Simple Detection of DNA Methyltransferase with an Integrated Padlock Probe
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Apparatus
2.3. MTase Activity Detection Procedures
2.4. Activity Detection of M.SssI with SYBRTM Gold
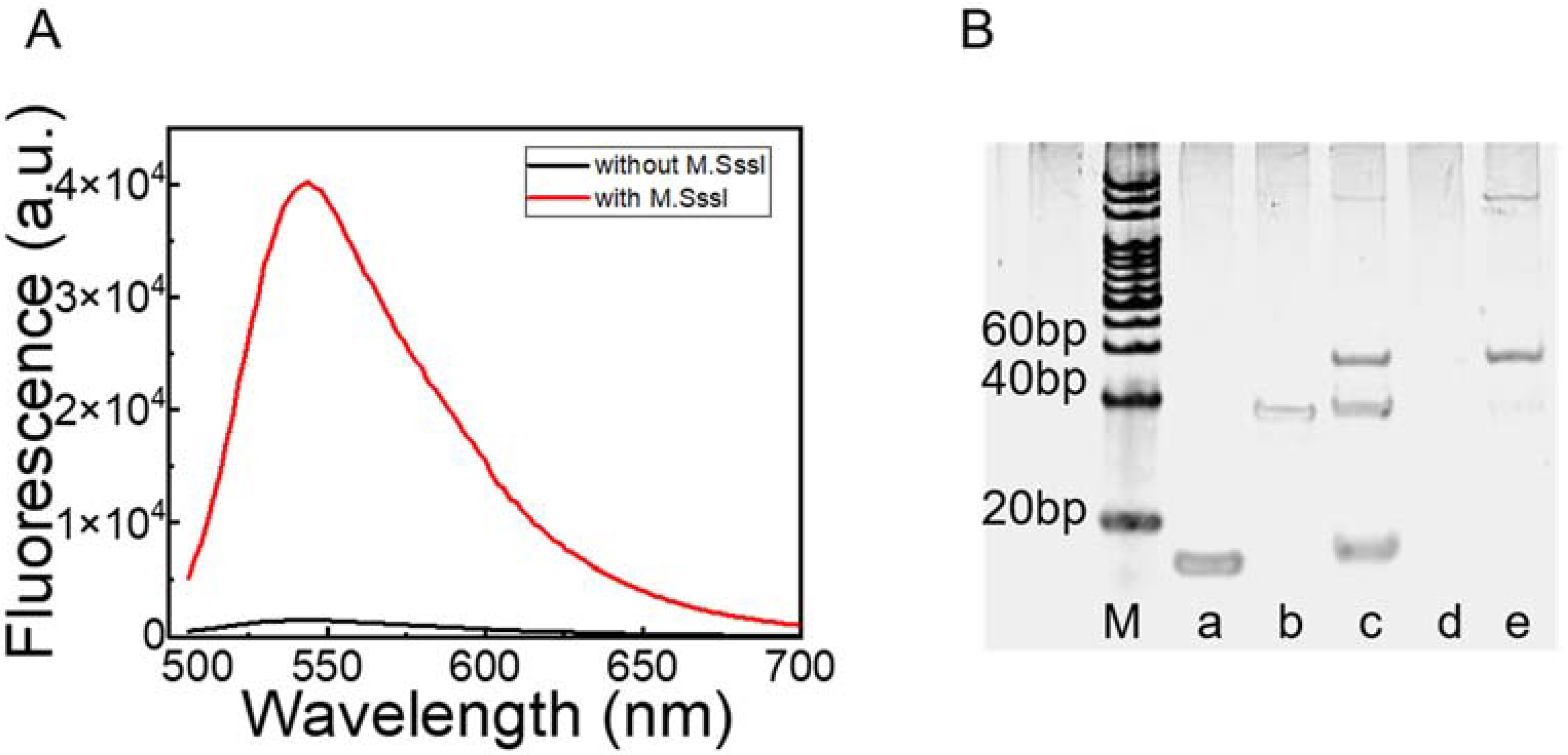
2.5. Gel Electrophoresis
2.6. Activity Detection of M.SssI in Human Healthy Serum and Selectivity of the Strategy
2.7. Evaluation of MTase Inhibitors
3. Results and Discussion
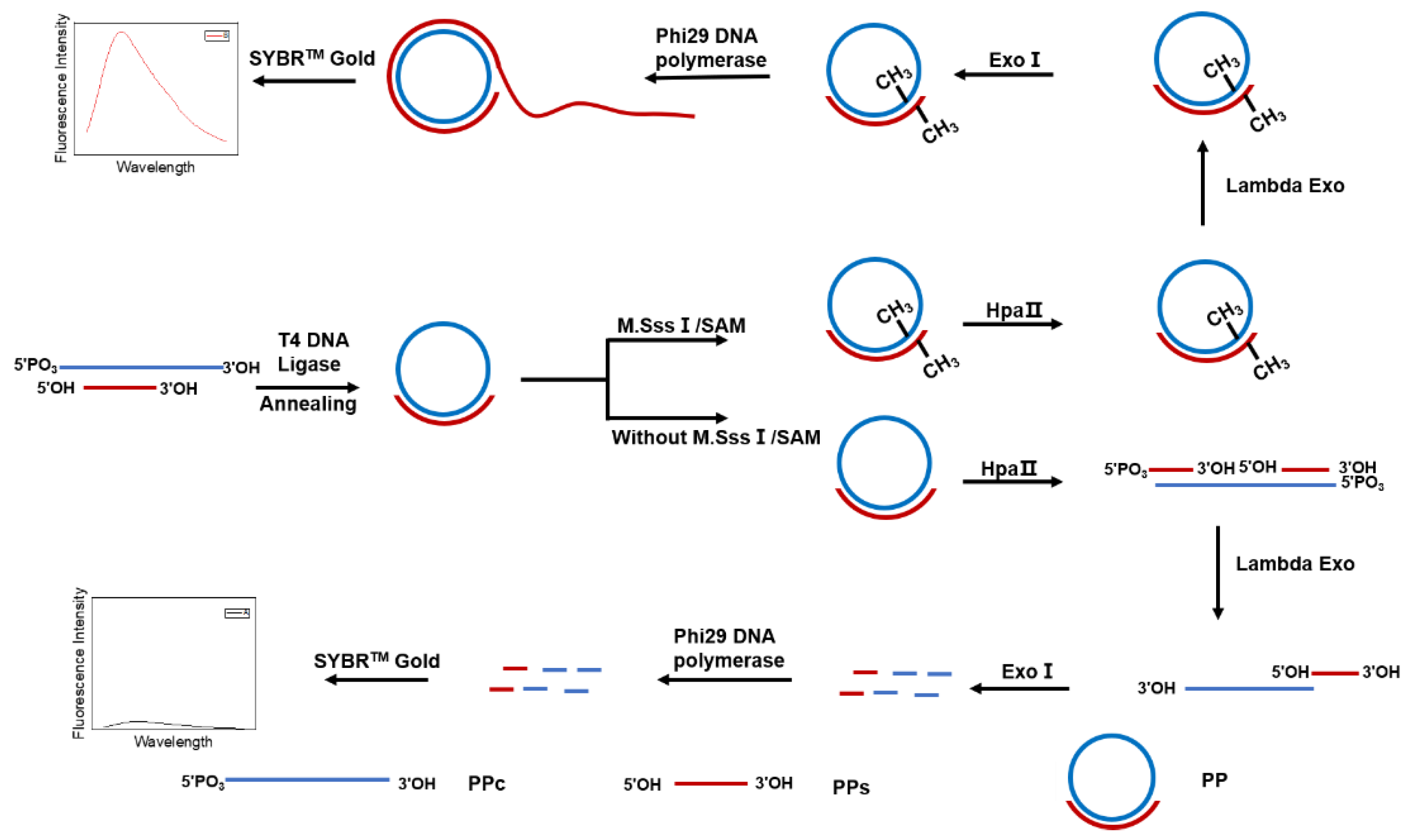
3.1. Scheme of M.SssI Activity Detection Using PP and RCA
3.2. Feasibility of M.SssI Verification Using PP and RCA
3.3. Optimization of the Experimental Conditions
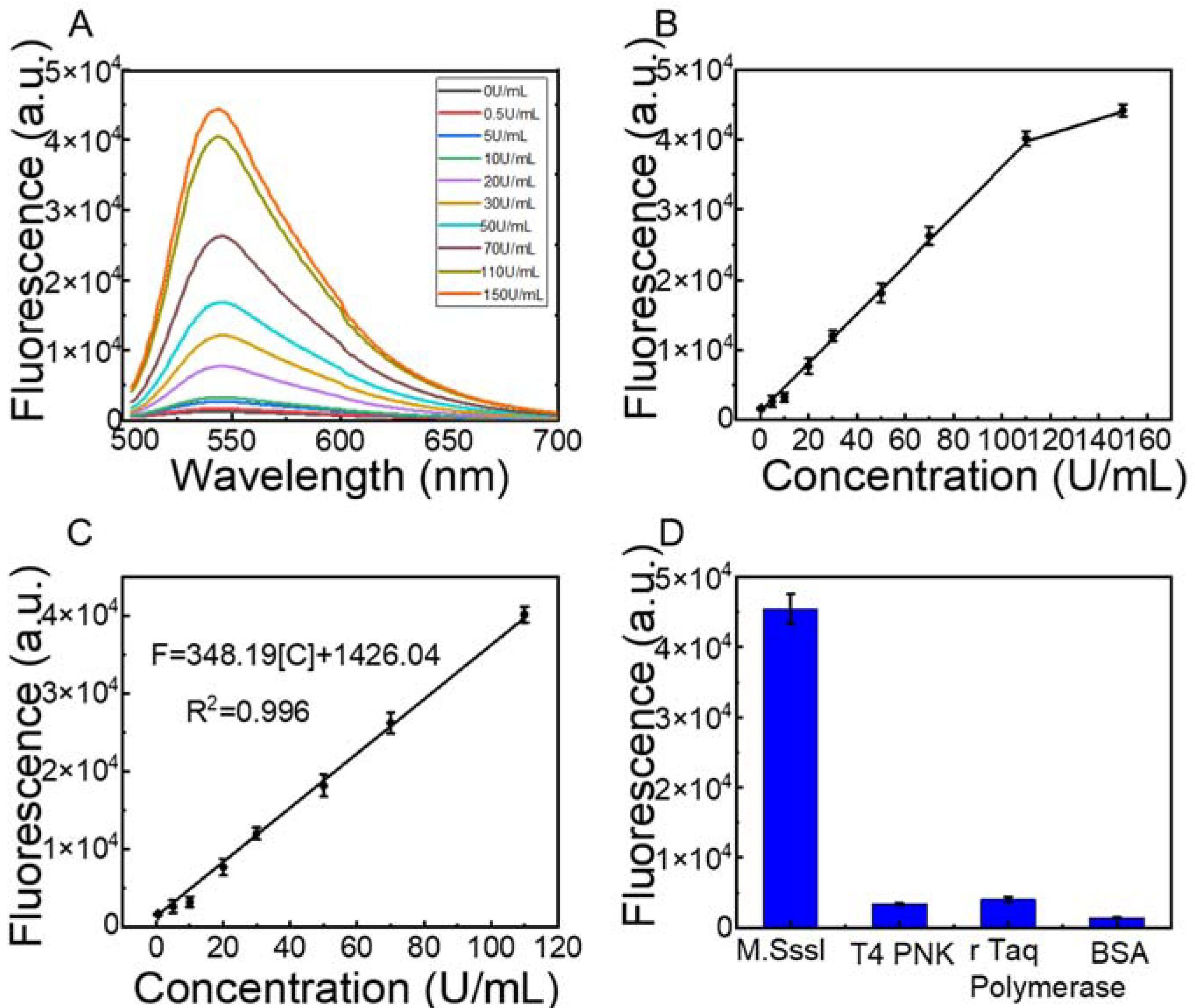
3.4. Sensitivity Detection of MTase and Selectivity of the Strategy
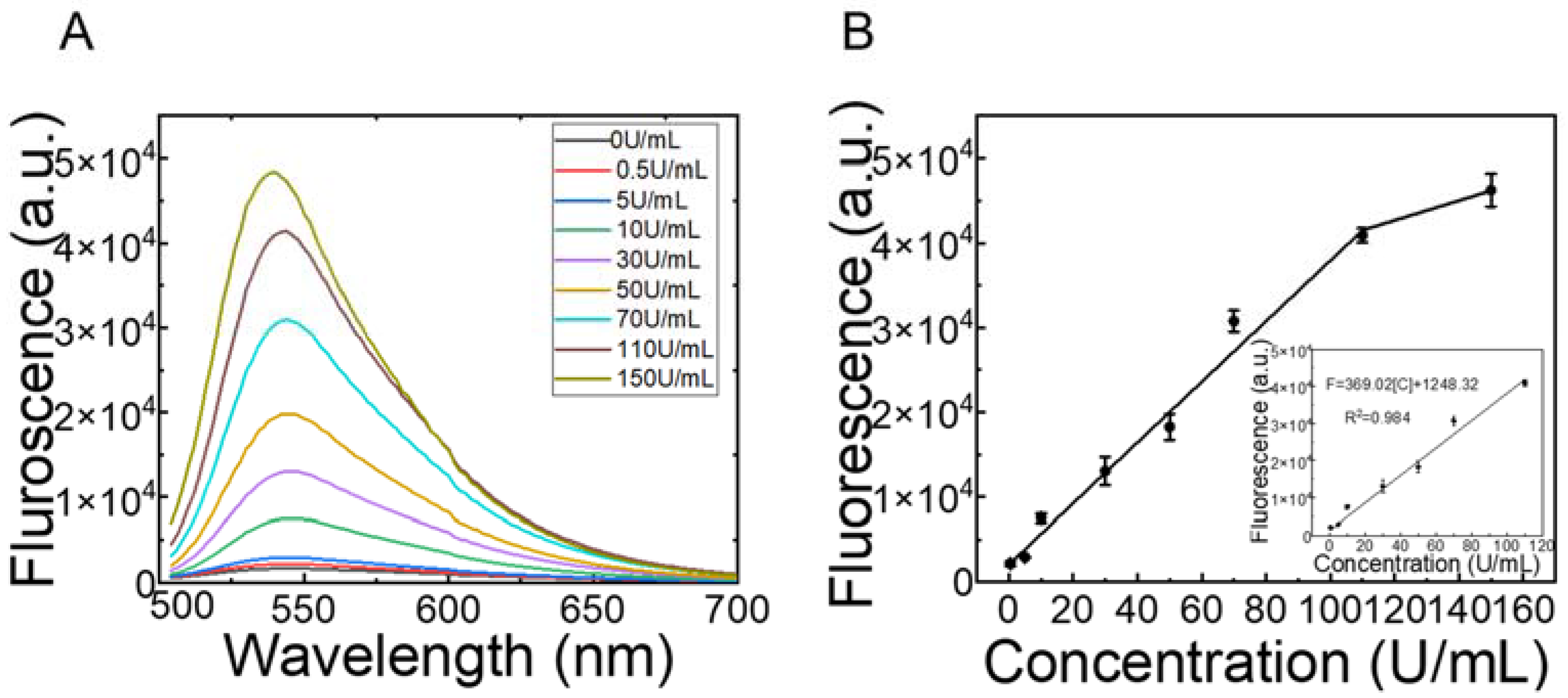
3.5. Detection of M.SssI Activity in 10% Human Serum
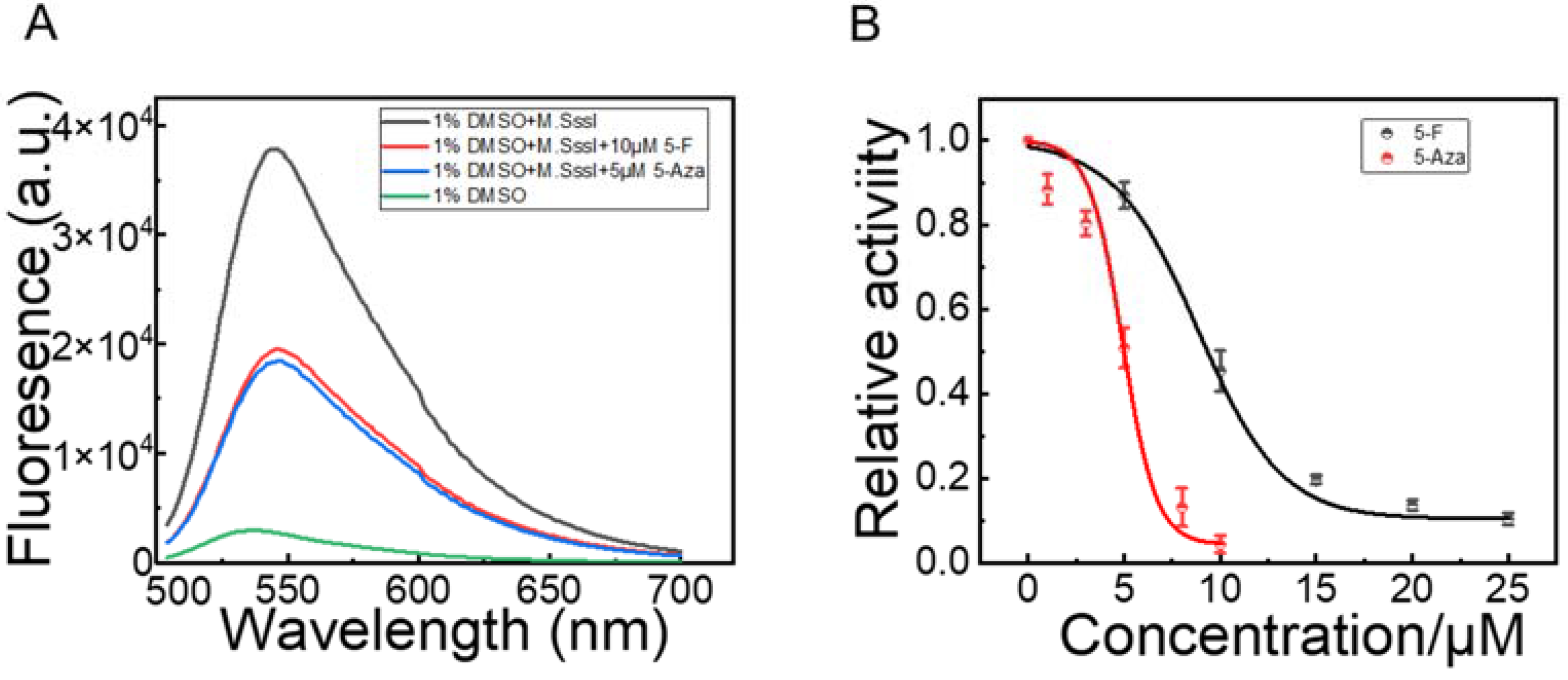
3.6. Inhibitory Activity Assay of M.SssI Inhibitors
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jurkowska, R.Z.; Jurkowski, T.P.; Jeltsch, A. Structure and Function of Mammalian DNA Methyltransferases. Chembiochem 2011, 12, 206–222. [Google Scholar] [CrossRef]
