Ultrasensitive Optical Fingerprinting of Biorelevant Molecules by Means of SERS-Mapping on Nanostructured Metasurfaces
Abstract
1. Introduction
2. Materials and Methods
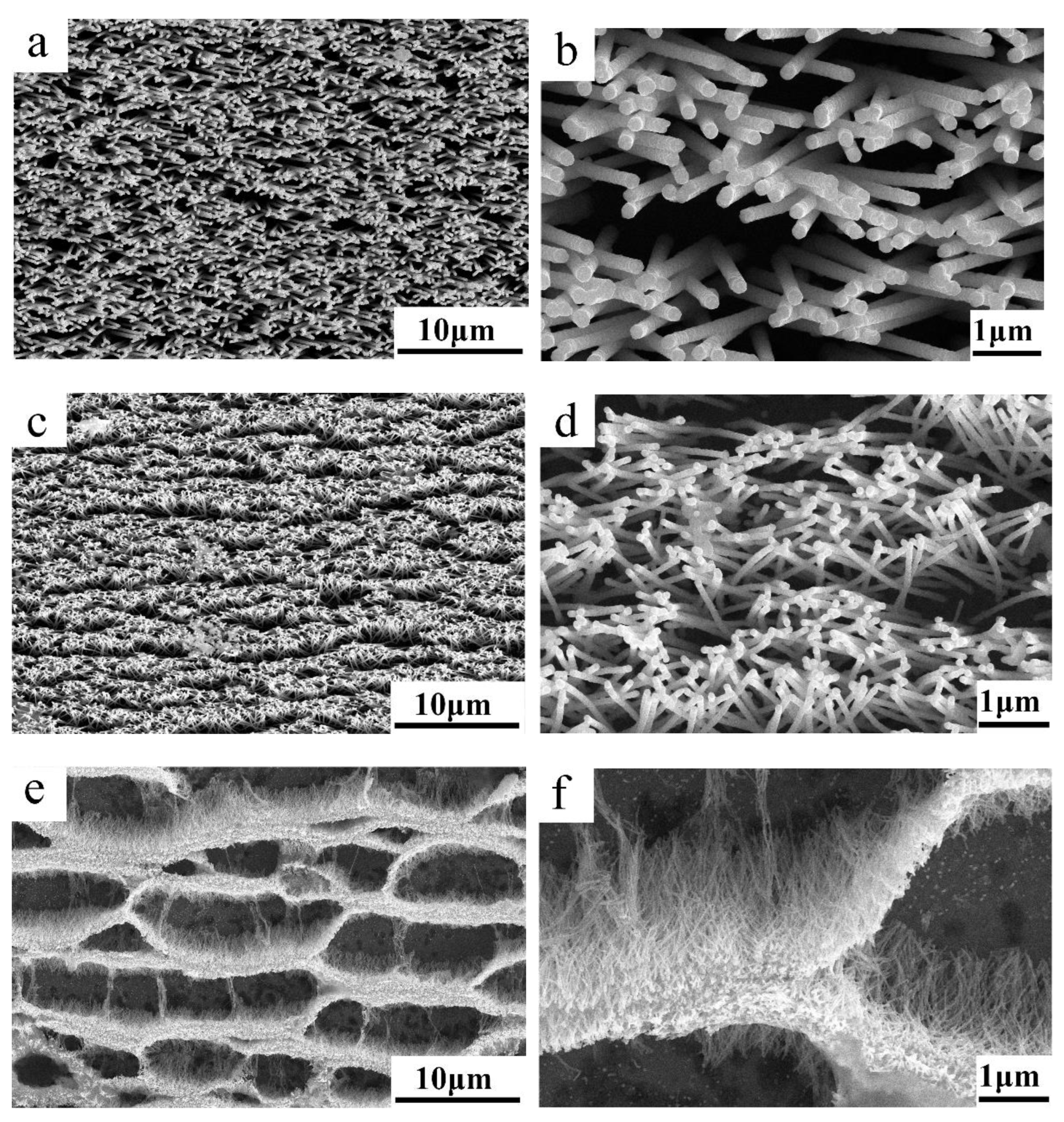
2.1. Prepare SERS Substrates
2.2. Molecules and Samples
2.3. SERS Spectroscopy and Mapping
3. Results and Discussion
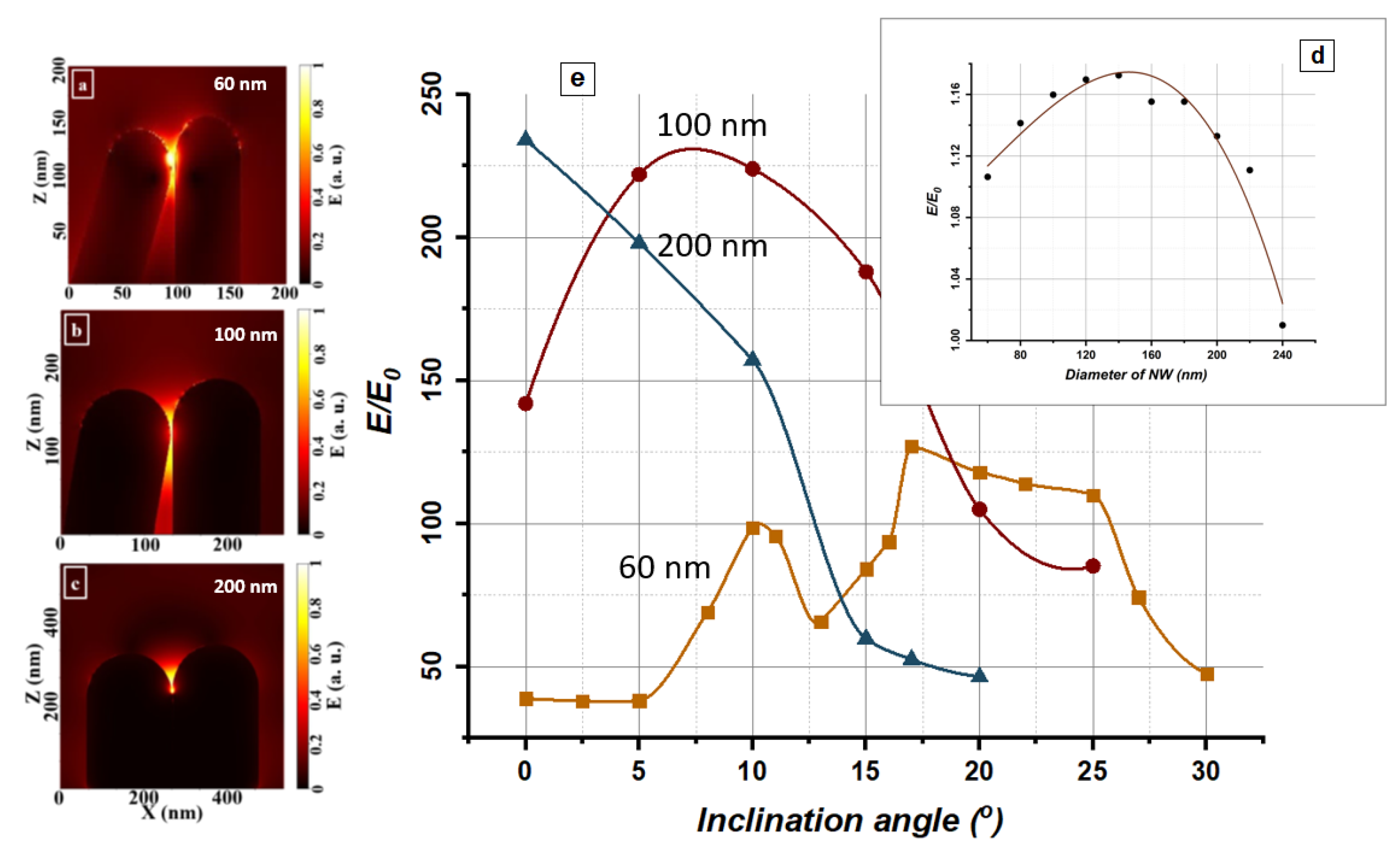
3.1. Physical Mechanisms of the SERS Enhancement Effect
3.2. SERS-Mapping with Small Molecules
3.3. SERS-Mapping with “Large” Molecules
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Li, L.; Lin, H.; Qiao, S.; Huang, Y.Z.; Li, J.Y.; Michon, J.; Gu, T.; Alosno-Ramos, C.; Vivien, L.; Yadav, A.; et al. Monolithically integrated stretchable photonics. Light Sci. Appl. 2018, 7, 17138. [Google Scholar] [CrossRef] [PubMed]
- Kurz, W.; Yetisen, A.K.; Kaito, M.V.; Fuchter, M.J.; Jakobi, M.; Elsner, M.; Koch, A.W. UV-Sensitive Wearable Devices for Colorimetric Monitoring of UV Exposure. Adv. Opt. Mater. 2020, 8, 1901969. [Google Scholar] [CrossRef]
- Khatua, S.; Orrit, M. Toward Single-Molecule Microscopy on a Smart Phone. ACS Nano 2013, 7, 8340–8343. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Acuna, G.; Kim, S.; Vietz, C.; Tseng, D.; Chae, J.; Shir, D.; Luo, W.; Tinnefeld, P.; Ozcan, A. Plasmonics Enhanced Smartphone Fluorescence Microscopy. Sci. Rep. 2017, 7, 2124. [Google Scholar] [CrossRef]
- Moore, T.J.; Moody, A.S.; Payne, T.D.; Sarabia, G.M.; Daniel, A.R.; Sharma, B. In Vitro and In Vivo SERS Biosensing for Disease Diagnosis. Biosensors 2018, 8, 46. [Google Scholar] [CrossRef]
- Wegner, K.; Just, S.; Gau, L.; Mueller, H.; Gerard, P.; Lepage, P.; Clavel, T.; Rohn, S. Rapid analysis of bile acids in different biological matrices using LC-ESI-MS/MS for the investigation of bile acid transformation by mammalian gut bacteria. Anal. Bioanal. Chem. 2017, 409, 1231–1245. [Google Scholar] [CrossRef]
- Gou, T.; Hu, J.M.; Wu, W.S.; Ding, X.; Zhou, S.F.; Fang, W.B.; Mu, Y. Smartphone-based mobile digital PCR device for DNA quantitative analysis with high accuracy. Biosens. Bioelectron. 2018, 120, 144–152. [Google Scholar] [CrossRef]
- Stedtfeld, R.D.; Tourlousse, D.M.; Seyrig, G.; Stedtfeld, T.M.; Kronlein, M.; Price, S.; Ahmad, F.; Gulari, E.; Tiedje, J.M.; Hashsham, S.A. Gene-Z: A device for point of care genetic testing using a smartphone. Lab Chip 2012, 12, 1454–1462. [Google Scholar] [CrossRef]
- Sauvage, J.P. From Chemical Topology to Molecular Machines (Nobel Lecture). Angew. Chem. Int. Ed. 2017, 56, 11080–11093. [Google Scholar] [CrossRef]
- Balzani, V.; Credi, A.; Raymo, F.M.; Stoddart, J.F. Artificial molecular machines. Angew. Chem. Int. Ed. 2000, 39, 3348–3391. [Google Scholar] [CrossRef]
