z.; Chen, M.; Wang, Y. A CRISPR-Cas12a-Based platform for ultrasensitive, rapid, and highly specific detection of Mycoplasma pneumonia in clinical application. Front. Bioeng. Biotechnol. 2023, 11, 1022066. [Google Scholar] [CrossRef]
- Xia, X.; Ma, B.; Zhang, T.; Lu, Y.; Khan, M.R.; Hu, Y.; Lei, C.; Deng, S.; He, Q.; He, G.; et al. G-Quadruplex-Probing CRISPR-Cas12 Assay for Label-Free Analysis of Foodborne Pathogens and Their Colonization In Vivo. ACS Sens. 2021, 6, 3295–3302. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Ye, Q.; Chen, M.; Zhou, B.; Zhang, J.; Pang, R.; Xue, L.; Wang, J.; Zeng, H.; Wu, S.; et al. An ultrasensitive CRISPR/Cas12a based electrochemical biosensor for Listeria monocytogenes detection. Biosens. Bioelectron. 2021, 179, 113073. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, J.; Li, S.; Zhu, X.; Wang, X.; Huang, J.; Yang, X.; Tai, J. LAMP-CRISPR-Cas12-based diagnostic platform for detection of Mycobacterium tuberculosis complex using real-time fluorescence or lateral flow test. Mikrochim. Acta 2021, 188, 347. [Google Scholar] [CrossRef]
- Lin, C.; Zhou, J.; Gao, N.; Liu, R.; Li, G.; Wang, J.; Lu, G.; Shen, J. Establishing a pulmonary aspergillus fumigatus infection diagnostic platform based on RPA-CRISPR-Cas12a. World J. Microbiol. Biotechnol. 2024, 40, 116. [Google Scholar] [CrossRef]
- Pakdeerat, S.; Boonklang, P.; Angchagun, K.; Chomkatekaew, C.; Apichaidejudom, N.; Dokket, Y.; Faosap, A.; Wongsuwan, G.; Wuthiekanun, V.; Aramrueung, P.; et al. Benchmarking CRISPR-BP34 for point-of-care melioidosis detection in low-income and middle-income countries: A molecular diagnostics study. Lancet Microbe 2024, 5, e379–e389. [Google Scholar] [CrossRef]
- Muriuki, R.; Ndichu, M.; Githigia, S.; Svitek, N. Novel CRISPR-Cas-powered pen-side test for East Coast fever. Int. J. Parasitol. 2024, 54, 507–521. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, X.; Li, L.; Cao, L.; Zhao, Z.; Huang, T.; Li, J.; Zhang, X.; Cao, S.; Zhang, N.; et al. Establishment of an ultrasensitive and visual detection platform for Neospora caninum based-on the RPA-CRISPR/Cas12a system. Talanta 2024, 269, 125413. [Google Scholar] [CrossRef]
- Sun, H.; Fan, J.; Chu, H.; Gao, Y.; Fang, J.; Wu, Q.; Ding, H.; Zhuo, X.; Kong, Q.; Lv, H.; et al. RPA-CRISPR/Cas12a-LFA combined with a digital visualization instrument to detect Toxoplasma gondii in stray dogs and cats in Zhejiang province, China. Microbiol. Spectr. 2024, 12, e0399823. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, H.; Chen, X.; Wang, L.; Abulaizi, W.; Yang, Y.; Li, B.; Wang, C.; Bai, X. Agarose Hydrogel-Boosted One-Tube RPA-CRISPR/Cas12a Assay for Robust Point-of-Care Detection of Zoonotic Nematode Anisakis. J. Agric. Food Chem. 2024, 72, 8257–8268. [Google Scholar] [CrossRef]
- Sutipatanasomboon, A.; Wongsantichon, J.; Sakdee, S.; Naksith, P.; Watthanadirek, A.; Anuracpreeda, P.; Blacksell, S.D.; Saisawang, C. RPA-CRISPR/Cas12a assay for the diagnosis of bovine Anaplasma marginale infection. Sci. Rep. 2024, 14, 7820. [Google Scholar] [CrossRef]