- Novak, P.; Jensen, T.J.; Garbe, J.C.; Stampfer, M.R.; Futscher, B.W. Stepwise DNA Methylation Changes Are Linked to Escape from Defined Proliferation Barriers and Mammary Epithelial Cell Immortalization. Cancer Res. 2009, 69, 5251–5258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodger, E.J.; Chatterjee, A.; Stockwell, P.A.; Eccles, M.R. Characterisation of DNA methylation changes in EBF3 and TBC1D16 associated with tumour progression and metastasis in multiple cancer types. Clin. Epigenetics 2019, 11, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Spainhour, J.C.G.; Lim, H.S.; Yi, S.V.; Qiu, P. Correlation Patterns Between DNA Methylation and Gene Expression in The Cancer Genome Atlas. Cancer Inform. 2019, 18, 1176935119828776. [Google Scholar] [CrossRef]
- VanderKraats, N.D.; Hiken, J.F.; Decker, K.F.; Edwards, J.R. Discovering high-resolution patterns of differential DNA methylation that correlate with gene expression changes. Nucleic Acids Res. 2013, 41, 6816–6827. [Google Scholar] [CrossRef]
- Sanchez-Romero, M.A.; Cota, I.; Casadesus, J. DNA methylation in bacteria: From the methyl group to the methylome. Curr. Opin. Microbiol. 2015, 25, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.-M.; Zhong, X.-L.; Wen, S.-H.; Zhang, L.; Liang, R.-P.; Qiu, J.-D. Colorimetric detection of methyltransferase activity based on the enhancement of CoOOH nanozyme activity by ssDNA. Sens. Actuators B-Chem. 2019, 281, 1073–1079. [Google Scholar] [CrossRef]
