Quantitation of MicroRNA-155 in Human Cells by Heterogeneous Enzyme-Linked Oligonucleotide Assay Coupled with Mismatched Catalytic Hairpin Assembly Reaction
Abstract
:1. Introduction
2. Experimental
2.1. Reagents, Materials, and Equipment
2.2. Measurement of MicroRNA Concentration
2.3. Purification and Quantitation of Cellular MicroRNAs
3. Results and Discussion
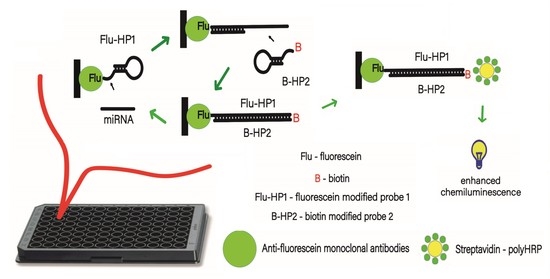
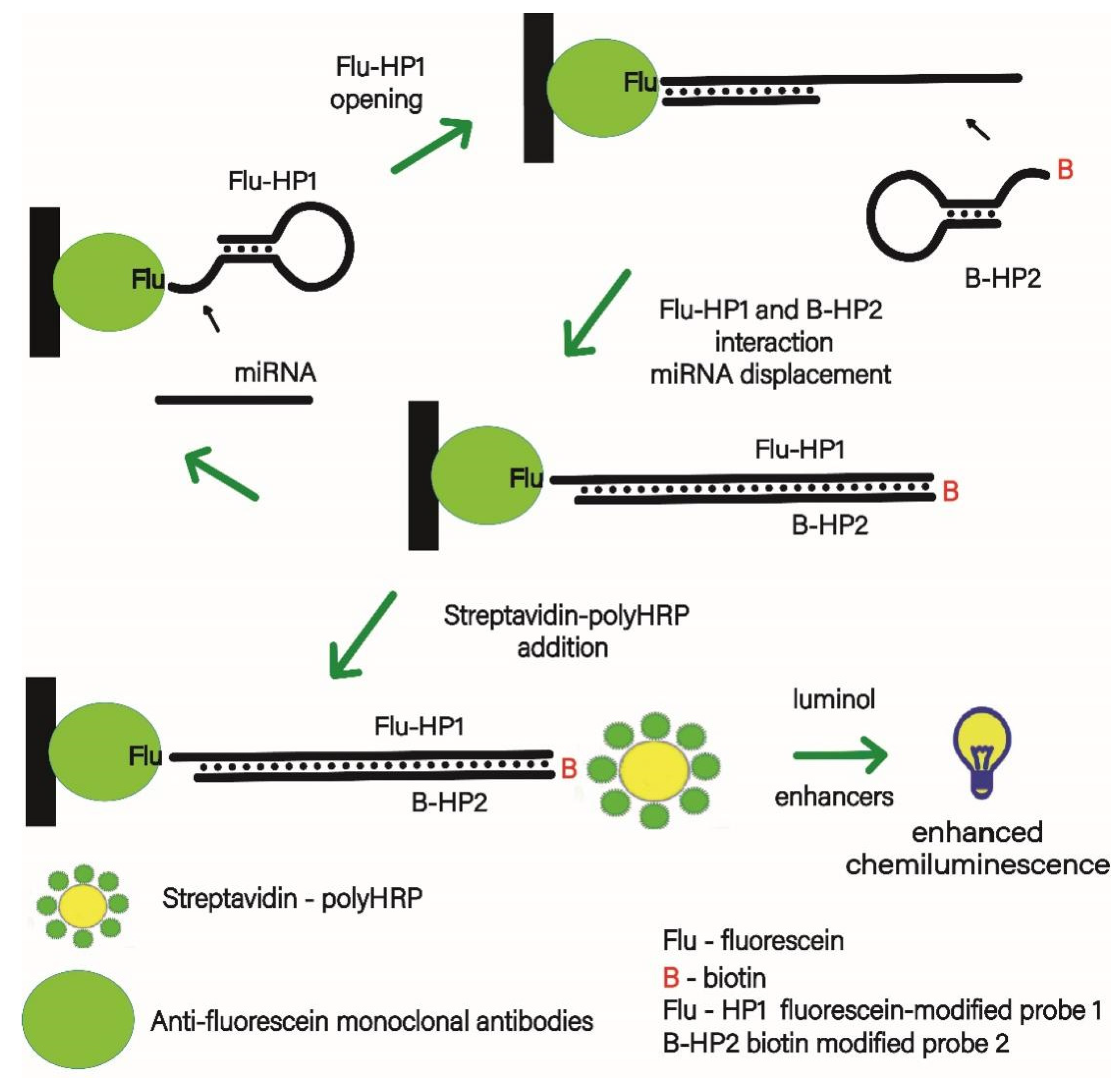
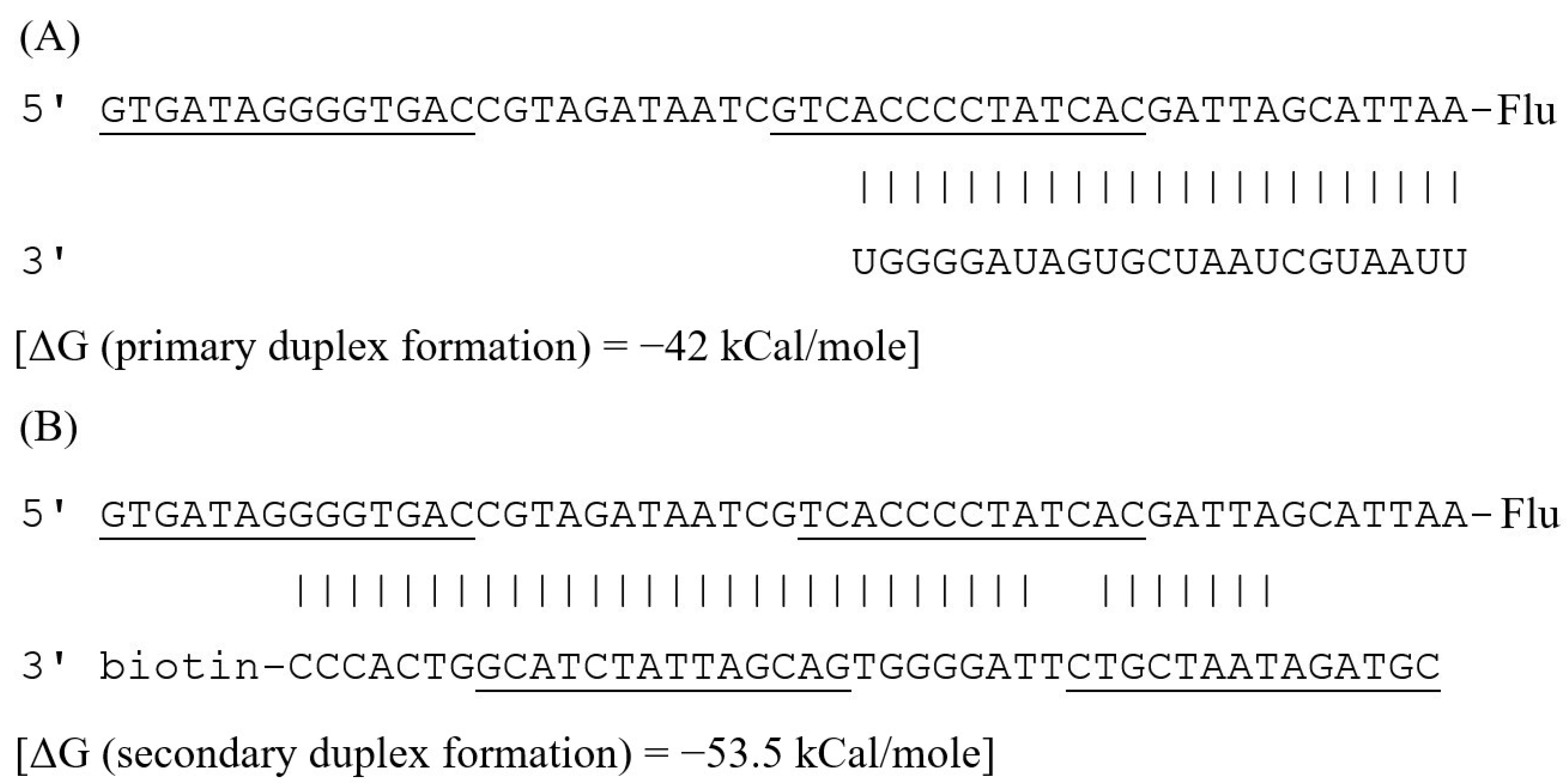
3.1. Design of the MicroRNA Assay
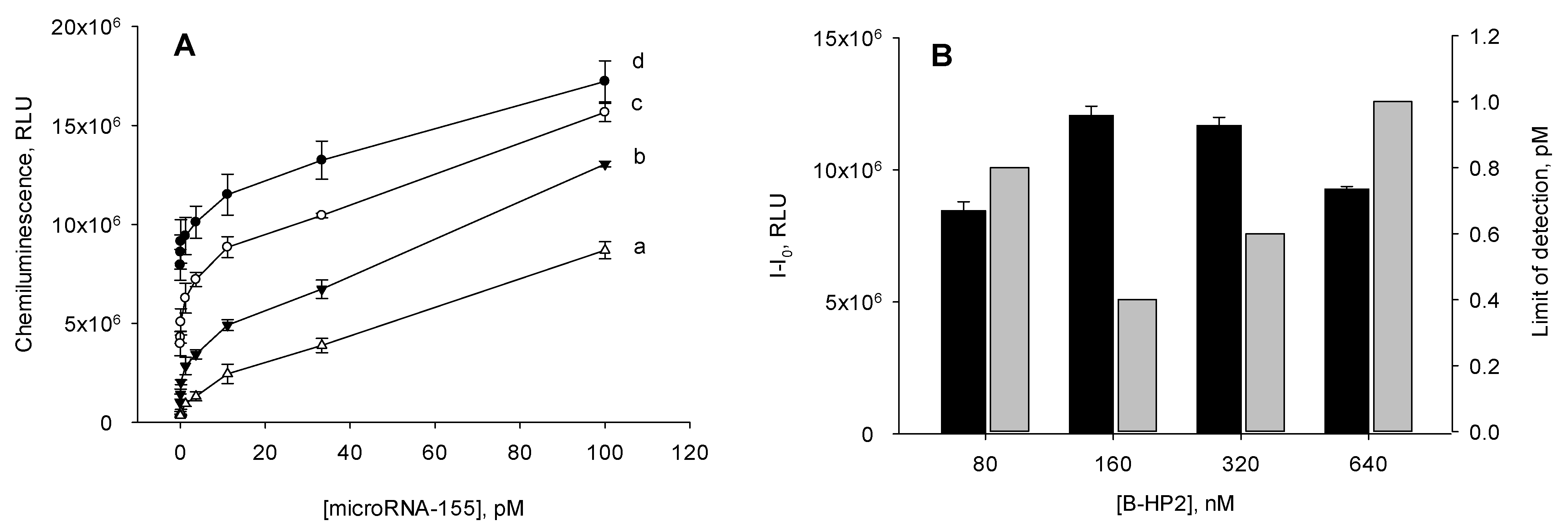
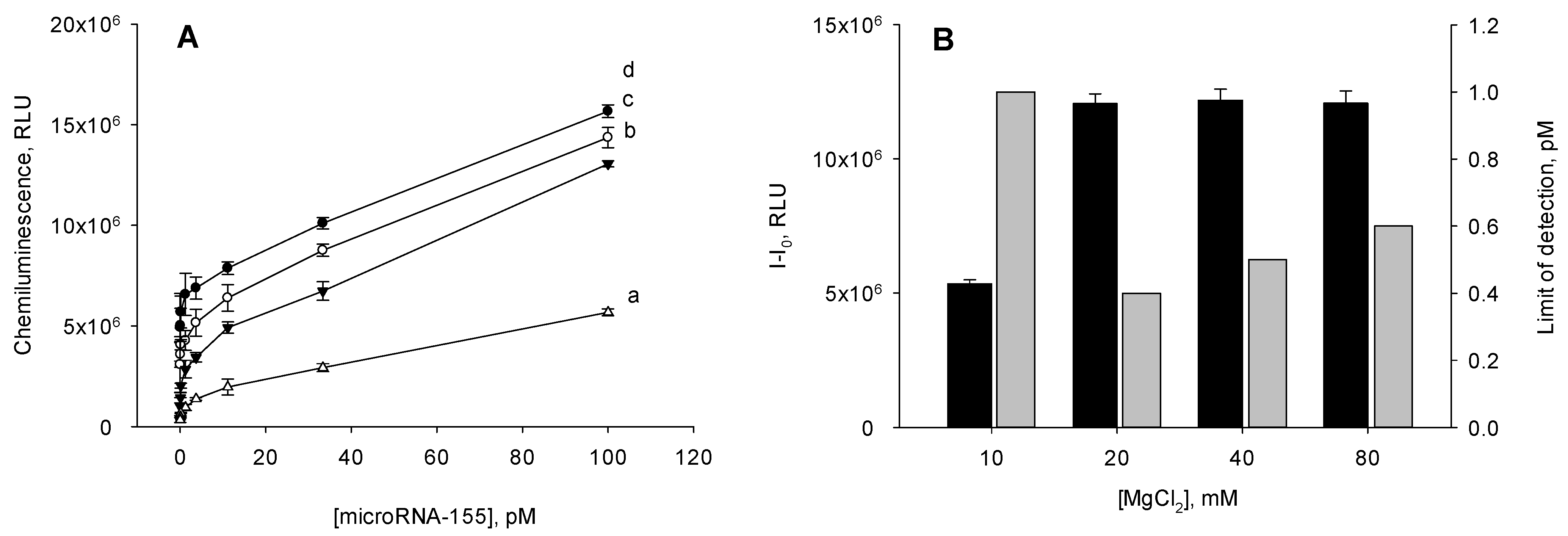
3.2. Optimization of the Assay Conditions for MicroRNA-155 Detection
3.3. Analytical Parameters of the Heterogenous mCHA-Based Assay of MicroRNA-155
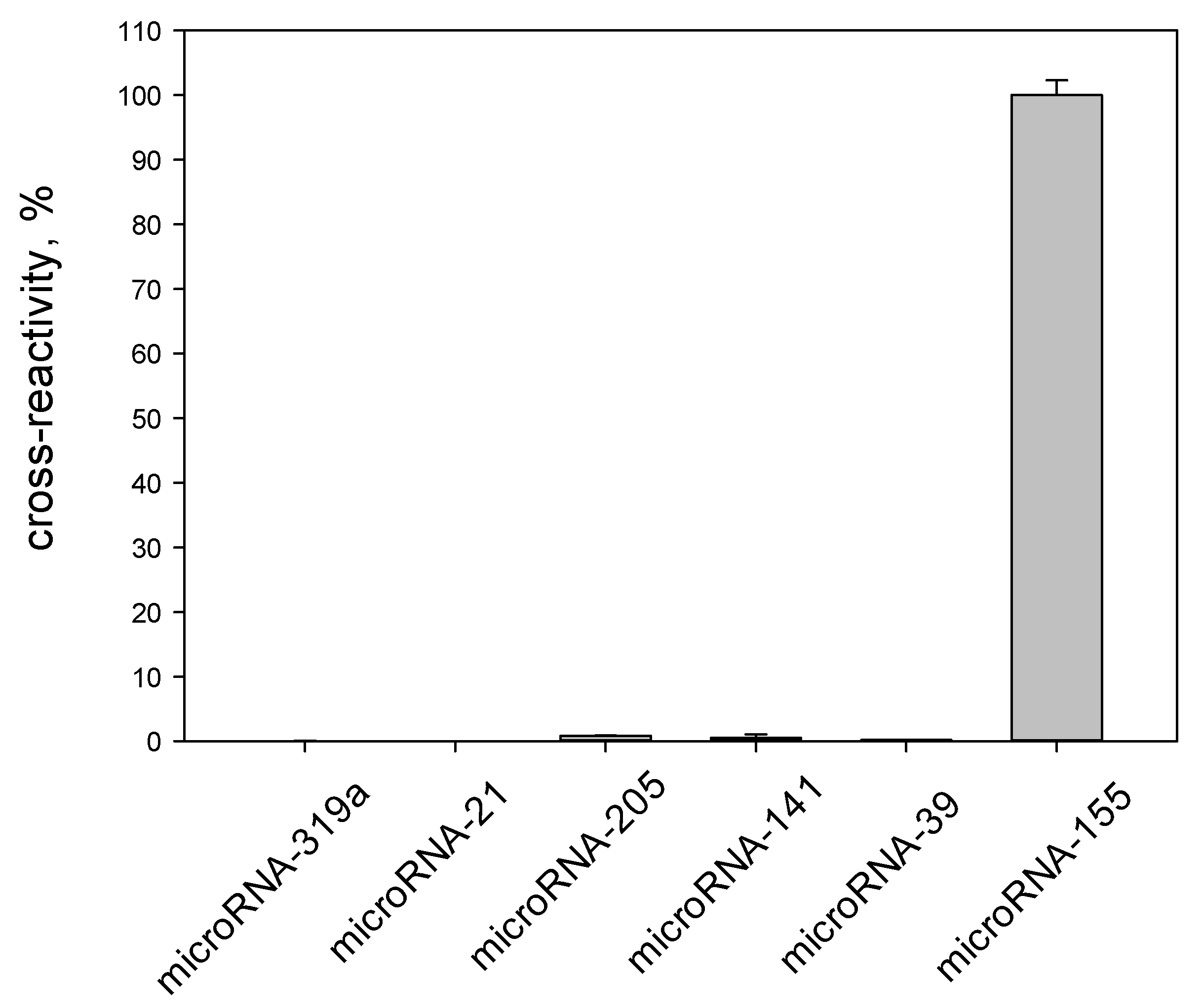
3.4. Specificity
3.5. Detection of MicroRNAs in Human Cells
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pritchard, C.C.; Cheng, H.H.; Tewari, M. MicroRNA profiling: Approaches and considerations. Nat. Rev. Gen. 2012, 13, 358–369. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. MiRBase: From miRNA sequences to function. Nucl. Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Garcia, I.; Miska, E.A. MicroRNA functions in animal development and human disease. Development 2005, 132, 4653–4662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plotnikova, O.; Baranova, A.; Skoblov, M. Comprehensive analysis of human microRNA–mRNA interactome. Front. Gen. 2019, 10, 933. [Google Scholar] [CrossRef] [PubMed]
- Due, H.; Svendsen, P.; Bødker, J.S.; Schmitz, A.; Bøgsted, M.; Johnsen, H.E.; El-Galaly, T.C.; Stidsholt Roug, A.; Dybkær, K. MiR-155 as a Biomarker in B-Cell Malignancies. Biomed. Res. Intern. 2016, 2016, 9513037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Williams, Z.; Ben-Dov, I.Z.; Elias, R.; Mihailovic, A.; Brown, M.; Rosenwaks, Z.; Tuschl, T. Comprehensive profiling of circulating miRNA via small RNA sequencing of cDNA libraries reveals biomarker potential and limitations. Proc. Natl. Acad. Sci. USA 2013, 110, 4255–4260. [Google Scholar] [CrossRef] [Green Version]
- Labib, M.; Berezovski, M.V. Electrochemical sensing of miRNAs: Avenues and paradigms. Biosens. Bioelectron. 2015, 68, 83–94. [Google Scholar] [CrossRef]