- Phuphisut, O.; Poodeepiyasawat, A.; Yoonuan, T.; Watthanakulpanich, D.; Thawornkuno, C.; Reamtong, O.; Sato, M.; Adisakwattana, P. Ov-RPA-CRISPR/Cas12a assay for the detection of Opisthorchis viverrini infection in field-collected human feces. Parasit. Vectors 2024, 17, 80. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Li, L.; Li, J.; Li, X.; Li, S.; Wang, X.; Zhang, N.; Yu, Y.; Zhang, X.; Zhao, Z.; et al. Rapid, sensitive, and visual detection of Clonorchis sinensis with an RPA-CRISPR/Cas12a-based dual readout portable platform. Int. J. Biol. Macromol. 2023, 249, 125967. [Google Scholar] [CrossRef] [PubMed]
- Shao, H.; Jian, J.; Peng, D.; Yao, K.; Abdulsalam, S.; Huang, W.; Kong, L.; Li, C.; Peng, H. Recombinase Polymerase Amplification Coupled with CRISPR-Cas12a Technology for Rapid and Highly Sensitive Detection of Heterodera avenae and Heterodera filipjevi. Plant Dis. 2023, 107, 1365–1376. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Li, J.; Liu, Y.; Cheng, W.; Huang, H.; Liang, X.; Huang, W.; Lin, L.; Zheng, Y.; Chen, W.; et al. Rapid and Ultrasensitive Detection of Plasmodium spp. Parasites via the RPA-CRISPR/Cas12a Platform. ACS Infect. Dis. 2023, 9, 1534–1545. [Google Scholar] [CrossRef]
- Cherkaoui, D.; Mesquita, S.G.; Huang, D.; Lugli, E.B.; Webster, B.L.; McKendry, R.A. CRISPR-assisted test for Schistosoma haematobium. Sci. Rep. 2023, 13, 4990. [Google Scholar] [CrossRef]
- Dueñas, E.; Nakamoto, J.A.; Cabrera-Sosa, L.; Huaihua, P.; Cruz, M.; Arévalo, J.; Milón, P.; Adaui, V. Novel CRISPR-based detection of Leishmania species. Front. Microbiol. 2022, 13, 958693. [Google Scholar] [CrossRef]
- Xu, S.; Wang, X.; Wu, C.; Zhu, X.; Deng, X.; Wu, Y.; Liu, M.; Huang, X.; Wu, L.; Huang, H. MscI restriction enzyme cooperating recombinase-aided isothermal amplification for the ultrasensitive and rapid detection of low-abundance EGFR mutations on microfluidic chip. Biosens. Bioelectron. 2024, 247, 115925. [Google Scholar] [CrossRef]
- Chen, M.; Zhang, C.; Hu, Z.; Li, Z.; Li, M.; Wu, L.; Zhou, M.; Liang, D. CRISPR/Cas12a-Based Ultrasensitive and Rapid Detection of JAK2 V617F Somatic Mutation in Myeloproliferative Neoplasms. Biosensors 2021, 11, 247. [Google Scholar] [CrossRef]
- Guan, X.; Zhao, J.; Sha, Z.; Liang, Y.; Huang, J.; Zhang, J.; Sun, S. CRISPR/Cas12a and aptamer-chemiluminescence based analysis for the relative abundance determination of tumor-related protein positive exosomes for breast cancer diagnosis. Biosens. Bioelectron. 2024, 259, 116380. [Google Scholar] [CrossRef]
- Zhu, F.; Yu, H.; Zhao, Q. CRISPR/Cas12a-Amplified Aptamer Switch Microplate Assay for Small Molecules. Anal. Chem. 2024, 96, 6853–6859. [Google Scholar] [CrossRef]
- Wei, G.; Peng, Z.; Liu, J.; Yang, K.; Zhao, C.; Xie, W.; Huang, T.; Liu, J.; Li, J.; An, G. Accurate Identification and Early Diagnosis of Osteosarcoma through CRISPR-Cas12a-Based Average Telomerase Activity Detection. ACS Synth. Biol. 2021, 10, 2409–2416. [Google Scholar] [CrossRef] [PubMed]