- Cui, L.; Zhao, M.-H.; Li, C.-C.; Wang, Q.; Luo, X.; Zhang, C.-Y. A Host-Guest Interaction-Based and Metal-Organic Gel-Based Biosensor with Aggregation-Induced Electrochemiluminescence Enhancement for Methyltransferase Assay. Anal. Chem. 2021, 93, 2974–2981. [Google Scholar] [CrossRef]
- Li, Y.; Wang, L.; Ding, C.; Luo, X. Highly selective ratiometric electrogenerated chemiluminescence assay of DNA methyltransferase activity via polyaniline and anti-fouling peptide modified electrode. Biosens. Bioelectron. 2019, 142, 111553. [Google Scholar] [CrossRef]
- Tian, R.; Liu, D.; Weng, T.; Yin, Y.; Xie, W.; Yin, B.; Shi, B.; Tlili, C.; Wang, D. DNA-functionalized biosensor for amplifying signal detection of DNA methyltransferase activity. J. Electroanal. Chem. 2021, 891, 115260. [Google Scholar] [CrossRef]
- Dadmehr, M.; Karimi, M.A.; Korouzhdehi, B. A signal-on fluorescence based biosensing platform for highly sensitive detection of DNA methyltransferase enzyme activity and inhibition. Spectrochim. Acta Part A-Mol. Biomol. Spectrosc. 2020, 228, 117731. [Google Scholar] [CrossRef] [PubMed]
- Kermani, H.A.; Hosseini, M.; Dadmehr, M.; Hosseinkhani, S.; Ganjali, M.R. DNA methyltransferase activity detection based on graphene quantum dots using fluorescence and fluorescence anisotropy. Sens. Actuators B-Chem. 2017, 241, 217–223. [Google Scholar] [CrossRef]