- Lee, J.; Crampton, K.T.; Tallarida, N.; Apkarian, V.A. Visualizing vibrational normal modes of a single molecule with atomically confined light. Nature 2019, 568, 78–82. [Google Scholar] [CrossRef] [PubMed]
- Sheremet, E.; Milekhin, A.G.; Rodriguez, R.D.; Weiss, T.; Nesterov, M.; Rodyakina, E.E.; Gordan, O.D.; Sveshnikova, L.L.; Duda, T.A.; Gridchin, V.A.; et al. Surface- and tip-enhanced resonant Raman scattering from CdSe nanocrystals. Phys. Chem. Chem. Phys. 2015, 17, 21198–21203. [Google Scholar] [CrossRef] [PubMed]
- Karimullin, K.R.; Arzhanov, A.I.; Eremchev, I.Y.; Kulnitskiy, B.A.; Surovtsev, N.V.; Naumov, A.V. Combined photon-echo, luminescence and Raman spectroscopies of layered ensembles of colloidal quantum dots. Laser Phys. 2019, 29, 124009. [Google Scholar] [CrossRef]
- Jäckel, F.; Kinkhabwala, A.A.; Moerner, W.E. Bowtie Nanoantennas as Substrates for Electrochemical Surface-Enhanced Raman Scattering (SERS). In Proceedings of the Frontiers in Optics 2007, San Jose, CA, USA, 16–20 September 2007. [Google Scholar]
- Kim, S.; Xuan, Y.; Drachev, V.P.; Varghese, L.T.; Fan, L.; Qi, M.; Webb, K.J. Nanoimprinted plasmonic nanocavity arrays. Opt. Express 2013, 21, 15081–15089. [Google Scholar] [CrossRef] [PubMed]
- Leopold, N.; Lendl, B. A New Method for Fast Preparation of Highly Surface-Enhanced Raman Scattering (SERS) Active Silver Colloids at Room Temperature by Reduction of Silver Nitrate with Hydroxylamine Hydrochloride. J. Phys. Chem. B 2003, 107, 5723–5727. [Google Scholar] [CrossRef]
- Milekhin, I.A.; Anikin, K.V.; Rahaman, M.; Rodyakina, E.E.; Duda, T.A.; Saidzhonov, B.M.; Vasiliev, R.B.; Dzhagan, V.M.; Milekhin, A.G.; Batsanov, S.A.; et al. Resonant plasmon enhancement of light emission from CdSe/CdS nanoplatelets on Au nanodisk arrays. J. Chem. Phys. 2020, 153, 164708. [Google Scholar] [CrossRef]
- Khlebtsov, B.N.; Khanadeev, V.A.; Panfilova, E.V.; Bratashov, D.N.; Khlebtsov, N.G. Gold nanoisland films as reproducible SERS substrates for highly sensitive detection of fungicides. ACS Appl. Mater. Interfaces 2015, 7, 6518–6529. [Google Scholar] [CrossRef]
- Melnik, N.N.; Sherestnev, I.A.; Tregulov, V.V. Research of the silver nanoparticles, deposited on the surface of a single crystal and porous silicon by the chemical method. Bull. Russ. Acad. Sci. Phys. 2021, 85, 1287–1290. [Google Scholar] [CrossRef]
- He, J.; Li, X.; Li, J. Facile construction of silver nanocubes/graphene oxide composites for highly sensitive SERS detection of multiple organic contaminants by a portable Raman spectrometer. J. Environ. Chem. Eng. 2022, 10, 108278. [Google Scholar] [CrossRef]
