Nanopore Technology for the Application of Protein Detection
Abstract
1. Introduction
2. Detection of Conformational Changes in Protein Space
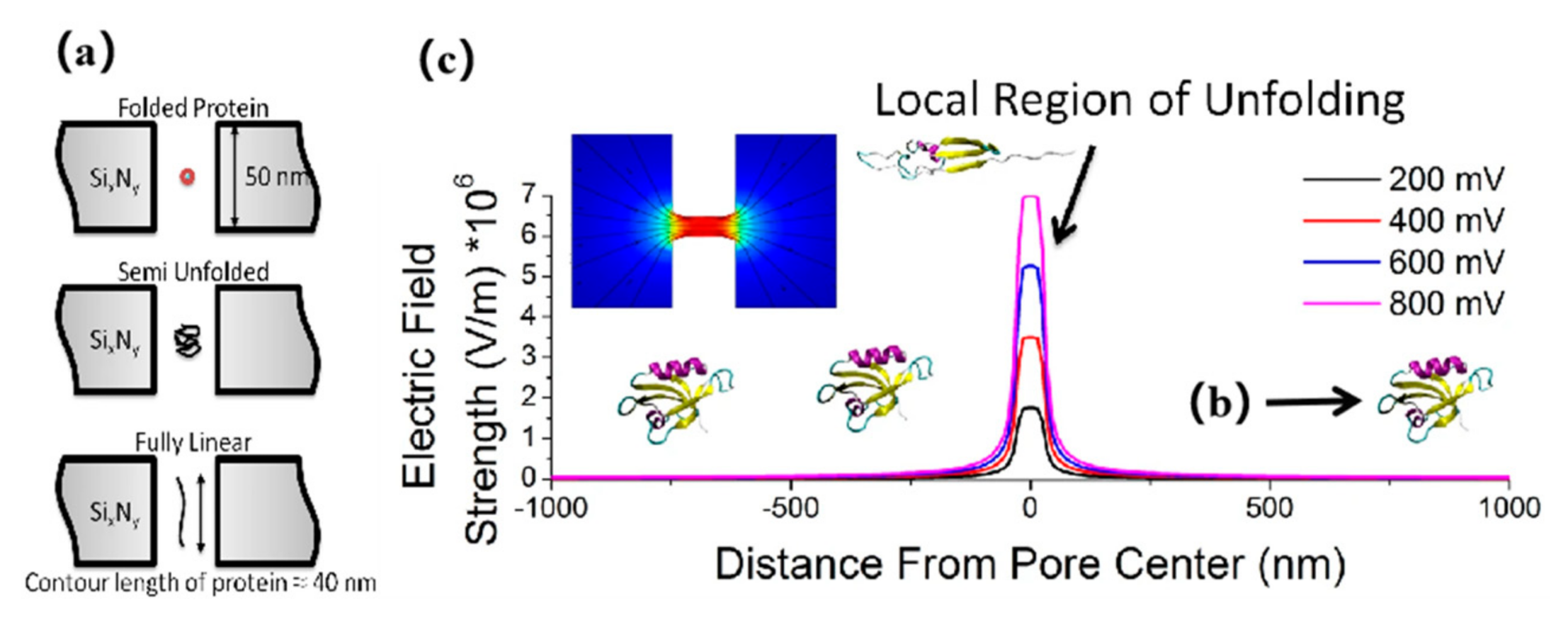
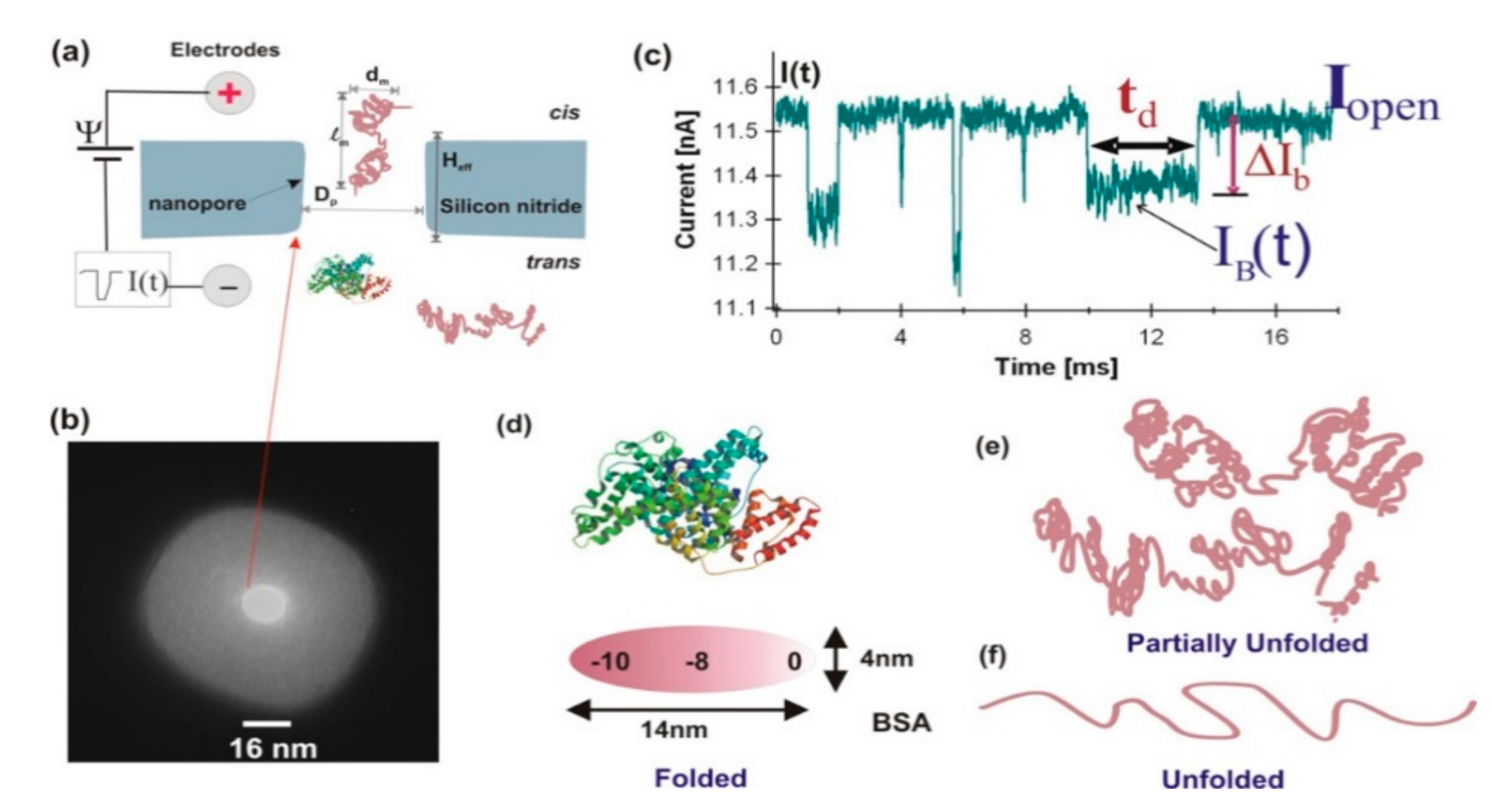
2.1. Characterization of Protein Folding/Unfolding
2.2. Characterization of Protein Size and Shape Aggregation
2.3. Characterization of Induced Protein Conformational Changes
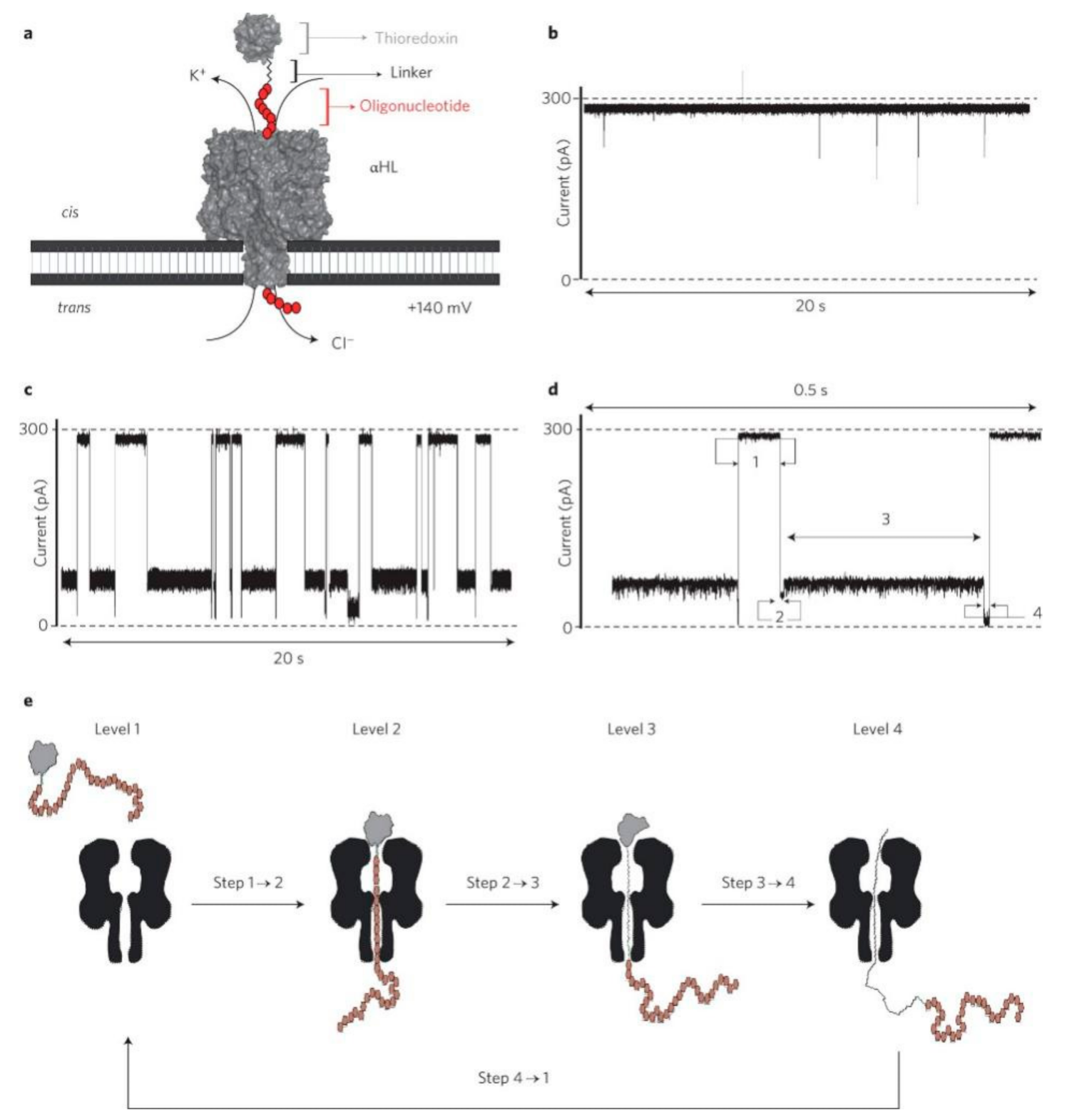
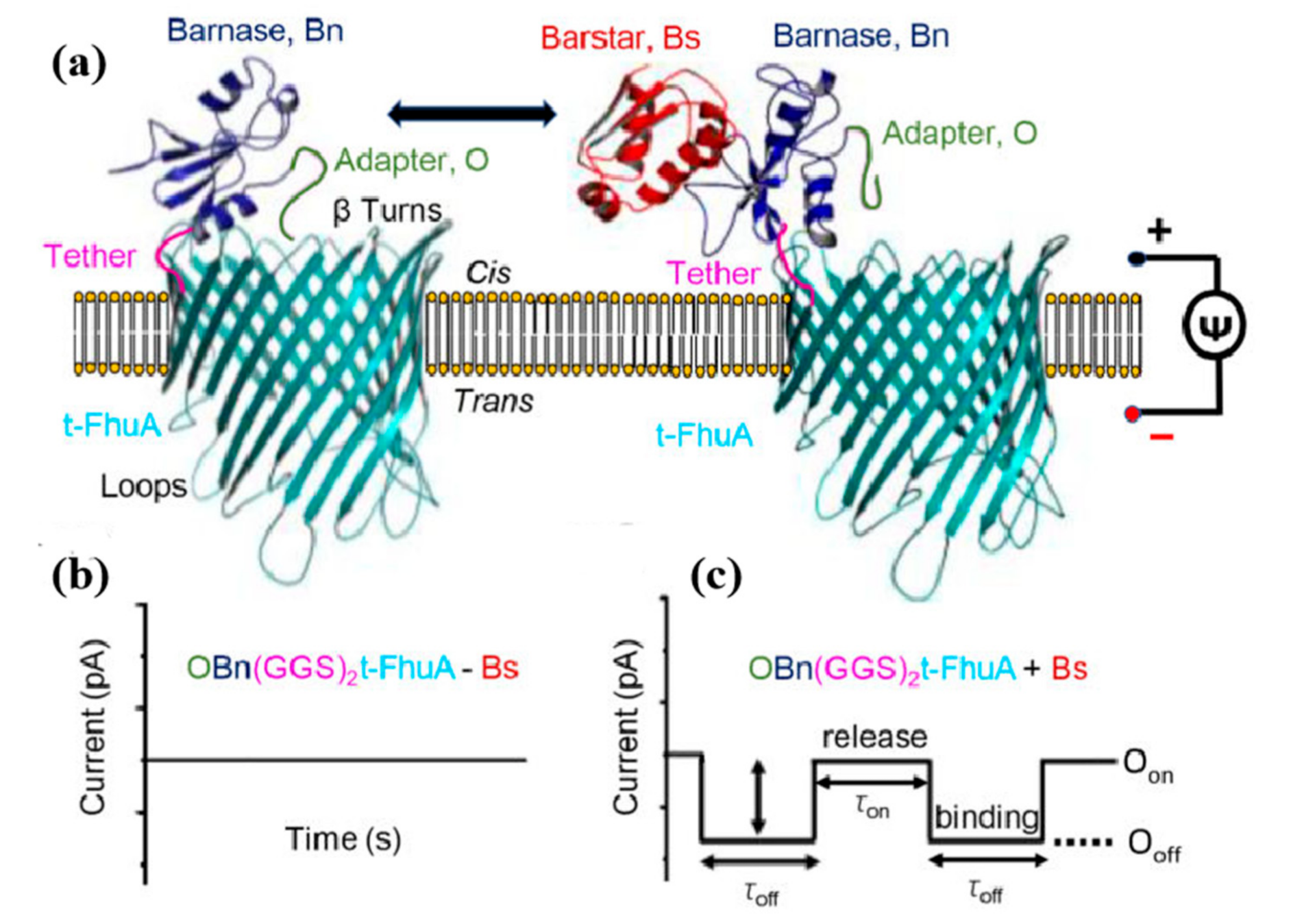
3. Detection of Protein-Protein Interactions
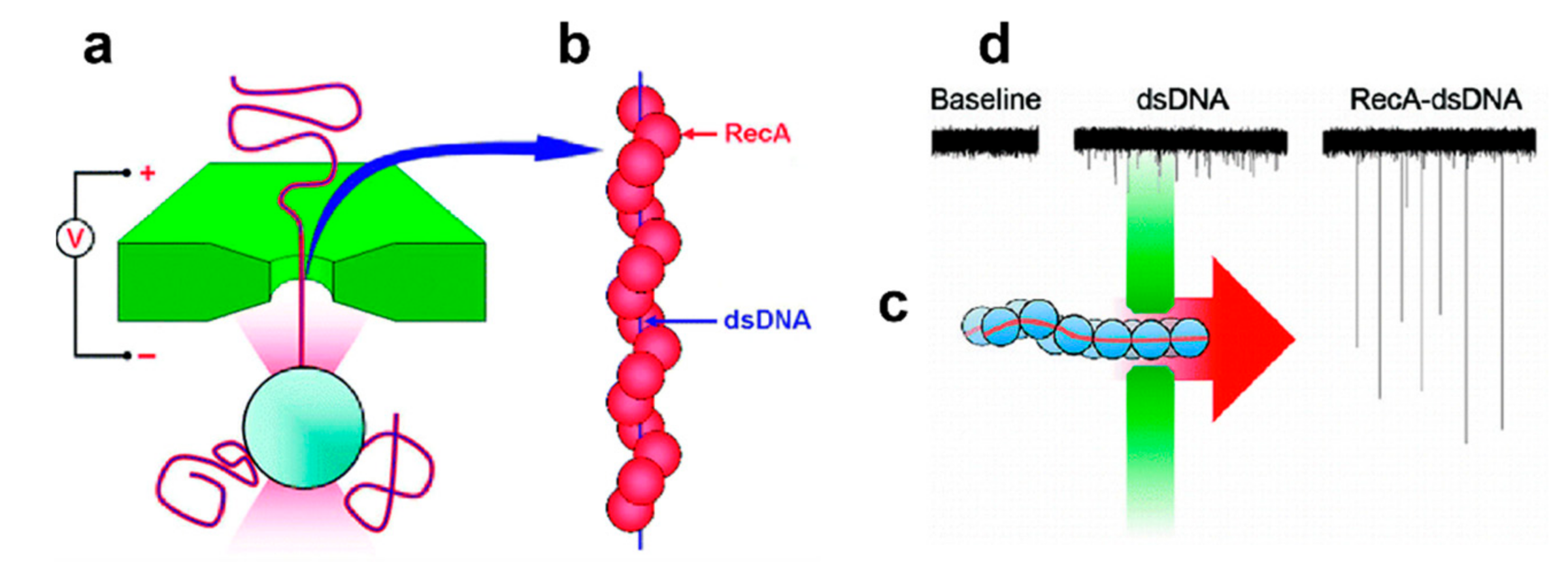
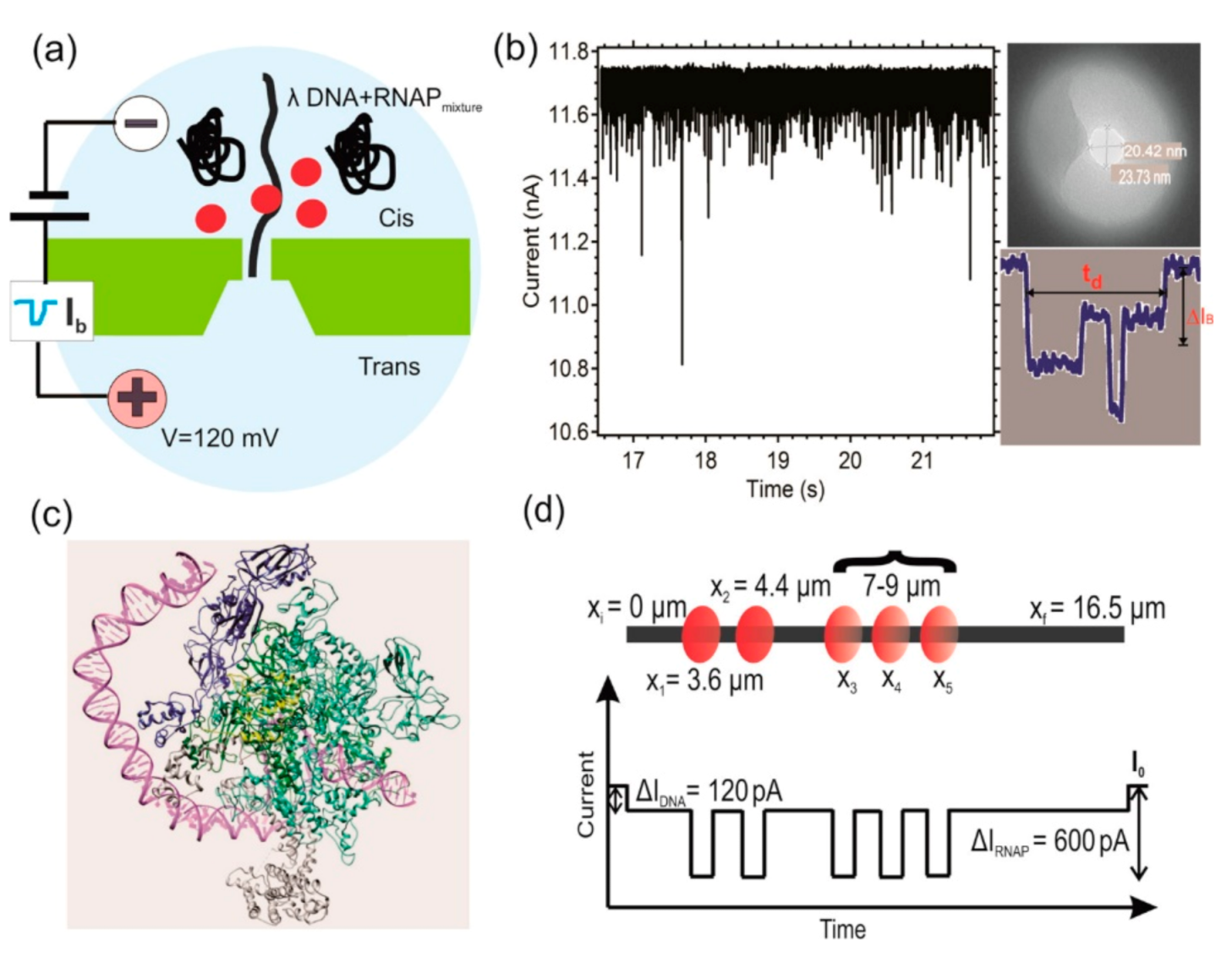
4. Detection of Protein-DNA Interactions
5. Conclusions and Outlook
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ishijima, A.; Kojima, H.; Higuchi, H.; Harada, Y.; Funatsu, T.; Yanagida, T. Multiple- and single-molecule analysis of the actomyosin motor by nanometer-piconewton manipulation with a microneedle: Unitary steps and forces. Biophys. J. 1996, 70, 383–400. [Google Scholar] [CrossRef]
- Spitaleri, A.; Garoli, D.; Schütte, M.; Lehrach, H.; Rocchia, W.; De Angelis, F. Adaptive nanopores: A bioinspired label-free approach for protein sequencing and identification. Nano Res. 2020, 14, 328–333. [Google Scholar] [CrossRef]
- Thakur, A.K.; Movileanu, L. Real-time measurement of protein-protein interactions at single-molecule resolution using a biological nanopore. Nat. Biotechnol. 2018, 37, 96–104. [Google Scholar] [CrossRef]
- Restrepo-Pérez, L.; Huang, G.; Bohländer, P.R.; Worp, N.; Eelkema, R.; Maglia, G.; Joo, C.; Dekker, C. Correction to resolving chemical modifications to a single amino acid within a peptide using a biological nanopore. ACS Nano 2020, 13, 2021. [Google Scholar] [CrossRef]
- Howorka, S.; Siwy, Z.S. Reading amino acids in a nanopore. Nat. Biotechnol. 2020, 38, 159–160. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wang, Y.; Liu, Y.; Wang, H.; Li, W.; Tang, P.; Weng, T.; Zhou, S.; Liang, L.; Yuan, J.; et al. Reduction chemistry-assisted nanopore determination method for immunoglobulin isotypes. Nanoscale 2020, 12, 19711–19718. [Google Scholar] [CrossRef]
- Tian, H.; Xie, W.; He, S.; Zhou, D.; Fang, S.; Liang, L.; Wang, D. Investigation of the adsorption behavior of BSA with tethered lipid layer-modified solid-state nanopores in a wide pH range. RSC Adv. 2019, 9, 15431–15436. [Google Scholar] [CrossRef]
- Ren, R.; Wang, X.; Cai, S.; Zhang, Y.; Korchev, Y.; Ivanov, A.P.; Edel, J.B. Back cover: Selective sensing of proteins using aptamer functionalized nanopore extended field-effect transistors (small methods 11/2020). Small Methods 2020, 4, 2070044. [Google Scholar] [CrossRef]