- Hu, J.; Liu, Y.; Zhang, C.Y. Construction of a single quantum dot nanosensor with the capability of sensing methylcytosine sites for sensitive quantification of methyltransferase. Nanoscale 2020, 12, 4519–4526. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Sun, S.; Tian, X.; Qiu, J.-G.; Jiang, B.; Zhang, C.Y. A dumbbell probe-based dual signal amplification strategy for sensitive detection of multiple DNA methyltransferases. Chem. Commun. 2020, 56, 13627–13630. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Liu, F.; Fan, T.; Ren, Y.; Jiang, Y. Rapid detection of methyltransferases utilizing dumbbell DNA-templated copper nanoparticles. Sens. Actuators B-Chem. 2018, 276, 499–506. [Google Scholar] [CrossRef]
- Chen, L.P.; Zhang, Y.; Xia, Q.; Luo, F.; Guo, L.H.; Qiu, B.; Lin, Z.Y. Fluorescence biosensor for DNA methyltransferase activity and related inhibitor detection based on methylation-sensitive cleavage primer triggered hyperbranched rolling circle ampli fication. Anal. Chim. Acta 2020, 1122, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.-L.; Xue, X.-X.; Deng, X.-M.; Jian, Y.-T.; Luo, J.; Jiang, M.-M.; Zheng, X.-J. Chemiluminescence strategy induced by HRP-sandwich structure based on strand displacement for sensitive detection of DNA methyltransferase. Microchem. J. 2020, 158, 105183. [Google Scholar] [CrossRef]