- Tang, Y.; Liu, M.; Zhao, Z.; Li, Q.; Liang, X.; Tian, J.; Zhao, S. Fluorometric determination of microRNA-122 by using ExoIII-aided recycling amplification and polythymine induced formation of copper nanoparticles. Microchim. Acta 2019, 186, 133. [Google Scholar] [CrossRef]
- Chen, S.; Zhao, J.; Xu, C.; Sakharov, I.Y.; Zhao, S. Absolute Quantification of MicroRNAs in a Single Cell with CL Detection Based on Rolling Circle Amplification on a Microchip Platform. Anal. Chem. 2021, 93, 9218–9225. [Google Scholar] [CrossRef]
- Bodulev, O.L.; Sakharov, I.Y. Modern Methods for Assessment of microRNAs. Biochemistry 2022, 87, 425–442. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Chen, F.; Li, Q.; Wang, L.; Fan, C. Isothermal Amplification of Nucleic Acids. Chem. Rev. 2015, 115, 12491–12545. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Xu, M.; Tian, X.; Cai, S.; Zeng, S. Research advances in the detection of miRNA. J. Pharm. Anal. 2019, 9, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Hou, J.; Jin, W.; Li, J.; Yue, Y.; Jin, H.; Wang, X. Increased circulating microRNA-155 as a potential biomarker for breast cancer screening: A meta-analysis. Molecules 2014, 19, 