- Wang, X.; Cheng, H.; Min, Y.; Li, X.; You, L.; Li, J. Fe3O4@m-ZrO2@Ag ternary magnetic nanocomposites for sensitive SERS sensing and photocatalytic removal of Cr(VI) and organic dyes. Compos. Part B Eng. 2022, 239, 109959. [Google Scholar] [CrossRef]
- Chikkaraddy, R.; Xomalis, A.; Jakob, L.A.; Baumberg, J.J. Mid-infrared-perturbed molecular vibrational signatures in plasmonic nanocavities. Light Sci. Appl. 2022, 11, 19. [Google Scholar] [CrossRef]
- Etchegoin, P.; Cohen, L.F.; Hartigan, H.; Brown, R.J.C.; Milton, M.J.T.; Gallop, J.C. Electromagnetic contribution to surface enhanced Raman scattering revisited. J. Chem. Phys. 2003, 119, 5281–5289. [Google Scholar] [CrossRef]
- Le Ru, E.C.; Etchegoin, P.G. Quantifying SERS enhancements. MRS Bull. 2013, 38, 631–640. [Google Scholar] [CrossRef]
- Barbillon, G. Latest Novelties on Plasmonic and Non-Plasmonic Nanomaterials for SERS Sensing. Nanomaterials 2020, 10, 1200. [Google Scholar] [CrossRef]
- Du, Y.; Shi, L.; He, T.; Sun, X.; Mo, Y. SERS enhancement dependence on the diameter and aspect ratio of silver-nanowire array fabricated by anodic aluminium oxide template. Appl. Surf. Sci. 2008, 255, 1901–1905. [Google Scholar] [CrossRef]
- Wu, G.; Cao, F.; Zhao, P.; Li, Z.; Yu, N.; Wang, Z.; Zhang, X.; Hu, Y.; Zhang, X.; Gu, H. Influence of Structural Parameters on the Surface Enhanced Raman Scattering of Au Nanoarrays. J. Nanosci. Nanotechnol. 2019, 19, 5317–5322. [Google Scholar] [CrossRef]
- Yang, J.; Song, G.; Zhou, L.; Wang, X.; You, L.; Li, J. Highly sensitively detecting tetramethylthiuram disulfide based on synergistic contribution of metal and semiconductor in stable Ag/TiO2 core-shell SERS substrates. Appl. Surf. Sci. 2021, 539, 147744. [Google Scholar] [CrossRef]
- Radziuk, D.; Moehwald, H. Prospects for plasmonic hot spots in single molecule SERS towards the chemical imaging of live cells. Phys. Chem. Chem. Phys. 2015, 17, 21072–21093. [Google Scholar] [CrossRef]
- Kovalets, N.P.; Kozhina, E.P.; Razumovskaya, I.V.; Bedin, S.A.; Piryazev, A.A.; Grigoriev, Y.V.; Naumov, A.V. Toward single-molecule surface-enhanced Raman scattering with novel type of metasurfaces synthesized by crack-stretching of metallized track-etched membranes. J. Chem. Phys. 2022, 156, 034902. [Google Scholar] [CrossRef]
- Benz, F.; Chikkaraddy, R.; Salmon, A.; Ohadi, H.; de Nijs, B.; Mertens, J.; Carnegie, C.; Bowman, R.W.; Baumberg, J.J. SERS of Individual Nanoparticles on a Mirror: Size Does Matter, but so Does Shape. J. Phys. Chem. Lett. 2016, 7, 2264–2269. [Google Scholar] [CrossRef]
- Koker, T.; Tang, N.; Tian, C.; Zhang, W.; Wang, X.; Martel, R.; Pinaud, F. Cellular imaging by targeted assembly of hot-spot SERS and photoacoustic nanoprobes using split-fluorescent protein scaffolds. Nat. Commun. 2018, 9, 607. [Google Scholar] [CrossRef] [PubMed]