- Tang, J.; Wu, J.; Zhu, R.; Wang, Z.; Zhao, C.; Tang, P.; Xie, W.; Wang, D.; Liang, L. Reversible photo-regulation on the folding/unfolding of telomere G-quadruplexes with solid-state nanopores. Analyst 2021, 146, 655–663. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Zhang, B.; Liang, L.; Wang, S.; Zhang, L.; Wang, L.; Cui, H.L.; Zhou, Y.; Wang, D. A solid-state nanopore-based single-molecule approach for label-free characterization of plant polysaccharides. Plant. Commun. 2021, 2, 100106. [Google Scholar] [CrossRef]
- Thakur, A.K.; Movileanu, L. Single-molecule protein detection in a biofluid using a quantitative nanopore sensor. ACS Sens. 2019, 4, 2320–2326. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Y.; Chen, X.; Guan, X.; Wang, L. Analysis with biological nanopore: On-pore, off-pore strategies and application in biological fluids. Talanta 2021, 223, 121684. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Wang, L.; Chen, X.; Guan, X. Label-free nanopore single-molecule measurement of trypsin activity. ACS Sens. 2016, 1, 607–613. [Google Scholar] [CrossRef]
- Wang, Y.; Guan, X.; Zhang, S.; Liu, Y.; Wang, S.; Fan, P.; Du, X.; Yan, S.; Zhang, P.; Chen, H.-Y.; et al. Structural-profiling of low molecular weight RNAs by nanopore trapping/translocation using Mycobacterium smegmatis porin A. Nat. Commun. 2021, 12, 3368. [Google Scholar] [CrossRef] [PubMed]
- Gundlach, J.H. Mycobacterium smegmatis porin A (MspA) nanopore–based DNA sequencing using phage DNA polymerase and blocking oligomers. Sci. Exch. 2012, 5, 375. [Google Scholar] [CrossRef][Green Version]
- Ouldali, H.; Sarthak, K.; Ensslen, T.; Piguet, F.; Manivet, P.; Pelta, J.; Behrends, J.C.; Aksimentiev, A.; Oukhaled, A. Electrical recognition of the twenty proteinogenic amino acids using an aerolysin nanopore. Nat. Biotechnol. 2019, 38, 176–181. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Haque, F.; Rychahou, P.G.; Evers, B.M.; Guo, P. Engineered nanopore of Phi29 DNA-packaging motor for real-time detection of single colon cancer specific antibody in serum. ACS Nano 2013, 7, 9814–9822. [Google Scholar] [CrossRef]
- Biesemans, A.; Soskine, M.; Maglia, G. A protein rotaxane controls the translocation of proteins across a ClyA nanopore. Nano Lett. 2015, 15, 6076–6081. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.; Yamazaki, H.; Ren, R.; Wanunu, M.; Ivanov, A.P.; Edel, J.B. Solid-state nanopore sensors. Nat. Rev. Mater. 2020, 5, 931–951. [Google Scholar] [CrossRef]
- Wu, L.; Wu, H.; Liu, L.; Ye, X.; Liu, Q. Fabrication and characterization of solid nanopores with silicon nitride membranes by focus ion beam. Acta Biophys. Sin. 2013, 29, 203. [Google Scholar] [CrossRef]
- Yen, P.; Wang, C.; Hwang, G.-J.; Chou, Y.C. Gate effects on DNA translocation through silicon dioxide nanopore. Rev. Sci. Instrum. 2012, 83, 13770–13773. [Google Scholar] [CrossRef]
- Lee, K.H.; Huang, Y.P.; Wong, C.C. Nanotip fabrication by anodic aluminum oxide templating. Electrochim. Acta 2011, 56, 2394–2398. [Google Scholar] [CrossRef]
- Zhang, Z.-Y.; Deng, Y.-S.; Tian, H.-B.; Yan, H.; Cui, H.-L.; Wang, D.-Q. Noise analysis of monolayer graphene nanopores. Int. J. Mol. Sci. 2018, 19, 2639. [Google Scholar] [CrossRef]