- Habimana, J.D.; Mukama, O.; Chen, G.; Chen, M.; Amissah, O.B.; Wang, L.; Liu, Y.; Sun, Y.; Li, A.L.; Deng, S.; et al. Harnessing enhanced CRISPR/Cas12a trans-cleavage activity with extended reporters and reductants for early diagnosis of Helicobacter pylori, the causative agent of peptic ulcers and stomach cancer. Biosens. Bioelectron. 2023, 222, 114939. [Google Scholar] [CrossRef]
- Jiang, C.; Zheng, X.; Lin, L.; Li, X.; Li, X.; Liao, Y.; Jia, W.; Shu, B. CRISPR Cas12a-mediated amplification-free digital DNA assay improves the diagnosis and surveillance of Nasopharyngeal carcinoma. Biosens. Bioelectron. 2023, 237, 115546. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, Y.; Wan, Y.; Li, M.; Xu, J.; Wang, Q.; Wu, D. Stimuli-responsive incremental DNA machine auto-catalyzed CRISPR-Cas12a feedback amplification permits ultrasensitive molecular diagnosis of esophageal cancer-related microRNA. Talanta 2024, 271, 125675. [Google Scholar] [CrossRef]
- Gao, Y.; Zhang, X.; Wang, X.; Sun, R.; Li, Y.; Li, J.; Quan, W.; Yao, Y.; Hou, Y.; Li, D.; et al. The clinical value of rapidly detecting urinary exosomal lncRNA RMRP in bladder cancer with an RT-RAA-CRISPR/Cas12a method. Clin. Chim. Acta 2024, 562, 119855. [Google Scholar] [CrossRef]
- Luo, B.; Zhou, J.; Zhan, X.; Ying, B.; Lan, F.; Wu, Y. Visual and colorimetric detection of microRNA in clinical samples based on strand displacement amplification and nanozyme-mediated CRISPR-Cas12a system. Talanta 2024, 277, 126310. [Google Scholar] [CrossRef]
- Luo, B.; Zhou, J.; Zhan, X.; Ying, B.; Lan, F.; Wu, Y. Smartphone-Based Free-to-Total Prostate Specific Antigen Ratio Detection System Using a Colorimetric Reaction Integrated with Proximity-Induced Bio-Barcode and CRISPR/Cas12a Assay. Small 2024, 20, e2310212. [Google Scholar] [CrossRef]
- Wan, X.; Chen, J.; Wu, Y.; Chen, Z.; Liu, Y.; Li, T.; Sun, J.; Zhang, T.; Zhou, F.; Huang, X.; et al. Rapid and Sensitive Diagnosis of Leber Hereditary Optic Neuropathy Variants Using CRISPR/Cas12a Detection. J. Mol. Diagn. 2023, 25, 540–554. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Li, Z.; Chen, M.; Hu, Z.; Wu, L.; Zhou, M.; Liang, D. Cas12a and Lateral Flow Strip-Based Test for Rapid and Ultrasensitive Detection of Spinal Muscular Atrophy. Biosensors 2021, 11, 154. [Google Scholar] [CrossRef]
- Mu, X.; Li, J.; Xiao, S.; Huang, Y.; Zhao, S.; Tian, J. CRISPR/Cas12a-mediated DNA-AgNC label-free logical gate for multiple microRNAs’ assay. Mikrochim. Acta 2024, 191, 376. [Google Scholar] [CrossRef]
- Zhuang, S.; Hu, T.; Zhou, X.; Zhou, H.; He, S.; Li, J.; Qiu, L.; Zhang, Y.; Xu, Y.; Pei, H.; et al. meHOLMES: A CRISPR-cas12a-based method for rapid detection of DNA methylation in a sequence-independent manner. Heliyon 2024, 10, e24574. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, S.; Hu, T.; Zhou, H.; He, S.; Li, J.; Zhang, Y.; Gu, D.; Xu, Y.; Chen, Y.; Wang, J. CRISPR-HOLMES-based NAD(+) detection. Front. Bioeng. Biotechnol. 2024, 12, 1355640. [Google Scholar] [CrossRef] [PubMed]
- Diener, C.; Keller, A.; Meese, E. Emerging concepts of miRNA therapeutics: From cells to clinic. Trends Genet. 2022, 38, 613–626. [Google Scholar] [CrossRef]