6282–6293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, H.; Fang, C.; Nam, S.; Cai, Q.; Long, X. The clinicopathological significance of microRNA-155 in breast cancer: A meta-analysis. BioMed Res. Int. 2014, 2014, 724209. [Google Scholar] [CrossRef]
- Jiang, S.; Zhang, H.W.; Lu, M.H.; He, X.H.; Li, Y.; Gu, H.; Liu, M.F.; Wang, E.D. MicroRNA-155 Functions as an OncomiR in Breast Cancer by Targeting the Suppressor of Cytokine Signaling 1 Gene. Cancer Res. 2010, 70, 31193127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Wang, M.; Lin, G.; Sun, S.; Li, X.; Qi, J.; Li, J. Serum MicroRNA-155 as a Potential Biomarker to Track Disease in Breast Cancer. PLoS ONE 2012, 7, e47003. [Google Scholar] [CrossRef]
- Han, F.; Zhao, J. Colorimetric Fluoride-Anion Sensor Based on Intramolecular Hydrogen Bonding and Enol-Keto Tautomerization of a Phenothiazine Derivative. Helv. Chim. Acta 2008, 91, 635–645. [Google Scholar] [CrossRef]
- Solovjev, A.M.; Galkin, I.I.; Pletjushkina, O.Y.; Medvedko, A.V.; Zhao, S.; Sakharov, I.Y. Isothermal chemiluminescent assay based on circular stand-displacement polymerization reaction amplification for cel-miRNA-39-3p determination in cell extracts. Int. J. Biol. Macromol. 2021, 182, 987–992. [Google Scholar] [CrossRef]