- Zhou, Z.; Hu, Y.; Wang, H.; Xu, Z.; Wang, W.; Bai, X.; Shan, X.; Lu, X. DNA translocation through hydrophilic nanopore in hexagonal boron nitride. Sci. Rep. 2013, 3, 3287. [Google Scholar] [CrossRef]
- Shi, J.; Hou, J.; Fang, Y. Recent advances in nanopore-based nucleic acid analysis and sequencing. Microchim. Acta 2015, 183, 925–939. [Google Scholar] [CrossRef]
- Cao, C.; Li, M.-Y.; Cirauqui, N.; Wang, Y.-Q.; Dal Peraro, M.; Tian, H.; Long, Y.-T. Mapping the sensing spots of aerolysin for single oligonucleotides analysis. Nat. Commun. 2018, 9, 2823. [Google Scholar] [CrossRef] [PubMed]
- Rauf, S.; Zhang, L.; Ali, A.; Ahmad, J.; Liu, Y.; Li, J. Nanopore-based, label-free, and real-time monitoring assay for DNA methyltransferase activity and inhibition. Anal. Chem. 2017, 89, 13252–13260. [Google Scholar] [CrossRef]
- Cao, C.; Ying, Y.-L.; Hu, Z.-L.; Liao, D.-F.; Tian, H.; Long, Y.-T. Discrimination of oligonucleotides of different lengths with a wild-type aerolysin nanopore. Nat. Nanotechnol. 2016, 11, 713–718. [Google Scholar] [CrossRef] [PubMed]
- Haque, F.; Li, J.; Wu, H.C.; Liang, X.J.; Guo, P. Solid-state and biological nanopore for real-time sensing of single chemical and sequencing of DNA. Nano Today 2013, 8, 56–74. [Google Scholar] [CrossRef]
- Kennedy, E.; Dong, Z.; Tennant, C.; Timp, G. Reading the primary structure of a protein with 0.07 nm 3 resolution using a subnanometre-diameter pore. Nat. Nanotechnol. 2016, 11, 968–976. [Google Scholar] [CrossRef]
- Larimi, M.G.; Mayse, L.A.; Movileanu, L. Interactions of a polypeptide with a protein nanopore under crowding conditions. ACS Nano 2019, 13, 4469–4477. [Google Scholar] [CrossRef]
- Waduge, P.; Hu, R.; Bandarkar, P.; Yamazaki, H.; Cressiot, B.; Zhao, Q.; Whitford, P.C.; Wanunu, M. Nanopore-Based Measurements of protein size, fluctuations, and conformational changes. ACS Nano 2017, 11, 5706–5716. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yao, X.F.; Wang, H.Y. Protein detection through single molecule nanopore. Chin. J. Anal. Chem. 2018, 46, e1838–e1846. [Google Scholar] [CrossRef]
- Chau, C.C.; Radford, S.E.; Hewitt, E.W.; Actis, P. Macromolecular crowding enhances the detection of DNA and proteins by a solid-state nanopore. Nano Lett. 2020, 20, 5553–5561. [Google Scholar] [CrossRef]
- Oukhaled, A.; Cressiot, B.; Bacri, L.; Pastoriza-Gallego, M.; Betton, J.-M.; Bourhis, E.; Jede, R.; Gierak, J.; Auvray, L.; Pelta, J. Dynamics of completely unfolded and native proteins through solid-state nanopores as a function of electric driving force. ACS Nano 2011, 5, 3628–3638. [Google Scholar] [CrossRef]
- Steinbock, L.J.; Krishnan, S.; Bulushev, R.D.; Borgeaud, S.; Blokesch, M.; Feletti, L.; Radenovic, A. Probing the size of proteins with glass nanopores. Nanoscale 2014, 6, 14380–14387. [Google Scholar] [CrossRef]