- Mosier-Boss, P.A. Review of SERS Substrates for Chemical Sensing. Nanomaterials 2017, 7, 142. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Irannejad, M.; Cui, B. Bowtie Nanoantenna with Single-Digit Nanometer Gap for Surface-Enhanced Raman Scattering (SERS). Plasmonics 2014, 10, 831–837. [Google Scholar] [CrossRef]
- Gupta, N.; Dhawan, A. Bridged-bowtie and cross bridged-bowtie nanohole arrays as SERS substrates with hotspot tunability and multi-wavelength SERS response. Opt. Express 2018, 26, 17899–17915. [Google Scholar] [CrossRef] [PubMed]
- Schönenberger, C.; van der Zande, B.M.I.; Fokkink, L.G.J.; Henny, M.; Schmid, C.; Krüger, M.; Bachtold, A.; Huber, R.; Birk, H.; Staufer, U. Template Synthesis of Nanowires in Porous Polycarbonate Membranes: Electrochemistry and Morphology. J. Phys. Chem. B 1997, 101, 5497–5505. [Google Scholar] [CrossRef]
- Liu, J.W.; Wang, J.L.; Huang, W.R.; Yu, L.; Ren, X.F.; Wen, W.C.; Yu, S.H. Ordering Ag nanowire arrays by a glass capillary: A portable, reusable and durable SERS substrate. Sci. Rep. 2012, 2, 987. [Google Scholar] [CrossRef]
- Kozhina, E.P.; Bedin, S.A.; Nechaeva, N.L.; Podoynitsyn, S.N.; Tarakanov, V.P.; Andreev, S.N.; Grigoriev, Y.V.; Naumov, A.V. Ag-Nanowire Bundles with Gap Hot Spots Synthesized in Track-Etched Membranes as Effective SERS-Substrates. Appl. Sci. 2021, 11, 1375. [Google Scholar] [CrossRef]
- Schmidt, M.S.; Hubner, J.; Boisen, A. Large area fabrication of leaning silicon nanopillars for surface enhanced Raman spectroscopy. Adv. Mater. 2012, 24, OP11–OP18. [Google Scholar] [CrossRef]
- Simovski, C. Circuit Model of Plasmon-Enhanced Fluorescence. Photonics 2015, 2, 568–593. [Google Scholar] [CrossRef]
- Din, R.U.; Zeng, X.D.; Ge, G.Q.; Zubairy, M.S. Tunable surface plasmon-polaritons based on quantum coherence. Opt. Express 2019, 27, 322–336. [Google Scholar] [CrossRef]
- Anger, P.; Bharadwaj, P.; Novotny, L. Enhancement and quenching of single-molecule fluorescence. Phys. Rev. Lett. 2006, 96, 113002. [Google Scholar] [CrossRef]
- Zong, C.; Xu, M.X.; Xu, L.J.; Wei, T.; Ma, X.; Zheng, X.S.; Hu, R.; Ren, B. Surface-Enhanced Raman Spectroscopy for Bioanalysis: Reliability and Challenges. Chem. Rev. 2018, 118, 4946–4980. [Google Scholar] [CrossRef]
- Thoreson, M.D.; Drachev, V.P.; Khaliullin, E.N.; Nashine, V.; Ben-Amotz, D.; Davisson, V.J.; Shalaev, V.M. Adaptive SERS substrates for protein sensing. In Proceedings of the Nanosensing: Materials and Devices, Philadelphia, PA, USA, 25–28 October 2004. [Google Scholar]
- Li, J.; Wuethrich, A.; Sina, A.A.I.; Cheng, H.H.; Wang, Y.; Behren, A.; Mainwaring, P.N.; Trau, M. A digital single-molecule nanopillar SERS platform for predicting and monitoring immune toxicities in immunotherapy. Nat. Commun. 2021, 12, 1087. [Google Scholar] [CrossRef]