- Jones, B.T.; Han, J.; Zhang, H.; Hammer, R.E.; Evers, B.M.; Rakheja, D.; Acharya, A.; Mendell, J.T. Target-directed microRNA degradation regulates developmental microRNA expression and embryonic growth in mammals. Genes Dev. 2023, 37, 661–674. [Google Scholar] [CrossRef] [PubMed]
- Mulrane, L.; McGee, S.F.; Gallagher, W.M.; O’Connor, D.P. miRNA dysregulation in breast cancer. Cancer Res. 2013, 73, 6554–6562. [Google Scholar] [CrossRef]
- Shukla, K.K.; Misra, S.; Pareek, P.; Mishra, V.; Singhal, B.; Sharma, P. Recent scenario of microRNA as diagnostic and prognostic biomarkers of prostate cancer. Urol. Oncol. 2017, 35, 92–101. [Google Scholar] [CrossRef]
- Zhang, H.C.; Du, Y.; Chen, L.; Yuan, Z.Q.; Cheng, Y. MicroRNA schizophrenia: Etiology, biomarkers and therapeutic targets. Neurosci. Biobehav. Rev. 2023, 146, 105064. [Google Scholar] [CrossRef]
- Ma, C.; Yang, Z.; Wang, J.; She, H.; Tan, L.; Ye, Q.; Wang, F.; Feng, X.; Mo, X.; Liu, K.; et al. Exosomes miRNA-499a-5p targeted CD38 to alleviate anthraquinone induced cardiotoxicity: Experimental research. Int. J. Surg. 2024, 110, 1992–2006. [Google Scholar] [CrossRef]
- Baldasici, O.; Pileczki, V.; Cruceriu, D.; Gavrilas, L.I.; Tudoran, O.; Balacescu, L.; Vlase, L.; Balacescu, O. Breast Cancer-Delivered Exosomal miRNA as Liquid Biopsy Biomarkers for Metastasis Prediction: A Focus on Translational Research with Clinical Applicability. Int. J. Mol. Sci. 2022, 23, 9371. [Google Scholar] [CrossRef]
- Takeuchi, C.; Yamashita, S.; Liu, Y.Y.; Takeshima, H.; Sasaki, A.; Fukuda, M.; Hashimoto, T.; Naka, T.; Ishizu, K.; Sekine, S.; et al. Precancerous nature of intestinal metaplasia with increased chance of conversion and accelerated DNA methylation. Gut 2024, 73, 255–267. [Google Scholar] [CrossRef]
- Guo, L.; Hong, T.; Lee, Y.T.; Hu, X.; Pan, G.; Zhao, R.; Yang, Y.; Yang, J.; Cai, X.; Rivera, L.; et al. Perturbing TET2 condensation promotes aberrant genome-wide DNA methylation and curtails leukaemia cell growth. Nat. Cell. Biol. 2024, 26, 2154–2167. [Google Scholar] [CrossRef] [PubMed]
- LaFlamme, C.W.; Rastin, C.; Sengupta, S.; Pennington, H.E.; Russ-Hall, S.J.; Schneider, A.L.; Bonkowski, E.S.; Almanza Fuerte, E.P.; Allan, T.J.; Zalusky, M.P.; et al. Diagnostic utility of DNA methylation analysis in genetically unsolved pediatric epilepsies and CHD2 episignature refinement. Nat. Commun. 2024, 15, 6524. [Google Scholar] [CrossRef] [PubMed]
- Cao, G.; Yang, N.; Xiong, Y.; Shi, M.; Wang, L.; Nie, F.; Huo, D.; Hou, C. Completely Free from PAM Limitations: Asymmetric RPA with CRISPR/Cas12a for Nucleic Acid Assays. ACS Sens. 2023, 8, 4655–4663. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Cai, Z.; Zhou, Z.; Li, M.; Hong, W.; Zhou, W.; Yu, D.; Wei, P.; He, J.; Wang, Y.; et al. CASMART, a one-step CRISPR Cas12a-mediated isothermal amplification for rapid and high-resolution digital detection of rare mutant alleles. Biosens. Bioelectron. 2023, 222, 114956. [Google Scholar] [CrossRef]