- Heiranian, A.B.; Mohammad, F.; Min, K.; Aluru, N.R. Antibody subclass detection using graphene nanopores. J. Phys. Chem. Lett. 2017, 8, 1670–1676. [Google Scholar] [CrossRef]
- Movileanu, L. Interrogating single proteins through nanopores: Challenges and opportunities. Trends Biotechnol. 2009, 27, 333–341. [Google Scholar] [CrossRef]
- Movileanu, L.; Howorka, S.; Braha, O.; Bayley, H. Detecting protein analytes that modulate transmembrane movement of a polymer chain within a single protein pore. Nat. Biotechnol. 2000, 18, 1091–1095. [Google Scholar] [CrossRef] [PubMed]
- Niedzwiecki, D.J.; Lanci, C.J.; Shemer, G.; Cheng, P.S.; Saven, J.G.; Drndić, M. Observing changes in the structure and oligomerization state of a helical protein dimer using solid-state nanopores. ACS Nano 2015, 9, 8907–8915. [Google Scholar] [CrossRef] [PubMed]
- Tavassoly, O.; Kakish, J.; Nokhrin, S.; Dmitriev, O.; Lee, J.S. The use of nanopore analysis for discovering drugs which bind to α-synuclein for treatment of Parkinson’s disease. Eur. J. Med. Chem. 2014, 88, 42–54. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, Y.; Fu, Y.; Ying, C.; Feng, Y.; Huang, Q.; Wang, C.; Pei, D.-S.; Wang, D. Monitoring tetracycline through a solid-state nanopore sensor. Sci. Rep. 2016, 6, 27959. [Google Scholar] [CrossRef]
- Runtuwene, L.R.; Tuda, J.S.B.; Mongan, A.E.; Makalowski, W.; Frith, M.C.; Imwong, M.; Srisutham, S.; Thi, L.A.N.; Tuan, N.N.; Eshita, Y.; et al. Nanopore sequencing of drug-resistance-associated genes in malaria parasites, Plasmodium falciparum. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Plesa, C.; Ruitenberg, J.W.; Witteveen, M.J.; Dekker, C. Detection of individual proteins bound along dna using solid-state nanopores. Nano Lett. 2015, 15, 3153–3158. [Google Scholar] [CrossRef]
- Japrung, D.; Bahrami, A.; Nadzeyka, A.; Peto, L.; Bauerdick, S.; Edel, J.B.; Albrecht, T. SSB Binding to single-stranded DNA Probed using solid-state nanopore sensors. J. Phys. Chem. B 2014, 118, 11605–11612. [Google Scholar] [CrossRef]
- Han, A.; Creus, M.; Schürmann, G.; Linder, V.; Ward, T.R.; De Rooij, N.F.; Staufer, U. Label-Free detection of single protein molecules and protein−protein interactions using synthetic nanopores. Anal. Chem. 2008, 80, 4651–4658. [Google Scholar] [CrossRef] [PubMed]
- Darvish, A.; Lee, J.S.; Peng, B.; Saharia, J.; VenkatKalyana Sundaram, R.; Goyal, G.; Bandara, N.; Ahn, C.W.; Kim, J.; Dutta, P.; et al. Back cover: Mechanical characterization of HIV-1 with a solid-state nanopore sensor. Electrophoresis 2019, 40, 597–828. [Google Scholar] [CrossRef] [PubMed]
- Roozbahani, G.M.; Chen, X.; Zhang, Y.; Juarez, O.; Li, D.; Guan, X. Computation-assisted Nanopore detection of thorium ions. Anal. Chem. 2018, 90, 5938–5944. [Google Scholar] [CrossRef]
- Wang, G.; Zhao, Q.; Kang, X.; Guan, X. Probing mercury(II)–DNA Interactions by nanopore stochastic sensing. J. Phys. Chem. B 2013, 117, 4763–4769. [Google Scholar] [CrossRef]
