Colorimetric Detection of DNase Type I 3′OH DNA Ends Using an Isothermal Amplification-Assisted Paper-Based Analytical Device
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
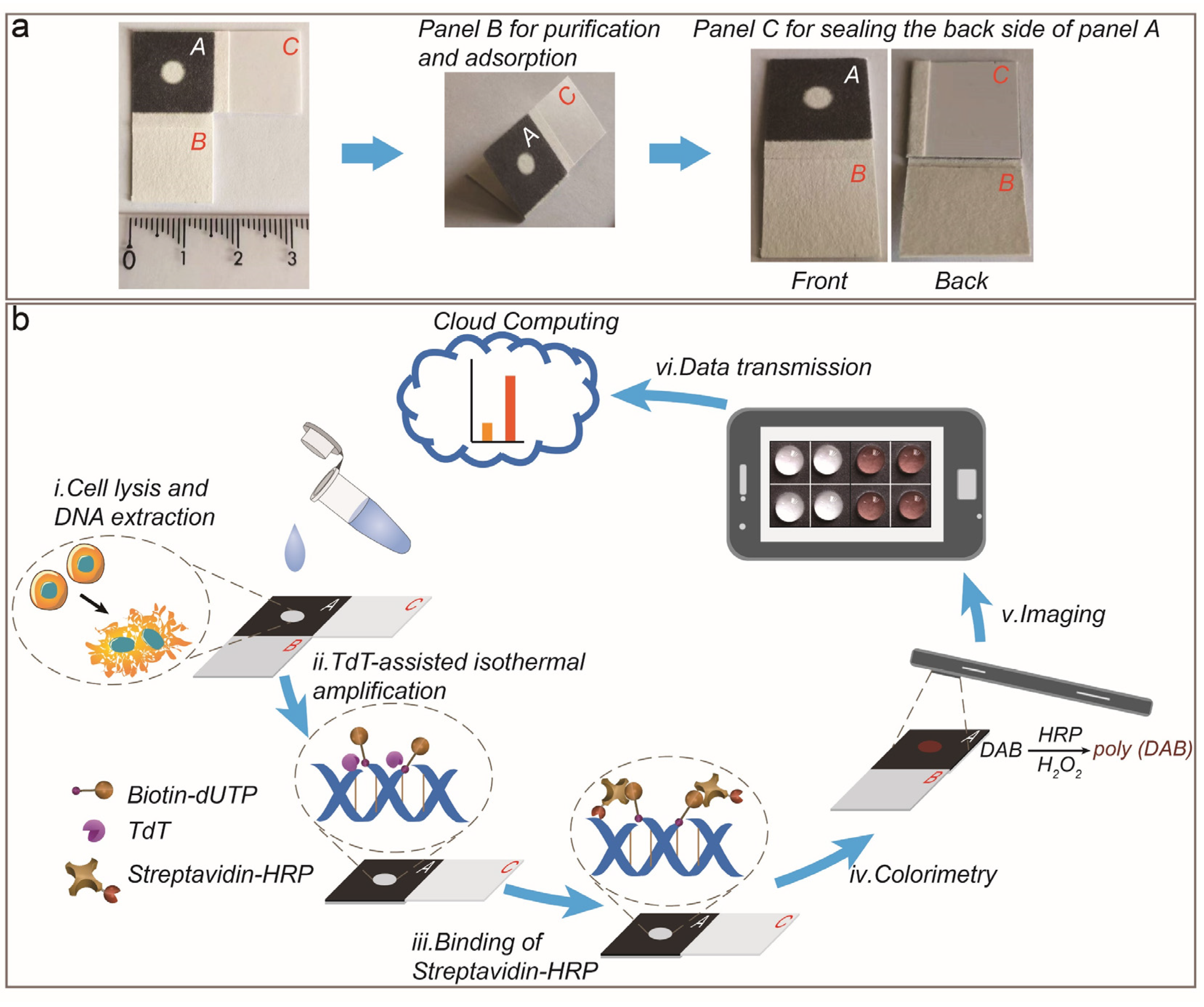
2.2. Fabrication of PAD
2.3. Detection of DNase Type I 3′OH DNA Ends by Standard TUNEL Assay
2.4. Colorimetric Detection of DNase Type I 3′OH DNA Ends by PAD
2.5. Analysis of Genotoxicity of Environmental Pollutants
2.6. Analysis of Genotoxicity of Wastewater Effluents
2.7. Comet Assay
3. Results and Discussion
3.1. Working Principle of PAD
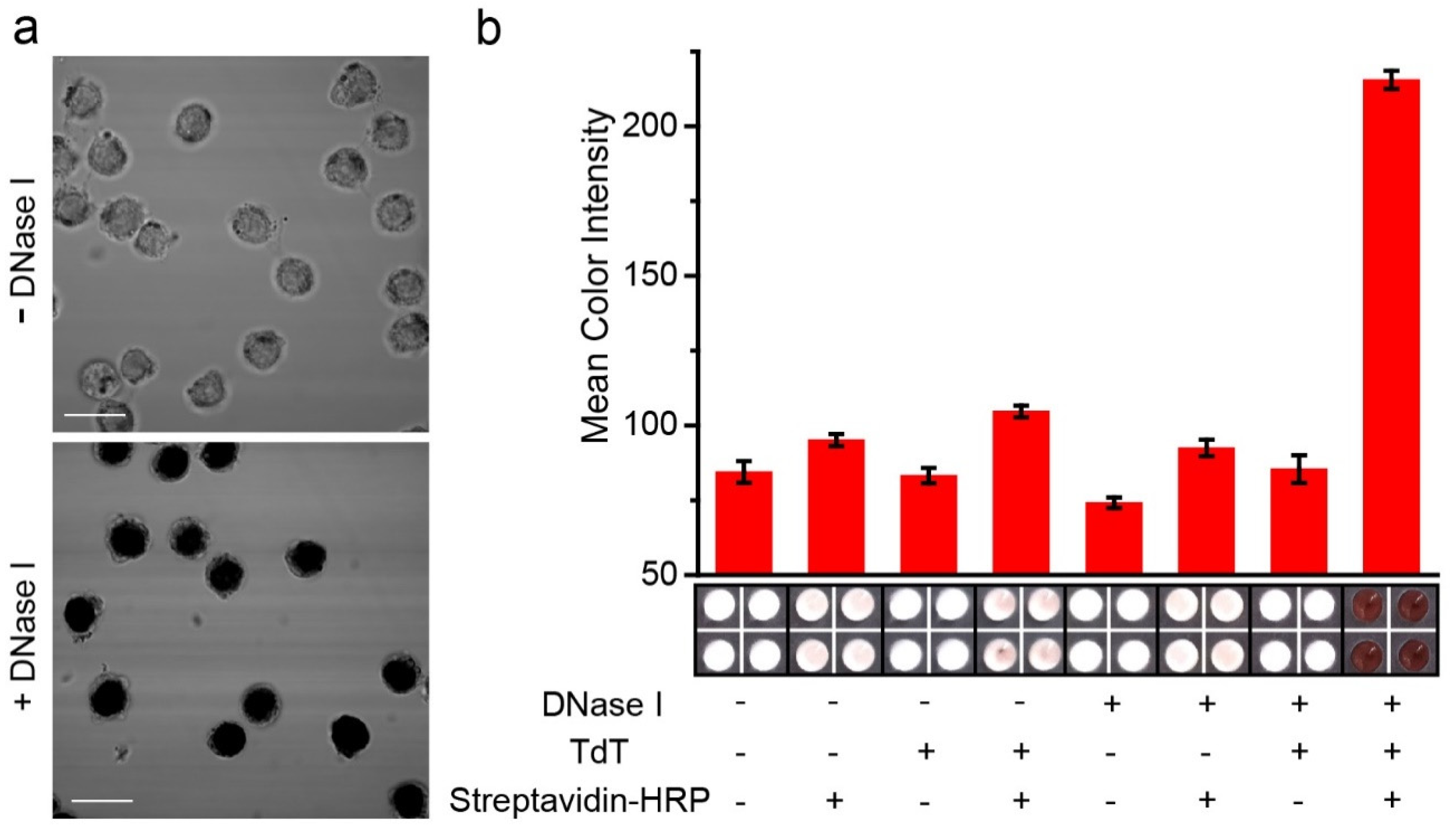
3.2. Detection of DNase Type I 3′OH DNA Ends by PAD
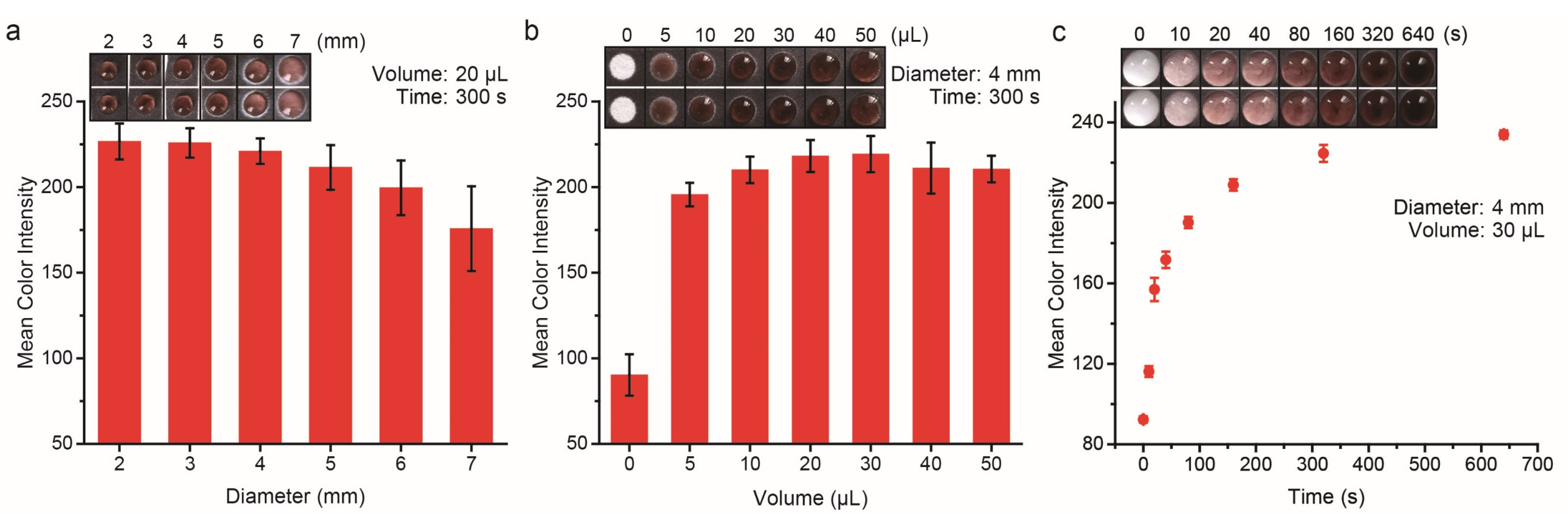
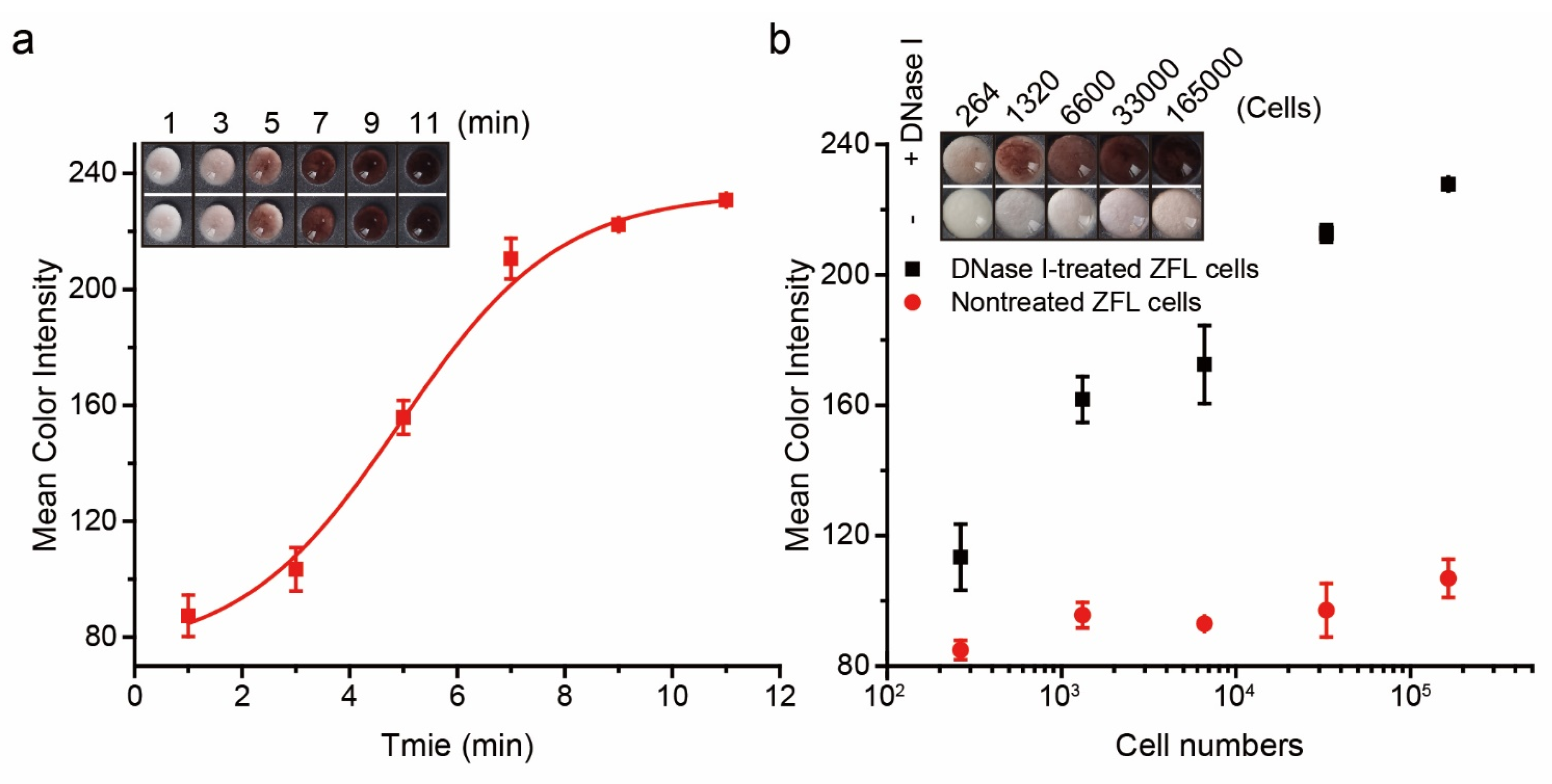
3.3. Evaluation of the Experimental Parameters
3.4. Assay Performance of PAD
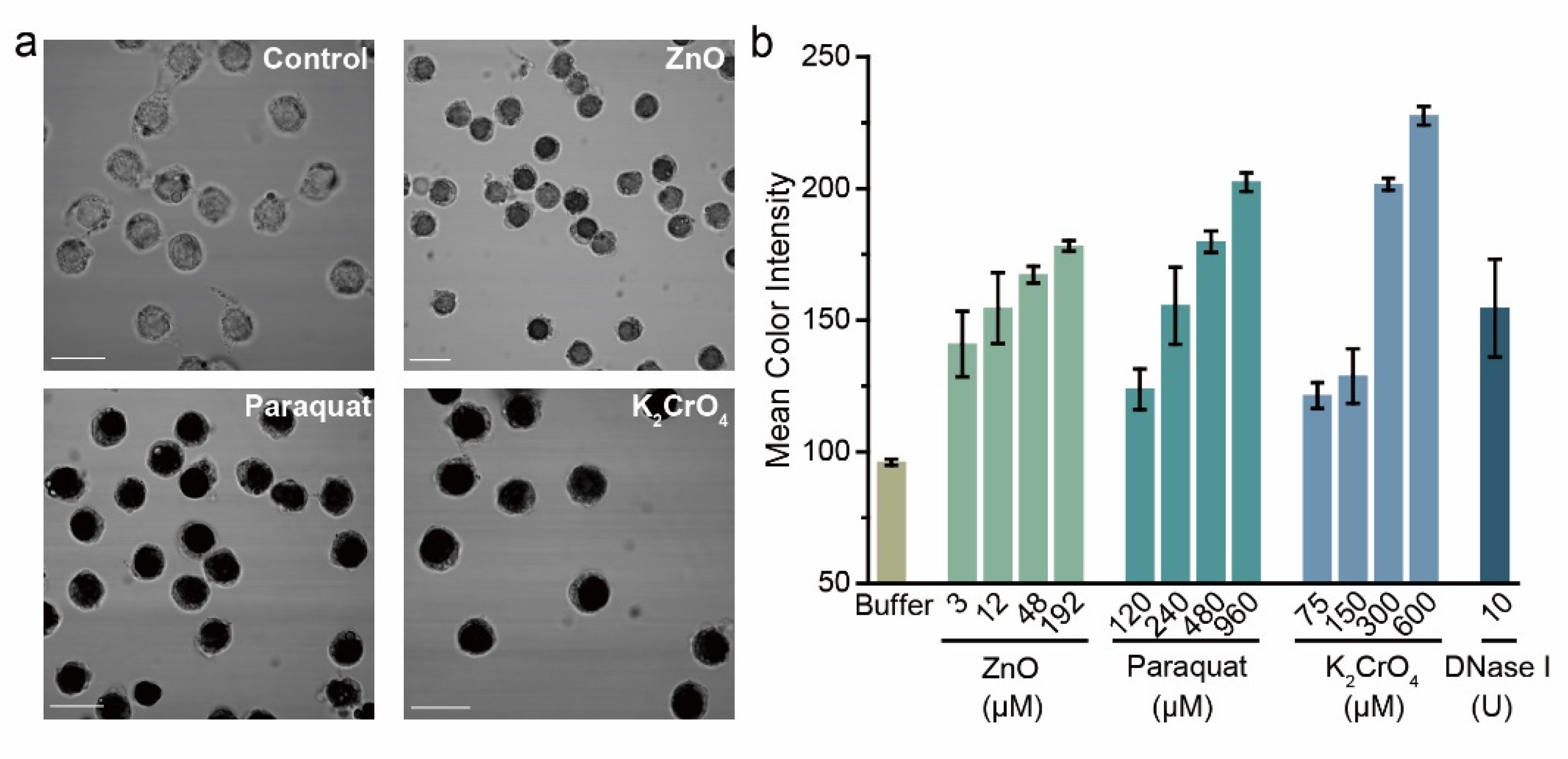
3.5. Detection of DNA Damages Induced by Environmental Genotoxic Agents
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hao, J.N.; Li, Y. Concurrent modulation of competitive mechanisms to design stimuli-responsive ln-MOFs: A light-operated dual-mode assay for oxidative DNA damage. Adv. Funct. Mater. 2019, 29, 1903058. [Google Scholar] [CrossRef]
- Ahn, J.M.; Hwang, E.T.; Youn, C.H.; Banu, D.L.; Kim, B.C.; Niazi, J.H.; Gu, M.B. Prediction and classification of the modes of genotoxic actions using bacterial biosensors specific for DNA damages. Biosens. Bioelectron. 2009, 25, 767–772. [Google Scholar] [CrossRef] [PubMed]