- Solovjev, A.M.; Kurzeev, S.A.; Sakharov, I.Y. Chemiluminescent microplate-based assay of DNA based on isothermal circular strand-displacement polymerization reaction (ICSDPR). Talanta 2020, 215, 120895. [Google Scholar] [CrossRef]
- Bodulev, O.L.; Zhao, S.; Sakharov, I.Y. Improving the sensitivity of the miRNA assay coupled with the mismatched catalytic hairpin assembly reaction by optimization of hairpin annealing conditions. Anal. Chem. 2021, 93, 6824–6830. [Google Scholar] [CrossRef]
- Sakharov, I.Y.; Demiyanova, A.S.; Gribas, A.V.; Uskova, N.A.; Efremov, E.E.; Vdovenko, M.M. 3-(10′-Phenothiazinyl)propionic acid is a potent primary enhancer of peroxidase-induced chemiluminescence and its application in sensitive ELISA of methylglyoxal-modified low density lipoprotein. Talanta 2013, 115, 414–417. [Google Scholar] [CrossRef]
- Jiang, Y.S.; Bhadra, S.; Li, B.; Ellington, A.D. Mismatches improve the performance of strand-displacement nucleic acid circuits. Angew. Chem. Int. Ed. Engl. 2014, 126, 1876–1879. [Google Scholar] [CrossRef] [Green Version]