- Wen, S.; Zeng, T.; Liu, L.; Zhao, K.; Zhao, Y.; Liu, X.; Wu, H.-C. Highly sensitive and selective DNA-based detection of mercury(II) with α-hemolysin nanopore. J. Am. Chem. Soc. 2011, 133, 18312–18317. [Google Scholar] [CrossRef]
- Yang, C.; Liu, L.; Zeng, T.; Yang, D.; Yao, Z.; Zhao, Y.; Wu, H.-C. Highly sensitive simultaneous detection of lead(II) and barium(II) with G-quadruplex DNA in α-hemolysin nanopore. Anal. Chem. 2013, 85, 7302–7307. [Google Scholar] [CrossRef] [PubMed]
- Takaya, D.; Niwa, H.; Mikuni, J.; Nakamura, K.; Handa, N.; Tanaka, A.; Yokoyama, S.; Honma, T. Protein ligand interaction analysis against new CaMKK2 inhibitors by use of X-ray crystallography and the fragment molecular orbital (FMO) method. J. Mol. Graph. Model. 2020, 99, 107599. [Google Scholar] [CrossRef]
- Kemmink, J.; Darby, N.J.; Creighton, T.E.; Dijkstra, K.; Scheek, R.M. Nuclear magnetic resonance characterization of the N-terminal thioredoxin-like domain of protein disulfide isomerase. Protein Sci. 1995, 4, 2587–2593. [Google Scholar] [CrossRef] [PubMed]
- Whitmore, L.; Wallace, B.A. Protein secondary structure analyses from circular dichroism spectroscopy: Methods and reference databases. Biopolymers 2008, 89, 392–400. [Google Scholar] [CrossRef]
- VD Dos Santos, A.C.; Heydenreich, R.; Derntl, C.; Mach-Aigner, A.R.; Mach, R.L.; Ramer, G.; Lendl, B. Nanoscale Infrared spectroscopy and chemometrics enable detection of intracellular protein distribution. Anal. Chem. 2020, 92, 15719–15725. [Google Scholar] [CrossRef]
- Ikai, A.; Afrin, R.; Sekiguchi, H. Pulling and pushing protein molecules by AFM. Curr. Nanosci. 2007, 3, 17–29. [Google Scholar] [CrossRef]
- Zhao, J. Studying the physics of charged macromolecules by single molecule fluorescence spectroscopy. J. Chem. Phys. 2020, 153, 170903. [Google Scholar] [CrossRef] [PubMed]
- Fezoui, Y.; Teplow, D.B. Kinetic studies of amyloid β-protein fibril assembly: Differential effects of α-helix stabilization. J. Biol. Chem. 2002, 277, 36948–36954. [Google Scholar] [CrossRef]
- Schnabel, J. Protein folding: The dark side of proteins. Nature 2010, 464, 828–829. [Google Scholar] [CrossRef]
- Trojanowski, J.Q.; Lee, V.M.Y. Parkinson’s disease and related synucleinopathies are a new class of nervous system amyloidoses. Neurotoxicology 2002, 23, 457–460. [Google Scholar] [CrossRef]
- Chandra, S.; Gallardo, G.; Fernández-Chacón, R.; Schlüter, O.M.; Südhof, T.C. Alpha-synuclein cooperates with CSPalpha in preventing neurodegeneration. Cell 2005, 123, 383–396. [Google Scholar] [CrossRef]
- Tyler, K.L. Creutzfeldt–Jakob disease. N. Engl. J. Med. 2003, 348, 681–682. [Google Scholar] [CrossRef]
- Cobb, N.J.; Surewicz, W.K. Prion Diseases and their biochemical mechanisms. Biochemistry 2009, 48, 2574–2585. [Google Scholar] [CrossRef]