- Shen, Y.T.; Yue, J.; Xu, W.Q.; Xu, S.P. Recent progress of surface-enhanced Raman spectroscopy for subcellular compartment analysis. Theranostics 2021, 11, 4872–4893. [Google Scholar] [CrossRef]
- Dochow, S.; Becker, M.; Spittel, R.; Beleites, C.; Stanca, S.; Latka, I.; Schuster, K.; Kobelke, J.; Unger, S.; Henkel, T.; et al. Raman-on-chip device and detection fibres with fibre Bragg grating for analysis of solutions and particles. Lab Chip 2013, 13, 1109–1113. [Google Scholar] [CrossRef]
- Etchegoin, P.G.; Le Ru, E.C.; Meyer, M. Evidence of natural isotopic distribution from single-molecule SERS. J. Am. Chem. Soc. 2009, 131, 2713–2716. [Google Scholar] [CrossRef]
- Le Ru, E.C.; Etchegoin, P.G. Single-molecule surface-enhanced Raman spectroscopy. Annu. Rev. Phys. Chem. 2012, 63, 65–87. [Google Scholar] [CrossRef]
- Cortes, E.; Etchegoin, P.G.; Le Ru, E.C.; Fainstein, A.; Vela, M.E.; Salvarezza, R.C. Strong correlation between molecular configurations and charge-transfer processes probed at the single-molecule level by surface-enhanced Raman scattering. J. Am. Chem. Soc. 2013, 135, 2809–2815. [Google Scholar] [CrossRef]
- Marr, J.M.; Schultz, Z.D. Imaging Electric Fields in SERS and TERS Using the Vibrational Stark Effect. J. Phys. Chem. Lett. 2013, 4, 3268–3272. [Google Scholar] [CrossRef]
- Lee, J.; Tallarida, N.; Chen, X.; Jensen, L.; Apkarian, V.A. Microscopy with a single-molecule scanning electrometer. Sci. Adv. 2018, 4, eaat5472. [Google Scholar] [CrossRef]
- Mockl, L.; Moerner, W.E. Super-resolution Microscopy with Single Molecules in Biology and Beyond-Essentials, Current Trends, and Future Challenges. J. Am. Chem. Soc. 2020, 142, 17828–17844. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Palla, M.; Bosco, F.G.; Rindzevicius, T.; Alstrom, T.S.; Schmidt, M.S.; Boisen, A.; Ju, J.; Lin, Q. Surface-enhanced Raman spectroscopy based quantitative bioassay on aptamer-functionalized nanopillars using large-area Raman mapping. ACS Nano 2013, 7, 5350–5359. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Pang, S.; Chen, J.; McLandsborough, L.; Nugen, S.R.; Fan, M.; He, L. Label-free mapping of single bacterial cells using surface-enhanced Raman spectroscopy. Analyst 2016, 141, 1356–1362. [Google Scholar] [CrossRef] [PubMed]
- Ilchenko, O.; Slipets, R.; Rindzevicius, T.; Durucan, O.; Morelli, L.; Schmidt, M.S.; Wu, K.; Boisen, A. Wide Line Surface-Enhanced Raman Scattering Mapping. Adv. Mater. Technol. 2020, 5, 1900999. [Google Scholar] [CrossRef]
- Lin, Y.K.; Tai, R.J.; Wei, S.C.; Luo, S.C. Electrochemical SERS on 2D Mapping for Metabolites Detection. Langmuir 2020, 36, 5990–5996. [Google Scholar] [CrossRef]