- Braunlin, W.H.; Giri, I.; Beadling, L.; Breslauer, K.J. Conformational screening of oligonucleotides by variable-temperature high performance liquid chromatography: Dissecting the duplex–hairpin–coil equilibria of d(CGCGAATTCGCG). Biopolymers 2004, 74, 221–231. [Google Scholar] [CrossRef]
- Bodulev, O.L.; Burkin, K.M.; Efremov, E.E.; Sakharov, I.Y. One-pot microplate-based chemiluminescent assay coupled with catalytic hairpin assembly amplification for DNA detection. Anal. Bioanal. Chem. 2020, 412, 5105–5111. [Google Scholar] [CrossRef]
- Špringer, T.; Šípová, H.; Vaisocherová, H.; Štěpánek, J.; Homola, J. Shielding effect of monovalent and divalent cations on solid-phase DNA hybridization: Surface plasmon resonance biosensor study. Nucl. Acids Res. 2010, 38, 7343–7351. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.N.; Yu, L.; Zhao, X.S.; Zhang, W.; Wan, J.; Yu, B. Establishment of plasma microRNA detection method by using Taqman probe based quantitative reverse transcription PCR. Cell. Mol. Biol. 2015, 61, 51–56. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bodulev, O.L.; Galkin, I.I.; Zhao, S.; Pletyushkina, O.Y.; Sakharov, I.Y. Quantitation of MicroRNA-155 in Human Cells by Heterogeneous Enzyme-Linked Oligonucleotide Assay Coupled with Mismatched Catalytic Hairpin Assembly Reaction. Biosensors 2022, 12, 570. https://doi.org/10.3390/bios12080570
Bodulev OL, Galkin II, Zhao S, Pletyushkina OY, Sakharov IY. Quantitation of MicroRNA-155 in Human Cells by Heterogeneous Enzyme-Linked Oligonucleotide Assay Coupled with Mismatched Catalytic Hairpin Assembly Reaction. Biosensors. 2022; 12(8):570. https://doi.org/10.3390/bios12080570
Chicago/Turabian StyleBodulev, Oleg L., Ivan I. Galkin, Shulin Zhao, Olga Y. Pletyushkina, and Ivan Y. Sakharov. 2022. "Quantitation of MicroRNA-155 in Human Cells by Heterogeneous Enzyme-Linked Oligonucleotide Assay Coupled with Mismatched Catalytic Hairpin Assembly Reaction" Biosensors 12, no. 8: 570. https://doi.org/10.3390/bios12080570
APA StyleBodulev, O. L., Galkin, I. I., Zhao, S., Pletyushkina, O. Y., & Sakharov, I. Y. (2022). Quantitation of MicroRNA-155 in Human Cells by Heterogeneous Enzyme-Linked Oligonucleotide Assay Coupled with Mismatched Catalytic Hairpin Assembly Reaction. Biosensors, 12(8), 570. https://doi.org/10.3390/bios12080570