- Gething, M.-J.; Sambrook, J. Protein folding in the cell. Nature 1992, 355, 33–45. [Google Scholar] [CrossRef]
- Tsai, C.J.; Kumar, S.; Ma, B.; Nussinov, R. Folding funnels, binding funnels, and protein function. Protein Sci. 1999, 8, 1181–1190. [Google Scholar] [CrossRef]
- Onuchic, J.N.; Luthey-Schulten, Z.; Wolynes, P.G. Theory of protein folding: The energy landscape perspective. Annu. Rev. Phys. Chem. 1997, 48, 545–600. [Google Scholar] [CrossRef] [PubMed]
- Mello, C.C.; Barrick, D. An experimentally determined protein folding energy landscape. Proc. Natl. Acad. Sci. USA 2004, 101, 14102–14107. [Google Scholar] [CrossRef] [PubMed]
- Iwaoka, M.; Wedemeyer, W.J.; Scheraga, H.A. Conformational unfolding studies of three-disulfide mutants of bovine pancreatic ribonuclease a and the coupling of proline isomerization to disulfide redox reactions. Biochemistry 1999, 38, 2805–2815. [Google Scholar] [CrossRef] [PubMed]
- Lucent, D.; Vishal, V.; Pande, V.S. Protein folding under confinement: A role for solvent. Proc. Natl. Acad. Sci. USA 2007, 104, 10430–10434. [Google Scholar] [CrossRef] [PubMed]
- Freedman, K.J.; Jürgens, M.; Prabhu, A.; Ahn, C.W.; Jemth, P.; Edel, J.B.; Kim, M.J. Chemical, thermal, and electric field induced unfolding of single protein molecules studied using nanopores. Anal. Chem. 2011, 83, 5137–5144. [Google Scholar] [CrossRef]
- Freedman, K.J.; Haq, S.R.; Edel, J.B.; Jemth, P.; Kim, M.J. Single molecule unfolding and stretching of protein domains inside a solid-state nanopore by electric field. Sci. Rep. 2013, 3, 1638. [Google Scholar] [CrossRef]
- Li, J.; Fologea, D.; Rollings, R.; Ledden, B. Characterization of protein unfolding with solid-state nanopores. Protein Pept. Lett. 2014, 21, 256–265. [Google Scholar] [CrossRef]
- Oukhaled, A.; Pastoriza-Gallego, M.; Bacri, L.; Mathé, J.; Auvray, L.; Pelta, J. Protein unfolding through nanopores. Protein Pept. Lett. 2014, 21, 266–274. [Google Scholar] [CrossRef]
- Rodriguez-Larrea, D.; Bayley, H. Multistep protein unfolding during nanopore translocation. Nat. Nanotechnol. 2013, 8, 288–295. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Larrea, D.; Bayley, H. Protein co-translocational unfolding depends on the direction of pulling. Nat. Commun. 2014, 5, 1–7. [Google Scholar] [CrossRef]
- Fologea, D.; Ledden, B.; McNabb, D.S.; Li, J. Electrical characterization of protein molecules by a solid-state nanopore. Appl. Phys. Lett. 2007, 91, 539011–539013. [Google Scholar] [CrossRef] [PubMed]
- Sha, J.; Si, W.; Xu, B.; Zhang, S.; Li, K.; Lin, K.; Shi, H.; Chen, Y. Identification of spherical and nonspherical proteins by a solid-state nanopore. Anal. Chem. 2018, 90, 13826–13831. [Google Scholar] [CrossRef]
- Giamblanco, N.; Coglitore, D.; Janot, J.M.; Coulon, P.E.; Charlot, B.; Balme, S. Detection of protein aggregate morphology through single antifouling nanopore. Sens. Actuators B Chem. 2018, 260, 736–745. [Google Scholar] [CrossRef]