- Palla, M.; Bosco, F.G.; Yang, J.; Rindzevicius, T.; Alstrom, T.S.; Schmidt, M.S.; Lin, Q.; Ju, J.; Boisen, A. Mathematical Model for Biomolecular Quantification Using Large-Area Surface-Enhanced Raman Spectroscopy Mapping. RSC Adv. 2015, 5, 85845–85853. [Google Scholar] [CrossRef]
- Fortuni, B.; Fujita, Y.; Ricci, M.; Inose, T.; Aubert, R.; Lu, G.; Hutchison, J.A.; Hofkens, J.; Latterini, L.; Uji, I.H. A novel method for in situ synthesis of SERS-active gold nanostars on polydimethylsiloxane film. Chem. Commun. 2017, 53, 5121–5124. [Google Scholar] [CrossRef]
- Han, X.X.; Rodriguez, R.S.; Haynes, C.L.; Ozaki, Y.; Zhao, B. Surface-enhanced Raman spectroscopy. Nat. Rev. Methods Prim. 2022, 1, 610–626. [Google Scholar] [CrossRef]
- Martin, C.R. Nanomaterials: A membrane-based synthetic approach. Science 1994, 266, 1961–1966. [Google Scholar] [CrossRef]
- Apel, P. Track etching technique in membrane technology. Radiat. Meas. 2001, 34, 559–566. [Google Scholar] [CrossRef]
- Apel, P.Y.; Dmitriev, S.N. Micro- and nanoporous materials produced using accelerated heavy ion beams. Adv. Nat. Sci. Nanosci. Nanotechnol. 2011, 2, 013002. [Google Scholar] [CrossRef]
- Razumovskaya, I.V.; Kovalets, N.P.; Bedin, S.A.; Grigor’ev, Y.V. Agglomeration of Nanowires on a Substrate for Surface-Enhanced Raman Scattering. J. Exp. Theor. Phys. 2021, 132, 818–823. [Google Scholar] [CrossRef]
- Kovalec, N.P.; Kozhina, E.P.; Doludenko, I.M.; Razumovskaya, I.V.; Bedin, S.A.; Grigoriev, Y.V.; Kanevsky, V.M. Agglomeration of Ensembles of Silver Nanowires, Obtained by the Method of Template Synthesis. Bull. Russ. Acad. Sci. Phys. 2021, 85, 854–857. [Google Scholar] [CrossRef]
- Langer, J.; Jimenez de Aberasturi, D.; Aizpurua, J.; Alvarez-Puebla, R.A.; Auguie, B.; Baumberg, J.J.; Bazan, G.C.; Bell, S.E.J.; Boisen, A.; Brolo, A.G.; et al. Present and Future of Surface-Enhanced Raman Scattering. ACS Nano 2020, 14, 28–117. [Google Scholar] [CrossRef]
- Li, C.; Huang, Y.; Li, X.; Zhang, Y.; Chen, Q.; Ye, Z.; Alqarni, Z.; Bell, S.E.J.; Xu, Y. Towards practical and sustainable SERS: A review of recent developments in the construction of multifunctional enhancing substrates. J. Mater. Chem. C 2021, 9, 11517–11552. [Google Scholar] [CrossRef]
- Sharma, B.; Frontiera, R.R.; Henry, A.I.; Ringe, E.; Van Duyne, R.P. SERS: Materials, applications, and the future. Mater. Today 2012, 15, 16–25. [Google Scholar] [CrossRef]
- Chan, T.Y.; Liu, T.Y.; Wang, K.S.; Tsai, K.T.; Chen, Z.X.; Chang, Y.C.; Tseng, Y.Q.; Wang, C.H.; Wang, J.K.; Wang, Y.L. SERS Detection of Biomolecules by Highly Sensitive and Reproducible Raman-Enhancing Nanoparticle Array. Nanoscale Res. Lett. 2017, 12, 344. [Google Scholar] [CrossRef]