- Wang, Z.Y.; Li, P.; Cui, L.; Xu, Q.; Zhang, C.Y. Construction of a Universal and Label-Free Chemiluminescent Sensor for Accurate Quantification of Both Bacteria and Human Methyltransferases. Anal. Chem. 2020, 92, 13573–13580. [Google Scholar] [CrossRef]
- Du, Y.-C.; Wang, S.-Y.; Li, X.-Y.; Wang, Y.-X.; Tang, A.-N.; Kong, D.-M. Terminal deoxynucleotidyl transferase-activated nicking enzyme amplification reaction for specific and sensitive detection of DNA methyltransferase and polynucleotide kinase. Biosens. Bioelectron. 2019, 145, 111700. [Google Scholar] [CrossRef]
- Huang, J.; Li, X.-Y.; Du, Y.-C.; Zhang, L.-N.; Liu, K.-K.; Zhu, L.-N.; Kong, D.-M. Sensitive fluorescent detection of DNA methyltransferase using nicking endonuclease-mediated multiple primers-like rolling circle amplification. Biosens. Bioelectron. 2017, 91, 417–423. [Google Scholar] [CrossRef]
- Chen, R.; Shi, H.; Meng, X.; Su, Y.; Wang, H.; He, Y. Dual-Amplification Strategy-Based SERS Chip for Sensitive and Reproducible Detection of DNA Methyltransferase Activity in Human Serum. Anal. Chem. 2019, 91, 3597–3603. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Cui, M.; Zhou, H.; Zhang, S. DNA-hybrid-gated functional mesoporous silica for sensitive DNA methyltransferase SERS detection. Chem. Commun. 2015, 51, 13983–13985. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.; Wei, Y.; Xu, J.; Yuan, R.; Xiang, Y. Coupling hybridization chain reaction with DNAzyme recycling for enzyme-free and dual amplified sensitive fluorescent detection of methyltransferase activity. Anal. Chim. Acta 2017, 949, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Pan, M.; Wei, J.; Liu, X.; Wang, F. Evaluation of DNA Methyltransferase Activity and Inhibition via Isothermal Enzyme-Free Concatenated Hybridization Chain Reaction. Acs Sens. 2017, 2, 932–939. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Ma, H.; Li, W.; Nie, Z.; Yao, S. An entropy-driven signal amplifying strategy for real-time monitoring of DNA methylation process and high-throughput screening of methyltransferase inhibitors. Anal. Chim. Acta 2017, 970, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Rouhanifard, S.H.; Mellis, I.A.; Dunagin, M.; Bayatpour, S.; Jiang, C.L.; Dardani, I.; Symmons, O.; Emert, B.; Torre, E.; Cote, A. ClampFISH detects indivicual nucleic acid molecules using click chemistry-based amplification. Nat. Biotechnol. 2019, 37, 84–89. [Google Scholar] [CrossRef]
- Deng, R.; Zhang, K.; Li, J. Isothermal Amplification for MicroRNA Detection: From the Test Tube to the Cell. Acc. Chem. Res. 2017, 50, 1059–1068. [Google Scholar] [CrossRef] [PubMed]
- Berman, B.P.; Weisenberger, D.J.; Laird, P.W. Locking in on the human methylome. Nat. Biotechnol. 2009, 27, 341–342. [Google Scholar] [CrossRef]