- Wei, R.; Tampé, R.; Rant, U. Stochastic Sensing of Proteins with Receptor-Modified Solid-State Nanopores. Nat. Nanotechnol. 2012, 7, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Yusko, E.C.; Bruhn, B.R.; Eggenberger, O.M.; Houghtaling, J.; Rollings, R.C.; Walsh, N.C.; Nandivada, S.; Pindrus, M.; Hall, A.R.; Sept, D.; et al. Real-time shape approximation and fingerprinting of single proteins using a nanopore. Nat. Nanotechnol. 2016, 12, 360–367. [Google Scholar] [CrossRef]
- Saharia, J.; Bandara, Y.M.N.D.Y.; Goyal, G.; Lee, J.S.; Karawdeniya, B.I.; Kim, M.J. Molecular-level profiling of human serum transferrin protein through assessment of nanopore-based electrical and chemical responsiveness. ACS Nano 2019, 13, 4246–4254. [Google Scholar] [CrossRef]
- Hu, G.; Fu, J.; Qiao, Y.; Meng, H.; Wang, Z.; Tu, J.; Lu, Z. Molecular dynamics discrimination of the conformational states of calmodulin through solid-state nanopores. Phys. Chem. Chem. Phys. 2020, 22, 19188–19194. [Google Scholar] [CrossRef] [PubMed]
- Chae, H.; Kwak, D.-K.; Lee, M.-K.; Chi, S.-W.; Kim, K.-B. Solid-state nanopore analysis on conformation change of p53TAD-MDM2 fusion protein induced by protein-protein interaction. Nanoscale 2018, 10, 17227–17235. [Google Scholar] [CrossRef]
- Chae, H.; Kwak, D.-K.; Lee, M.-K.; Chi, S.-W.; Kim, K.-B. Nanopore analysis on the drug-induced conformation change of a p53-linker-mouse double minute 2 protein complex. J. Nanosci. Nanotechnol. 2019, 20, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Uram, J.D.; Ke, K.; Hunt, A.J.; Mayer, M. Submicrometer pore-based characterization and quantification of antibody-virus interactions. Small 2006, 2, 967–972. [Google Scholar] [CrossRef]
- Freedman, K.J.; Bastian, A.R.; Chaiken, I.; Kim, M.J. Solid-state nanopore detection of protein complexes: Applications in healthcare and protein kinetics. Small 2013, 9, 750–759. [Google Scholar] [CrossRef] [PubMed]
- Kwak, D.-K.; Chae, H.; Lee, M.-K.; Ha, J.-H.; Goyal, G.; Kim, M.J.; Kim, K.-B.; Chi, S.-W. Probing the small-molecule inhibition of an anticancer therapeutic protein-protein interaction using a solid-state nanopore. Angew. Chem. Int. Ed. 2016, 55, 5713–5717. [Google Scholar] [CrossRef]
- Chuah, K.; Wu, Y.; Vivekchand, S.R.C.; Gaus, K.; Reece, P.J.; Micolich, A.P.; Gooding, J.J. Nanopore blockade sensors for ultrasensitive detection of proteins in complex biological samples. Nat. Commun. 2019, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Hall, A.R.; Van Dorp, S.; Lemay, S.G.; Dekker, C. Electrophoretic force on a protein-coated dna molecule in a solid-state nanopore. Nano Lett. 2009, 9, 4441–4445. [Google Scholar] [CrossRef]
- Smeets, R.M.M.; Kowalczyk, S.W.; Hall, A.R.; Dekker, N.H.; Dekker, C. Translocation of RecA-coated double-stranded DNA through solid-state nanopores. Nano Lett. 2009, 9, 3089–3095. [Google Scholar] [CrossRef]