- Hao, J.; Yang, T.; Liu, Y.; Jia, M.; Zeng, Z.; Xiong, W. Application of a lyophilized CRISPR/Cas12a and RPA assay for rapid detection of Actinobacillus pleuropneumoniae. Microchem. J. 2024, 206, 111443. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, L.; Yu, X.; Wang, G.; Pan, T.; Huang, Z.; Cui, T.; Huang, T.; Huang, Z.; Nie, L.; et al. Development of a rapid, sensitive detection method for SARS-CoV-2 and influenza virus based on recombinase polymerase amplification combined with CRISPR-Cas12a assay. J. Med. Virol. 2023, 95, e29215. [Google Scholar] [CrossRef]
- Yu, Y.; Li, Q.; Shi, W.; Yang, Y.; He, H.; Dai, J.; Mao, G.; Ma, Y. Programmable Aptasensor for Regulating CRISPR/Cas12a Activity. ACS Sens. 2024, 9, 244–250. [Google Scholar] [CrossRef]
- Lu, Z.; Ni, W.; Liu, N.; Jin, D.; Li, T.; Li, K.; Zhang, Y.; Yao, Q.; Zhang, G.-J. CRISPR/Cas12a-based fluorescence biosensor for detection of exosomal miR-21 derived from lung cancer. Microchem. J. 2023, 187, 108370. [Google Scholar] [CrossRef]
- Zhao, Z.; Wang, S.; Dong, Z.; Fan, Q.; Lei, R.; Kuang, R.; Zhang, Y. One-Step Reverse-Transcription Recombinase-Aided Amplification CRISPR/Cas12a-Based Lateral Flow Assay for Fast Field Screening and Accurate Differentiation of Four Major Tobamoviruses Infecting Tomato and Pepper. J. Agric. Food Chem. 2023, 71, 17025–17035. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Smith, B.M.; Jain, P.K. Enhancement of trans-cleavage activity of Cas12a with engineered crRNA enables amplified nucleic acid detection. Nat. Commun. 2020, 11, 4906. [Google Scholar] [CrossRef]
- Moon, J.; Liu, C. Asymmetric CRISPR enabling cascade signal amplification for nucleic acid detection by competitive crRNA. Nat. Commun. 2023, 14, 7504. [Google Scholar] [CrossRef] [PubMed]
- Rananaware, S.R.; Vesco, E.K.; Shoemaker, G.M.; Anekar, S.S.; Sandoval, L.S.W.; Meister, K.S.; Macaluso, N.C.; Nguyen, L.T.; Jain, P.K. Programmable RNA detection with CRISPR-Cas12a. Nat. Commun. 2023, 14, 5409. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.; Meng, Q.; Sun, B.; Zhao, B.; Dang, L.; Zhong, M.; Liu, S.; Xu, H.; Mei, H.; Liu, J.; et al. MeCas12a, a Highly Sensitive and Specific System for COVID-19 Detection. Adv. Sci. 2020, 7, 2001300. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Yin, K.; Li, Z.; Lalla, R.V.; Ballesteros, E.; Sfeir, M.M.; Liu, C. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat. Commun. 2020, 11, 4711. [Google Scholar] [CrossRef]


| Virus | Amplification | Visualization | Sensitivity | Detection Time | One/Two-Step | Reference |
|---|---|---|---|---|---|---|
| HPV16/18 | RPA | Fluorescence | / | 1 h | Two-step | [42] |
| SARS-CoV-2 | RPA | Fluorescence | 0.4 copies/μL | 50 min | Two-step | [43] |
| SARS-CoV-2 | LAMP | Fluorescence (portable device) | 10 copies/μL | 40 min | One-step (physically separated) | [44] |
| SARS-CoV-2 | RT-LAMP | Fluorescence (naked eye/blue light) | 5 copies/μL | 45 min | One-step (physically separated) | [45] |
| SARS-CoV-2 | RT-LAMP | Fluorescence (naked eye) | 30 copies/μL | 40 min | One-step (physically separated) | [46] |
| SARS-CoV-2 | RT-RPA | Fluorescence | 10 copies/μL | 30 min | One-step (optochemical control) | [47] |
| SARS-CoV-2 variants | RPA | Fluorescence | 30 copies/μL | 1 h | One-step (optochemical control) | [48] |