- Cao, A.; Zhang, C.Y. Sensitive and Label-Free DNA Methylation Detection by Ligation-Mediated Hyperbranched Rolling Circle Amplification. Anal. Chem. 2012, 84, 6199–6205. [Google Scholar] [CrossRef]
- Diep, D.; Plongthongkum, N.; Gore, A.; Fung, H.-L.; Shoemaker, R.; Zhang, K. Library-free methylation sequencing with bisulfite padlock probes. Nat. Methods 2012, 9, 270–272. [Google Scholar] [CrossRef]
- Zhao, H.; Lu, Z.-H. Detection of DNA methylation by hyperbranched rolling circle amplification and DNA microarray. Chin. Chem. Lett. 2014, 25, 1559–1564. [Google Scholar] [CrossRef]
- Huang, R.; He, L.; Li, S.; Liu, H.; Jin, L.; Chen, Z.; Zhao, Y.; Li, Z.; Deng, Y.; He, N. A simple fluorescence aptasensor for gastric cancer exosome detection based on branched rolling circle amplification. Nanoscale 2020, 12, 2445–2451. [Google Scholar] [CrossRef]
- Li, X.; Meng, M.; Zheng, L.; Xu, Z.; Song, P.; Yin, Y.; Eremin, S.A.; Xi, R. Chemiluminescence Immunoassay for S-Adenosylhomocysteine Detection and Its Application in DNA Methyltransferase Activity Evaluation and Inhibitors Screening. Anal. Chem. 2016, 88, 8556–8561. [Google Scholar] [CrossRef] [PubMed]
- Raddatz, G. The Intestinal Microbiota Programs DnA Methylation to Control Tissue Homeostasis and Inflammation. In Gene Expression Omnibus. Available online: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE137037 (accessed on 15 March 2022).