- Reifferscheid, G.; Buchinger, S. Cell-based genotoxicity testing. In Whole Cell Sensing System II; Belkin, S., Gu, M.B., Eds.; Springer: Berlin, Germany, 2009; pp. 85–111. [Google Scholar]
- Xue, W.; Zhao, D.; Zhang, Q.; Chang, Y.Y.; Liu, M. Origami paper-based analytical device for DNA damage analysis. Chem. Commun. 2021, 57, 11465–11468. [Google Scholar] [CrossRef] [PubMed]
- Tanner, N.A.; Zhang, Y.; Evans, T.C. Visual detection of isothermal nucleic acid amplification using pH-sensitive dyes. Biotechniques 2015, 58, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Asiello, P.J.; Baeumner, A.J. Miniaturized isothermal nucleic acid amplification, a review. Lab Chip 2011, 11, 1420–1430. [Google Scholar] [CrossRef]
- Rao, S.; Chen, Y.; Hong, T.; He, Z.; Guo, S.; Lai, H.; Guo, G.; Du, Y.; Zhou, X. Simultaneous and sensitive detection of multisite 5-methylcytosine including non-CpG sites at single-5mC-resolution. Anal. Chem. 2016, 88, 10547–10551. [Google Scholar] [CrossRef] [PubMed]
- Kurita, R.; Arai, K.; Nakamoto, K.; Kato, D.; Niwa, O. Determination of DNA methylation using electrochemiluminescence with surface accumulable coreactant. Anal. Chem. 2012, 84, 1799–1803. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, C.; Zhang, X.; Yang, W.; Wu, F.; Zou, G.; Weng, X.; Zhou, X. Gene specific-loci quantitative and single-base resolution analysis of 5-formylcytosine by compound-mediated polymerase chain reaction. Chem. Sci. 2018, 9, 3723–3728. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.X.; Chen, F.; Li, Q.; Wang, L.H.; Fan, C.H. Isothermal amplification of nucleic acids. Chem. Rev. 2015, 115, 12491–12545. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.D.; Zhang, Y.F.; Yu, Z.N.; Xu, Y.Z.; Liu, S.Y.; Dai, Z.; Zou, X.Y. A primer-initiated strand displacement amplification strategy for sensitive detection of 5-Hydroxymethylcytosine in genomic DNA. Chin. Chem. Lett. 2022, 33, 3777–3781. [Google Scholar] [CrossRef]
- Boonbanjong, P.; Tree-rattrakoon, K.; Waiwinya, W.; Pitikultham, P.; Japrung, D. Isothermal amplification technology for disease diagnosis. Biosensors 2022, 12, 677. [Google Scholar] [CrossRef] [PubMed]
- Jee-Woong, P. Principles and applications of loop-mediated isothermal amplification to point-of-care tests. Biosensors 2022, 12, 857. [Google Scholar]
- Wang, X.; Zhou, S.; Chu, C.; Yang, M.; Huo, D.; Hou, C. Dual methylation-sensitive restriction endonucleases coupling with an RPA-assisted CRISPR/Cas13a system (DESCS) for highly sensitive analysis of DNA methylation and its application for point of-care detection. ACS Sens. 2021, 6, 2419–2428. [Google Scholar] [CrossRef]