| SARS-CoV-2 | RT-RPA | Fluorescence (UV) | 5 aM | 50 min | One-step | [49] |
| ASFV | RPA | Fluorescence (blue light)/LFD | 6.8 copies/μL | 1 h | Two-step | [50] |
| ASFV | Amplification-free | magnetic-SERS nanoprobe | 10 fM | 2 h | Multi-step | [51] |
| ASFV | LAMP | Fluorescence | 1 copies/μL | 50 min | One-step (physically separated) | [52] |
| MPXV | PCSDA | Fluorescence | 2.8 × 10−4 ng/μL | 100 min | Two-step | [53] |
| MPXV | RPA | Fluorescence/LFA | 1 copies/μL | 45 min | Two-step | [54] |
| PCV3 | ERA | Fluorescence | 1.16 copies/μL | 1 h | Two-step | [55] |
| HSV-1 | Amplification-free | Electrochemical signal | 3 aM | 6.5 h | Multi-step | [56] |
| PRV | MIRA | Fluorescence (blue light) | 1.65 × 104 copies/μL | 25 min | Two-step | [57] |
| 12 respiratory pathogens * | RPA | Fluorescence | 2.5 × 100 copies/μL | 90 min | One-step (physically separated) | [58] |
| GBS, HPV16/18 | RPA | Fluorescence | 16.6 aM | 30 min | One-step | [59] |
| H1N1, H3N2, IVB, HRSV, SARS-CoV-2 variants | RPA | Fluorescence | 0.1 copies/μL | 1 h | One-step (microfluidic chip) | [60] |
| SARS-CoV-2 variants | RT-RPA | Fluorescence (UV/blue light) | 0.01 copies/μL | 1 h | Two-step | [61] |
| SARS-CoV-2/ASFV | RPA/RT-RPA | Fluorescence (portable device) | 8 copies/μL | 1.5 h | Two-step | [62] |
| IA, IB, RSV, SARS-CoV-2 | RPA | Fluorescence (portable device/naked eye) | 0.8 copies/μL | 1.5 h | One-step (microfluidic chip) | [63] |
| SARS-CoV-2 variant, IA, IB, RSV, IC | RT-RPA | Fluorescence (portable device) | 1 copies/μL | 40 min | One-step (physically separated) | [64] |
| Pathogens | Amplification | Visualization | Sensitivity | Detection Time | One/Two-Step | Reference |
|---|---|---|---|---|---|---|
| mycoplasma | RPA | Fluorescence (Blue Light) | 10 aM | 30 min | One-step (physically separated) | [68] |
| M. pneumoniae | ERA | Fluorescence/LFA | 1 copies/μL | 30 min | Two-step | [69] |
| M. pneumoniae | MCDA | Fluorescence | 50 fg | 50 min | Two-step | [70] |
| Salmonella enterica | LAMP | Fluorescence | 20 CFU | <1 h | Two-step | [71] |
| Listeria monocytogenes | RAA | Fluorescence | 0.68 aM | 40 min | Two-step | [72] |
| Mycobacterium tuberculosis | LAMP | Fluorescence/LFA | 50 fg | 1 h | Two-step | [73] |
| Aspergillus fumigatus | RPA | Fluorescence/LFA | 102 copies/µL | 40 min | Two-step | [74] |
| Burkholderia pseudomallei | RPA | LFA | 50 CFU/mL | 90 min | Two-step | [75] |
| Parasites | Amplification | Visualization | Sensitivity | Detection Time | One/Two-Step | Reference |
|---|---|---|---|---|---|---|
| Theileria parva | RPA | Fluorescence/LFA | 1 infected lymphocyte/3 μL | 80 min | Two-step | [76] |
| Neospora caninum | RPA | Fluorescence/LFA | 1 parasites/mL | 90 min | Two-step | [77] |
| Toxoplasma gondii | RPA | Fluorescence/LFA | 31 copies/μL | 55 min | Two-step | [78] |
| Anisakis | RPA | Fluorescence (naked eye/portable device) | 31.6 copies/μL | 80 min | One-step (physically separated) | [79] |
| Anaplasma marginale | RPA | Fluorescence/LFA | 4 copies/μL | <1 h | Two-step | [80] |
| Opisthorchis viverrini | RPA | Fluorescence (UV) | 1 ng | <1 h | Two-step | [81] |