- Kowalczyk, S.W.; Hall, A.R.; Dekker, C. Detection of local protein structures along DNA using solid-state nanopores. Nano Lett. 2010, 10, 324–328. [Google Scholar] [CrossRef]
- Spiering, A.; Getfert, S.; Sischka, A.; Reimann, P.; Anselmetti, D. Nanopore translocation dynamics of a single DNA-bound protein. Nano Lett. 2011, 11, 2978–2982. [Google Scholar] [CrossRef] [PubMed]
- Bradley, L.; Rollings, R.; Talaga, D.S.; Li, J. The Translocation Time of DNA and Protein Molecules in Solid-State Nanopores. Available online: https://www.researchgate.net/publication/253087222_The_translocation_time_of_DNA_and_protein_molecules_in_solid-state_nanopores (accessed on 16 July 2021).
- Raillon, C.; Cousin, P.; Traversi, F.; Garcia-Cordero, E.; Hernandez, N.; Radenovic, A. Nanopore detection of single molecule RNAP–DNA transcription complex. Nano Lett. 2012, 12, 1157–1164. [Google Scholar] [CrossRef]
- Van Meervelt, V.; Soskine, M.; Maglia, G. Detection of two isomeric binding configurations in a protein–aptamer complex with a biological nanopore. ACS Nano 2014, 8, 12826–12835. [Google Scholar] [CrossRef] [PubMed]
- Kong, J.; Bell, N.; Keyser, U. Quantifying protein concentration using designed DNA carriers and solid-state nanopores. Nano Lett. 2016, 16, 3557–3562. [Google Scholar] [CrossRef]
- Sze, J.Y.Y.; Ivanov, A.P.; Cass, A.E.G.; Edel, J.B. Single molecule multiplexed nanopore protein screening in human serum using aptamer modified DNA carriers. Nat. Commun. 2017, 8, 1552. [Google Scholar] [CrossRef] [PubMed]
- Celaya, G.; Perales-Calvo, J.; Muga, A.; Moro, F.; Rodriguez-Larrea, D. Label-free, multiplexed, single-molecule analysis of protein-DNA complexes with nanopores. ACS Nano 2017, 11, 5815–5825. [Google Scholar] [CrossRef] [PubMed]
- Kaur, H.; Nandivada, S.; Acharjee, M.C.; McNabb, D.S.; Li, J. Estimating RNA polymerase protein binding sites on λ DNA using solid-state nanopores. ACS Sens. 2019, 4, 100–109. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, X.; Xiang, Y.; Liu, Q.; Wang, L.; Ma, Q.; Ma, W.; Zeng, D.; Yin, Y.; Wang, D. Nanopore Technology for the Application of Protein Detection. Nanomaterials 2021, 11, 1942. https://doi.org/10.3390/nano11081942
Zeng X, Xiang Y, Liu Q, Wang L, Ma Q, Ma W, Zeng D, Yin Y, Wang D. Nanopore Technology for the Application of Protein Detection. Nanomaterials. 2021; 11(8):1942. https://doi.org/10.3390/nano11081942
Chicago/Turabian StyleZeng, Xiaoqing, Yang Xiang, Qianshan Liu, Liang Wang, Qianyun Ma, Wenhao Ma, Delin Zeng, Yajie Yin, and Deqiang Wang. 2021. "Nanopore Technology for the Application of Protein Detection" Nanomaterials 11, no. 8: 1942. https://doi.org/10.3390/nano11081942
APA StyleZeng, X., Xiang, Y., Liu, Q., Wang, L., Ma, Q., Ma, W., Zeng, D., Yin, Y., & Wang, D. (2021). Nanopore Technology for the Application of Protein Detection. Nanomaterials, 11(8), 1942. https://doi.org/10.3390/nano11081942