- Sun, Y.; Sun, Y.; Tian, W.; Liu, C.; Gao, K.; Li, Z. A novel restriction endonuclease GlaI for rapid and highly sensitive detection of DNA methylation coupled with isothermal exponential amplification reaction. Chem. Sci. 2018, 9, 1344–1351. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.P.; Hu, J.; Long, Y.; Zhang, C.Y. Sensitive detection of DNA methyltransferase using hairpin probe-based primer generation rolling circle amplification-induced chemiluminescence. Anal. Chem. 2013, 85, 6143–6150. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.F.; Wen, Y.Q.; Peng, H.Z.; Long, Y.T.; He, Y.; Huang, Q.; Li, D.; Fan, C.H. A methylation-stimulated DNA machine: An autonomous isothermal route to methyltransferase activity and inhibition analysis. Anal. Bioanal. Chem. 2011, 399, 3459–3464. [Google Scholar] [CrossRef]
- Samejima, K.; Earnshaw, W.C. Trashing the genome: The role of nucleases during apoptosis. Nat. Rev. Mol. Cell Biol. 2005, 6, 677–688. [Google Scholar] [CrossRef]
- van der Veen, S.; Tang, C.M. The BER necessities: The repair of DNA damage in human-adapted bacterial pathogens. Nat. Rev. Microbiol. 2015, 13, 83–94. [Google Scholar] [CrossRef]
- Baskin, D.S.; Widmayer, M.A.; Sharpe, M.A. Qantification of DNase type I ends, DNase type II ends, and modified bases using fluorescently labeled ddUTP, terminal deoxynucleotidyl transferase, and formamidopyrimidine-DNA glycosylase. Biotechniques 2010, 49, 505–512. [Google Scholar] [CrossRef]
- Beard, B.C.; Wilson, S.H.; Smerdon, M.J. Suppressed catalytic activity of base excision repair enzymes on rotationally positioned uracil in nucleosomes. Proc. Natl. Acad. Sci. USA 2003, 100, 7465–7470. [Google Scholar] [CrossRef]