| Clonorchis sinensis | RPA | Fluorescence/LFA | 1 copies/μL | <1 h | One-step (physically separated) | [82] |
| Heterodera avenae/Heterodera filipjevi | RPA | Fluorescence/LFA | 10−4 J2 | <1 h | Two-step | [83] |
| Plasmodium spp. | RPA | Fluorescence/LFA | 3.11–7.27 parasites/μL | <1 h | Two-step | [84] |
| Schistosoma haematobium | RPA | Fluorescence/LFA | 2 eggs | 70 min | One-step | [85] |
| Leishmania | PCR | Fluorescence | 1–42 parasites/106 human cells | <1 h | Two-step | [86] |
| Diseases/Biomarkers | Amplification | Visualization | Sensitivity | Detection Time | One/Two-Step | Reference |
|---|---|---|---|---|---|---|
| NSCLC | RAA | Fluorescence (blue light) | 10 copies/μL | <1 h | Two-step (microfluidic chip) | [87] |
| MPN | RPA | Fluorescence/LFA | 3 copies/μL | 1.5 h | Two-step | [88] |
| Breast cancer | Amplification-free | Fluorescence | 1.45 × 102/3.73 × 102 particles/μL | 1 h | Multi-step | [89] |
| CCBMH | Amplification-free | Fluorescence | AFB1: 31 pM Cd2+: 3.9 nM | <2 h | Multi-step | [90] |
| Osteosarcoma | Amplification-free | Fluorescence | 3 HeLa cells | 70 min | Multi-step | [91] |
| Stomach cancer | LAMP | Fluorescence/LFA | 43 aM | <1 h | One-step | [92] |
| NPC | Amplification-free | Fluorescence (inverted microscope) | 5 copies/μL | <2 h | One-step (microfluidic chip) | [93] |
| Esophageal cancer | Amplification-free | Fluorescence | 1.26 fM | <3 h | Multi-step | [94] |
| BCa | RT-RAA | Fluorescence | 0.1 copies/μL | 30 min | Twp-step | [95] |
| HCC | TTSD | Colorimetric assays | 0.5 pM | <2 h | Multi-step | [96] |
| Prostate cancer | Amplification-free | Colorimetric assays (portable device) | f-PSA: 0.04 ng/mL t-PSA: 0.06 ng/mL | <1 h | Multi-step | [97] |
| LHON | ERA | Fluorescence (blue light) | / | 30 min | Two-step | [98] |
| SMA | PCR/RPA | Fluorescence/LFA | 526 aM | 1.5 h | Two-step | [99] |
| miRNA-155/miRNA-141 | Amplification-free | Fluorescence | 84 fmol/L | <2 h | Multi-step | [100] |
| methylated DNA | PCR | Fluorescence/LFA | / | 6 h | Multi-step | [101] |
| NAD+ | Amplification-free | Fluorescence | 22.5 nM | 30 min | One-step | [102] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, K.; Zeng, Q.; Jiang, M.; Hu, Z.; Zhou, M.; Xia, K. CRISPR/Cas12a-Based Biosensing: Advances in Mechanisms and Applications for Nucleic Acid Detection. Biosensors 2025, 15, 360. https://doi.org/10.3390/bios15060360
Du K, Zeng Q, Jiang M, Hu Z, Zhou M, Xia K. CRISPR/Cas12a-Based Biosensing: Advances in Mechanisms and Applications for Nucleic Acid Detection. Biosensors. 2025; 15(6):360. https://doi.org/10.3390/bios15060360
Chicago/Turabian StyleDu, Kun, Qinlong Zeng, Mingjun Jiang, Zhiqing Hu, Miaojin Zhou, and Kun Xia. 2025. "CRISPR/Cas12a-Based Biosensing: Advances in Mechanisms and Applications for Nucleic Acid Detection" Biosensors 15, no. 6: 360. https://doi.org/10.3390/bios15060360
APA StyleDu, K., Zeng, Q., Jiang, M., Hu, Z., Zhou, M., & Xia, K. (2025). CRISPR/Cas12a-Based Biosensing: Advances in Mechanisms and Applications for Nucleic Acid Detection. Biosensors, 15(6), 360. https://doi.org/10.3390/bios15060360