- Pei, L.; Huang, Y.; Li, C.; Zhang, Y.; Rasco, B.A.; Lai, K. Detection of Triphenylmethane Drugs in Fish Muscle by Surface-Enhanced Raman Spectroscopy Coupled with Au-Ag Core-Shell Nanoparticles. J. Nanomater. 2014, 2014, 730915. [Google Scholar] [CrossRef]
- Zhou, B.; Shen, J.; Li, P.; Ge, M.; Lin, D.; Li, Y.Y.; Lu, J.; Yang, L. Gold Nanoparticle-Decorated Silver Needle for Surface-Enhanced Raman Spectroscopy Screening of Residual Malachite Green in Aquaculture Products. ACS Appl. Nano Mater. 2019, 2, 2752–2757. [Google Scholar] [CrossRef]
- De La Torre, G.; Claessens, C.G.; Torres, T. Phthalocyanines: Old dyes, new materials. Putting color in nanotechnology. Chem. Commun. 2007, 20, 2000–2015. [Google Scholar] [CrossRef]
- Kitching, H.; Kenyon, A.J.; Parkin, I.P. The interaction of gold and silver nanoparticles with a range of anionic and cationic dyes. Phys Chem Chem Phys 2014, 16, 6050–6059. [Google Scholar] [CrossRef] [PubMed]
- Menges, F. Spectragryph—Optical Spectroscopy Software, 1.2.15; Optical Spectroscopy Software: Oberstdorf, Germany, 2020. [Google Scholar]
- Tarakanov, V.P. Code KARAT in simulations of power microwave sources including Cherenkov plasma devices, vircators, orotron, E-field sensor, calorimeter etc. EPJ Web Conf. 2017, 149, 04024. [Google Scholar] [CrossRef]
- Wang, W.; Yang, Q.; Fan, F.; Xu, H.; Wang, Z.L. Light propagation in curved silver nanowire plasmonic waveguides. Nano Lett. 2011, 11, 1603–1608. [Google Scholar] [CrossRef] [PubMed]
- Neacsu, C.C.; Berweger, S.; Raschke, M.B. Tip-Enhanced Raman Imaging and Nanospectroscopy: Sensitivity, Symmetry, and Selection Rules. NanoBiotechnology 2009, 3, 172–196. [Google Scholar] [CrossRef][Green Version]
- Zhao, Y.; Song, Y.; Zhang, Y.; Fan, Y.; Lai, K. Ultra sensitive detection of malachite green in fish muscle with gold nanoparticles and graphene oxide hybrid as a substrate for surface enhanced Raman scattering. J. Food Meas. Charact. 2019, 14, 658–667. [Google Scholar] [CrossRef]
- Neacsu, C.C.; Dreyer, J.; Behr, N.; Raschke, M.B. Scanning-probe Raman spectroscopy with single-molecule sensitivity. Phys. Rev. B 2006, 73, 193406. [Google Scholar] [CrossRef]
- Arzhanov, A.I.; Savostianov, A.O.; Magaryan, K.A.; Karimullin, K.R.; Naumov, A.V. Photonics of semiconductor quantum dots: Basic aspects. Photonics Russ. 2021, 15, 622–641. [Google Scholar] [CrossRef]
- Arzhanov, A.I.; Savostianov, A.O.; Magaryan, K.A.; Karimullin, K.R.; Naumov, A.V. Photonics of semiconductor quantum dots: Applied aspects. Photonics Russ. 2022, 16, 96–112. [Google Scholar] [CrossRef]
- Neese, F. Software update: The ORCA program system, version 4.0. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2018, 8, e1327. [Google Scholar] [CrossRef]