- Wyatt, M.D.; Allan, J.M.; Lau, A.Y.; Ellenberger, T.E.; Sam-son, L.D. 3-Methyladenine DNA glycosylases: Structure, function, and biological importance. Bioessays 1999, 21, 668–676. [Google Scholar] [CrossRef]
- Zhang, Y.; Hua, R.N.; Zhang, C.Y. Analysis of the isolated and the clustered DNA damages by single-molecule counting. Anal. Chem. 2019, 91, 10381–10385. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Hua, R.N.; Zhang, C.Y. Integration of enzymatic labeling with single-molecule detection for sensitive quantification of diverse DNA damages. Anal. Chem. 2020, 92, 4700–4706. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, S.; Trung, T.Q.; Lee, N.E. Recent progress, challenges, and prospects of fully integrated mobile and wearable point-of-care testing systems for self-testing. Chem. Soc. Rev. 2020, 49, 1812–1866. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.Y.; Wang, T.; Chen, Z.; Jin, L.; Wu, Z.Z.; Yan, J.Q.; Zhao, X.N.; Cai, L.; Deng, Y.; Guo, Y.; et al. The point-of-care-testing of nucleic acids by chip, cartridge and paper sensors. Chin. Chem. Lett. 2021, 32, 3675–3686. [Google Scholar] [CrossRef]
- Huang, J.Y.; Lin, H.T.; Chen, T.H.; Chen, C.A.; Chang, H.T.; Chen, C.F. Signal amplified gold nanoparticles for cancer diagnosis on paper-based analytical devices. ACS Sens. 2018, 3, 174–182. [Google Scholar] [CrossRef]