- Yuan, F.-L.; Li, X.; Xu, R.-S.; Jiang, D.-L.; Zhou, X.-G. DNA Methylation: Roles in Rheumatoid Arthritis. Cell Biochem. Biophys. 2014, 70, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Feng, X.; Lv, Z.; Xu, X.; Lu, Y.; Wu, W.; Wu, H.; Liu, H.; Cao, L.; Ye, S. ACADS acts as a potential methylation biomarker associated with the proliferation and metastasis of hepatocellular carcinomas. Aging-Us 2019, 11, 8825–8844. [Google Scholar] [CrossRef] [PubMed]
- Nagahara, M.; Hoon, D.S.B. Sex Hormone Receptors and Epigenetics in Breast Cancer. In Female Sex Hormones and Cancers; George, G.C., Ed.; Nova Science Publishers: New York, NY, USA, 2010. [Google Scholar]
- Yang, M.; Chen, L.; Guo, L.; Qiu, B.; Lin, Z. High Sensitive Electrochemiluminescence Biosensor Based on Ru(phen)(3)(2+)-loaded Double Strand DNA as Signal Tags use to Detect DNA Methyltransferase Activity. Electroanalysis 2021, 34, 387–396. [Google Scholar] [CrossRef]
- Mao, X.; Wei, M.; Zhu, C.; Lu, J.; Gao, J.; Simon, A.J.; Shi, J.; Huang, Q.; Fan, C. Real Time in Vitro Regulation of DNA Methylation Using a 5-Fluorouracil Conjugated DNA-Based Stimuli-Responsive Platform. ACS Appl. Mater. Interfaces 2013, 5, 2604–2609. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Kang, T.; Zhu, J.; Wu, P.; Cai, C. Sensitivity-Improved SERS Detection of Methyltransferase Assisted by Plasmonically Engineered Nanoholes Array and Hybridization Chain Reaction. ACS Sens. 2020, 5, 3639–3648. [Google Scholar] [CrossRef]
- Zhang, Y.; Hao, L.; Zhao, Z.; Yang, X.; Wang, L.; Liu, S. Immuno-DNA binding directed template-free DNA extension and enzyme catalysis for sensitive electrochemical DNA methyltransferase activity assay and inhibitor screening. Analyst 2020, 145, 3064–3072. [Google Scholar] [CrossRef] [PubMed]
| Sample | Added (U/mL) | Measured (U/mL) a | Mean Recovery (%) b | RSD (%) c |
|---|---|---|---|---|
| 1 | 0.5 | 0.52 | 104.00 | 8.87 |
| 2 | 50 | 44.55 | 89.10 | 8.77 |
| 3 | 110 | 115.73 | 105.21 | 9.34 |
| Sample | Added (U/mL) | Measured (U/mL) a | Mean Recovery (%) b | RSD (%) c |
|---|---|---|---|---|
| 1 | 10 | 10.35 | 103.50 | 4.97 |
| 2 | 50 | 50.94 | 101.88 | 6.33 |
| 3 | 70 | 67.50 | 96.09 | 4.06 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Han, Y.; Zhou, F.; Fan, T.; Liu, F. Simple Detection of DNA Methyltransferase with an Integrated Padlock Probe. Biosensors 2022, 12, 569. https://doi.org/10.3390/bios12080569
Wang Y, Han Y, Zhou F, Fan T, Liu F. Simple Detection of DNA Methyltransferase with an Integrated Padlock Probe. Biosensors. 2022; 12(8):569. https://doi.org/10.3390/bios12080569
Chicago/Turabian StyleWang, Yuehua, Yingli Han, Fangyu Zhou, Tingting Fan, and Feng Liu. 2022. "Simple Detection of DNA Methyltransferase with an Integrated Padlock Probe" Biosensors 12, no. 8: 569. https://doi.org/10.3390/bios12080569
APA StyleWang, Y., Han, Y., Zhou, F., Fan, T., & Liu, F. (2022). Simple Detection of DNA Methyltransferase with an Integrated Padlock Probe. Biosensors, 12(8), 569. https://doi.org/10.3390/bios12080569