- Weigend, F.; Ahlrichs, R. Balanced basis sets of split valence, triple zeta valence and quadruple zeta valence quality for H to Rn: Design and assessment of accuracy. Phys. Chem. Chem. Phys. 2005, 7, 3297–3305. [Google Scholar] [CrossRef]
- Martynov, A.G.; Mack, J.; Ngoy, B.P.; Nyokong, T.; Gorbunova, Y.G.; Tsivadze, A.Y. Electronic structure and NH-tautomerism of a novel metal-free phenanthroline-annelated phthalocyanine. Dye. Pigment. 2017, 140, 469–479. [Google Scholar] [CrossRef]
- McKeown, N.B.; Makhseed, S.; Msayib, K.J.; Ooi, L.L.; Helliwell, M.; Warren, J.E. A phthalocyanine clathrate of cubic symmetry containing interconnected solvent-filled voids of nanometer dimensions. Angew. Chem. Int. Ed. 2005, 44, 7546–7549. [Google Scholar] [CrossRef]


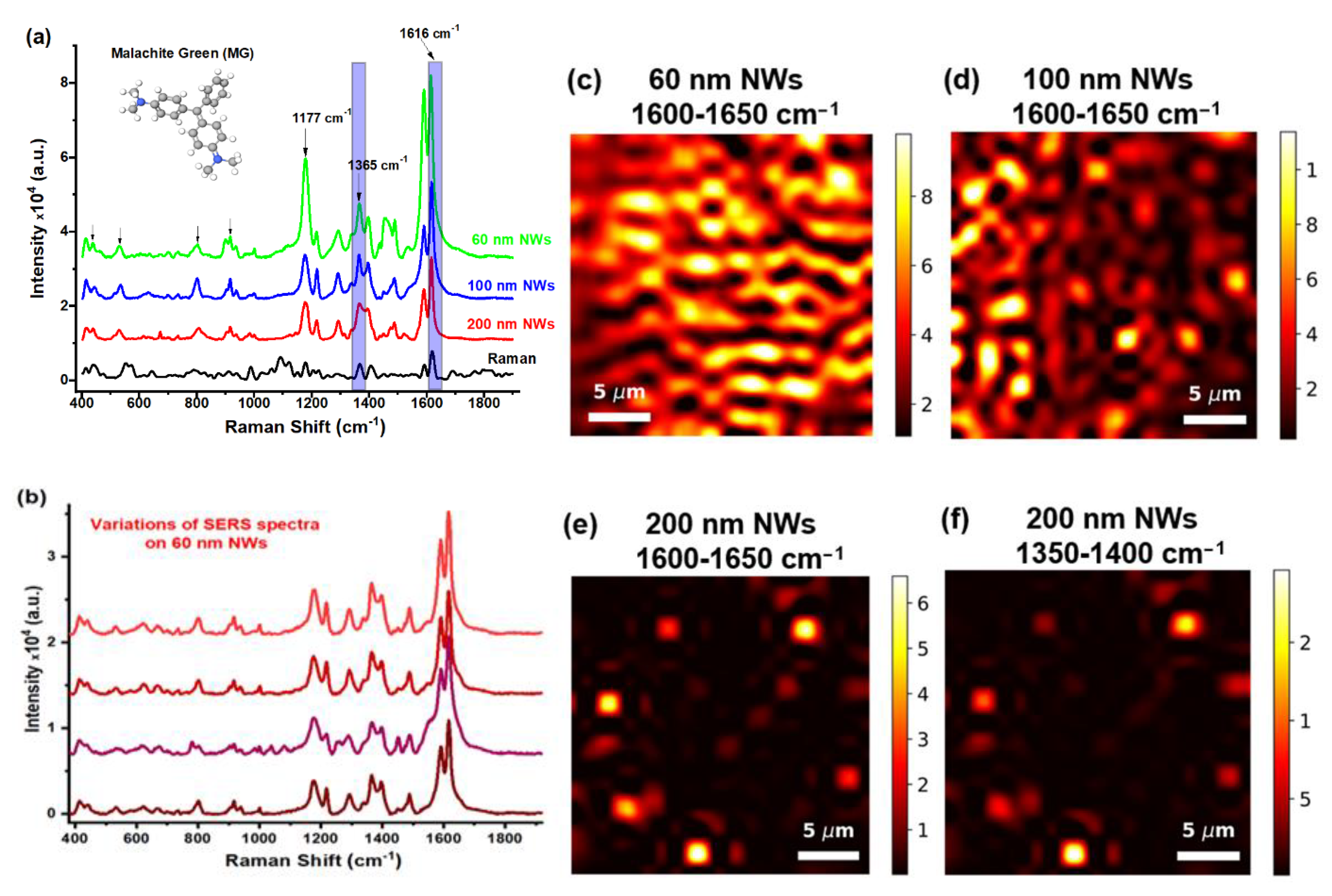

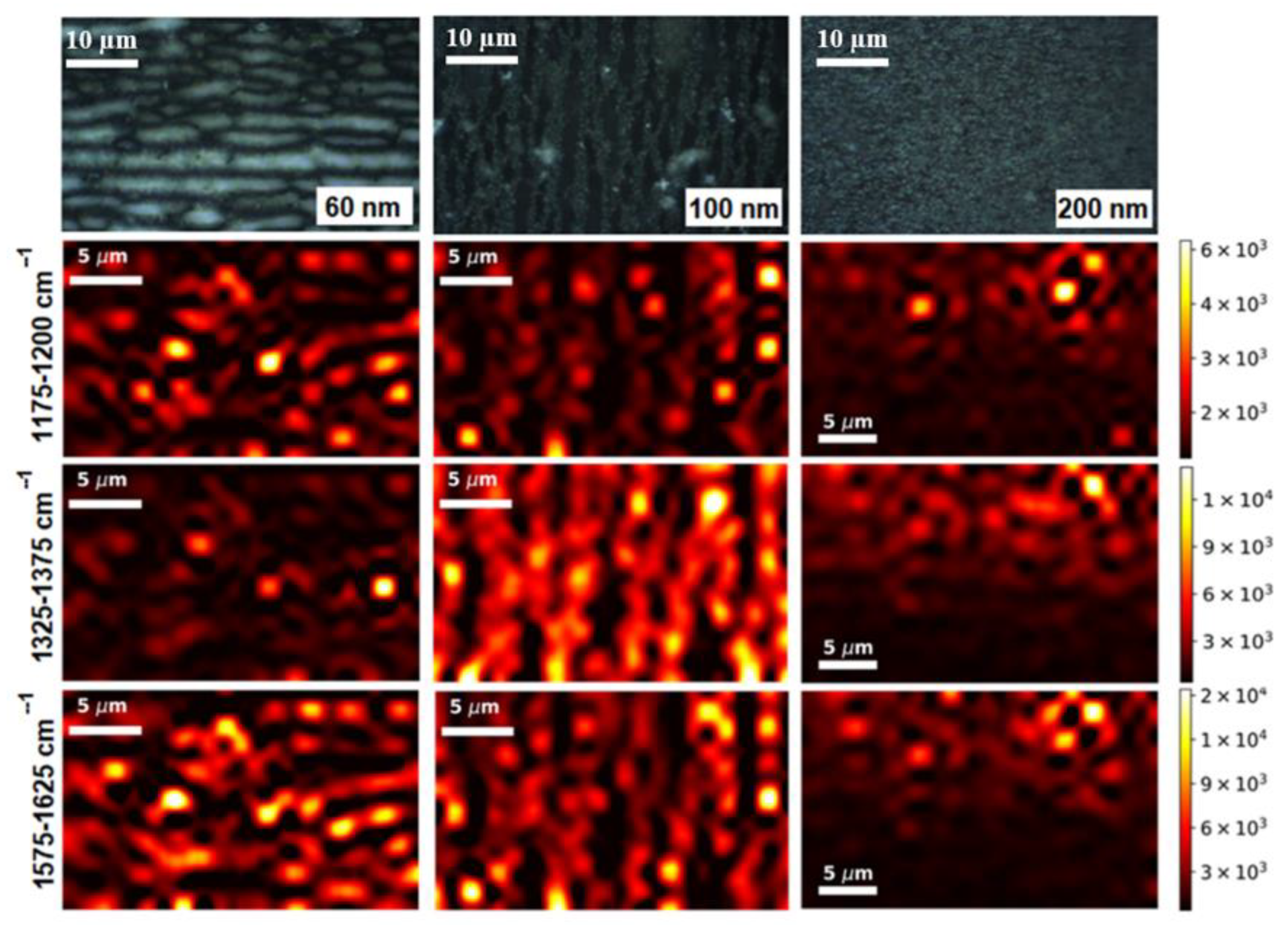
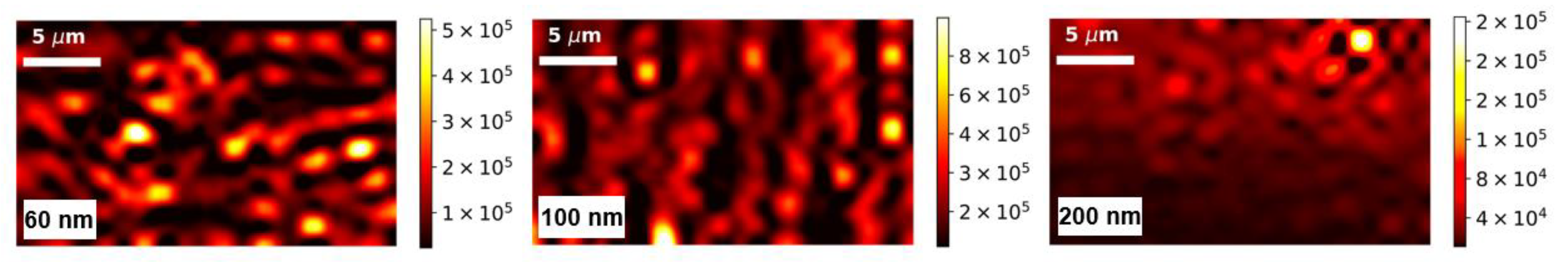

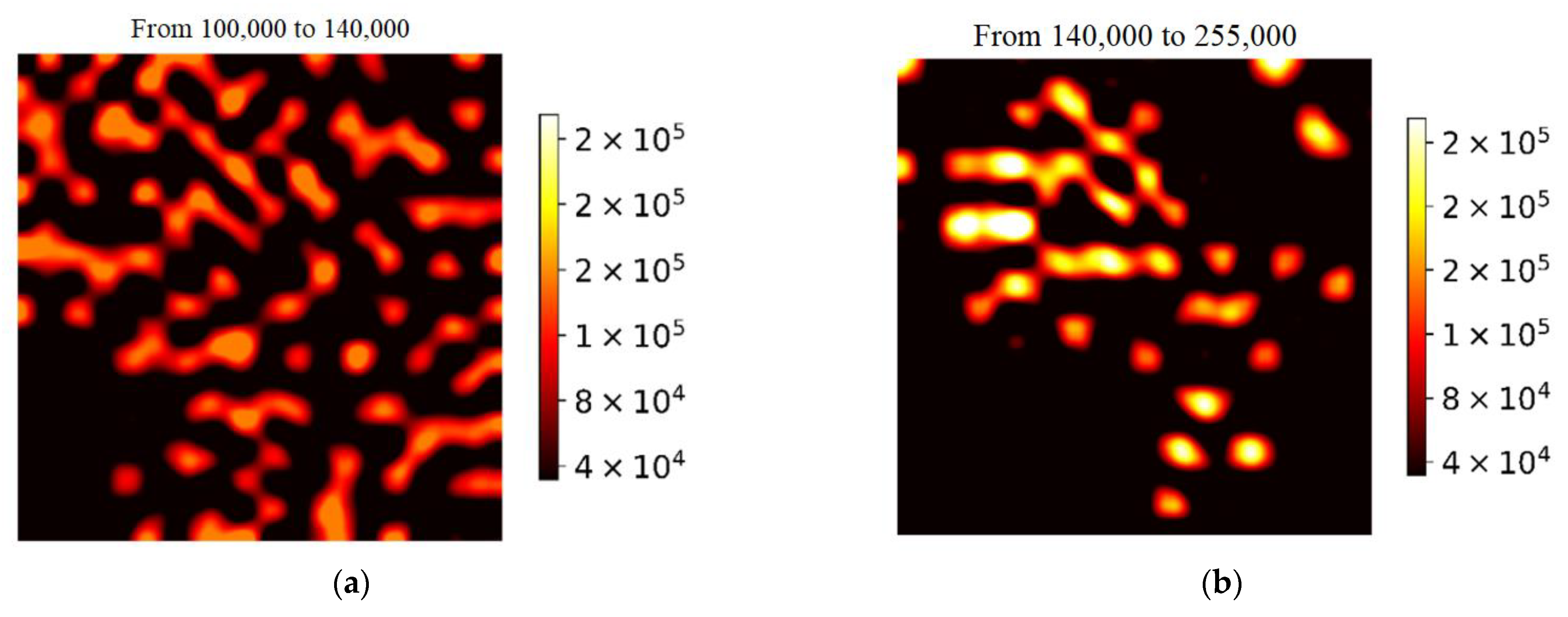
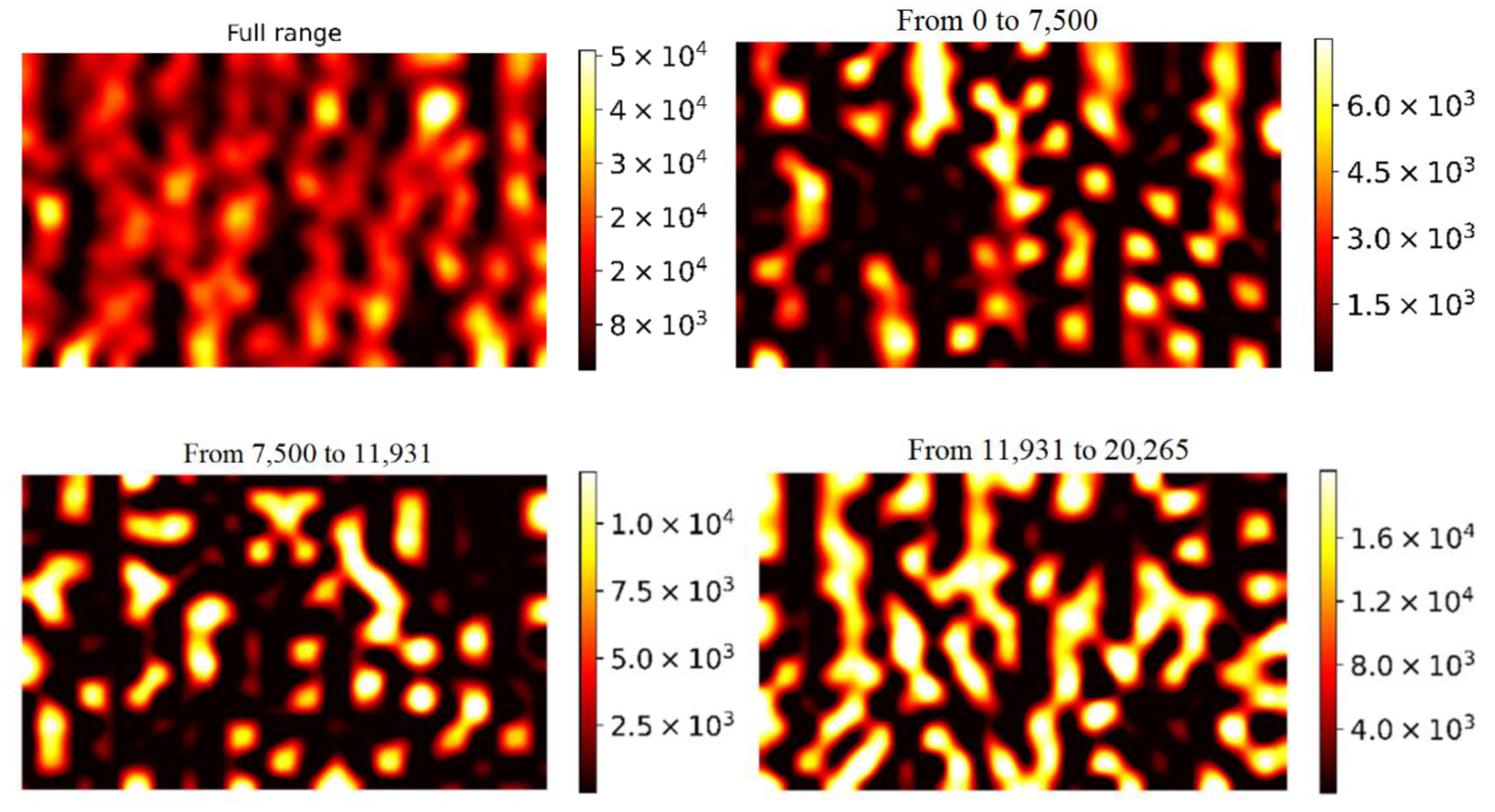

| Adsorbed Molecules | NW Diameter, nm | Noise Threshold | Enhancing Area, % | Mean | Median | η, % |
|---|---|---|---|---|---|---|
| MG | 60 | 87,764 | 59 | 100,722 | 98,750 | 1.96 |
| 100 | 5600 | 42 | 63,293 | 50,685 | 19.92 | |
| 200 | 3248 | 27 | 4884 | 2456 | 49.71 | |
| Phthalocyanine | 60 | 3989 | 63 | 6900 | 5188 | 24.81 |
| 100 | 6060 | 77 | 13,068 | 12,720 | 2.66 | |
| 200 | 963 | 61 | 1630 | 1251 | 23.25 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kozhina, E.; Bedin, S.; Martynov, A.; Andreev, S.; Piryazev, A.; Grigoriev, Y.; Gorbunova, Y.; Naumov, A. Ultrasensitive Optical Fingerprinting of Biorelevant Molecules by Means of SERS-Mapping on Nanostructured Metasurfaces. Biosensors 2023, 13, 46. https://doi.org/10.3390/bios13010046
Kozhina E, Bedin S, Martynov A, Andreev S, Piryazev A, Grigoriev Y, Gorbunova Y, Naumov A. Ultrasensitive Optical Fingerprinting of Biorelevant Molecules by Means of SERS-Mapping on Nanostructured Metasurfaces. Biosensors. 2023; 13(1):46. https://doi.org/10.3390/bios13010046
Chicago/Turabian StyleKozhina, Elizaveta, Sergey Bedin, Alexander Martynov, Stepan Andreev, Alexey Piryazev, Yuri Grigoriev, Yulia Gorbunova, and Andrey Naumov. 2023. "Ultrasensitive Optical Fingerprinting of Biorelevant Molecules by Means of SERS-Mapping on Nanostructured Metasurfaces" Biosensors 13, no. 1: 46. https://doi.org/10.3390/bios13010046
APA StyleKozhina, E., Bedin, S., Martynov, A., Andreev, S., Piryazev, A., Grigoriev, Y., Gorbunova, Y., & Naumov, A. (2023). Ultrasensitive Optical Fingerprinting of Biorelevant Molecules by Means of SERS-Mapping on Nanostructured Metasurfaces. Biosensors, 13(1), 46. https://doi.org/10.3390/bios13010046