- Zangheri, M.; Cevenini, L.; Anfossi, L.; Baggiani, C.; Simoni, P.; Di Nardo, F.; Roda, A. A simple and compact smartphone accessory for quantitative chemiluminescence-based lateral flow immunoassay for salivary cortisol detection. Biosens. Bioelectron. 2015, 64, 63–68. [Google Scholar] [CrossRef]
- Sicard, C.; Glen, C.; Aubie, B.; Wallace, D.; Jahanshahi-Anbuhi, S.; Pennings, K.; Daigger, G.T.; Pelton, R.; Brennan, J.D.; Filipe, C.D.M. Tools for water quality monitoring and mapping using paper-based sensors and cell phones. Water Res. 2015, 70, 360–369. [Google Scholar] [CrossRef]
- Lopez-Ruiz, N.; Curto, V.F.; Erenas, M.M.; Benito-Lopez, F.; Diamond, D.; Palma, A.J.; Capitan-Vallvey, L.F. Smartphone-based simultaneous pH and nitrite colorimetric determination for paper microfluidic devices. Anal. Chem. 2014, 86, 9554–9562. [Google Scholar] [CrossRef]
- OECD. Test No. 203: Fish, Acute Toxicity Test; OECD Guidelines for the Testing of Chemicals; OECD Publishing: Paris, France, 1992. [Google Scholar]
- Dinis-Oliveira, R.J.; Sousa, C.; Remiao, F.; Duarte, J.A.; Ferreira, R.; Navarro, A.S.; Bastos, M.L.; Carvalho, F. Sodium salicylate prevents paraquat-induced apoptosis in the rat lung. Free Radic. Biol. Med. 2007, 43, 48–61. [Google Scholar] [CrossRef]
- Depault, F.; Cojocaru, M.; Fortin, F.; Chakrabarti, S.; Lemieux, N. Genotoxic effects of chromium(VI) and cadmium(II) in human blood lymphocytes using the electron microscopy in situ end-labeling (EM-ISEL) assay. Toxicol. Vitr. 2006, 20, 513–518. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wu, H.F.; Hong, W.D.; Aguilar, Z.P.; Fu, F.; Xu, H.Y. The effect of reproductive toxicity induced by ZnO NPs in mice during early pregnancy through mitochondrial apoptotic pathway. Environ. Toxicol. 2021, 36, 1143–1151. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xue, W.; Song, K.; Chang, Y.; Liu, M. Colorimetric Detection of DNase Type I 3′OH DNA Ends Using an Isothermal Amplification-Assisted Paper-Based Analytical Device. Biosensors 2022, 12, 1012. https://doi.org/10.3390/bios12111012
Xue W, Song K, Chang Y, Liu M. Colorimetric Detection of DNase Type I 3′OH DNA Ends Using an Isothermal Amplification-Assisted Paper-Based Analytical Device. Biosensors. 2022; 12(11):1012. https://doi.org/10.3390/bios12111012
Chicago/Turabian StyleXue, Wei, Kaiyun Song, Yangyang Chang, and Meng Liu. 2022. "Colorimetric Detection of DNase Type I 3′OH DNA Ends Using an Isothermal Amplification-Assisted Paper-Based Analytical Device" Biosensors 12, no. 11: 1012. https://doi.org/10.3390/bios12111012
APA StyleXue, W., Song, K., Chang, Y., & Liu, M. (2022). Colorimetric Detection of DNase Type I 3′OH DNA Ends Using an Isothermal Amplification-Assisted Paper-Based Analytical Device. Biosensors, 12(11), 1012. https://doi.org/10.3390/bios12111012




