Force Dependence of Proteins’ Transition State Position and the Bell–Evans Model
Abstract
1. Introduction

2. Materials and Methods
2.1. Molecular Synthesis
2.2. Optical Tweezers Setup
3. Results
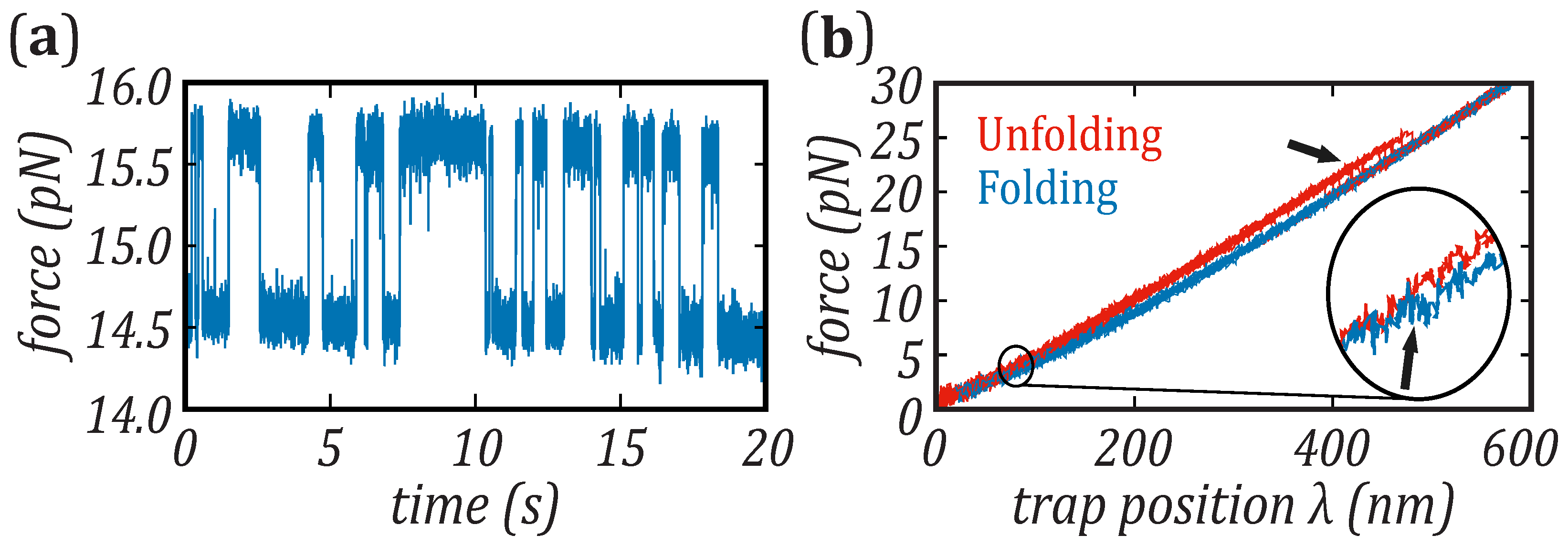
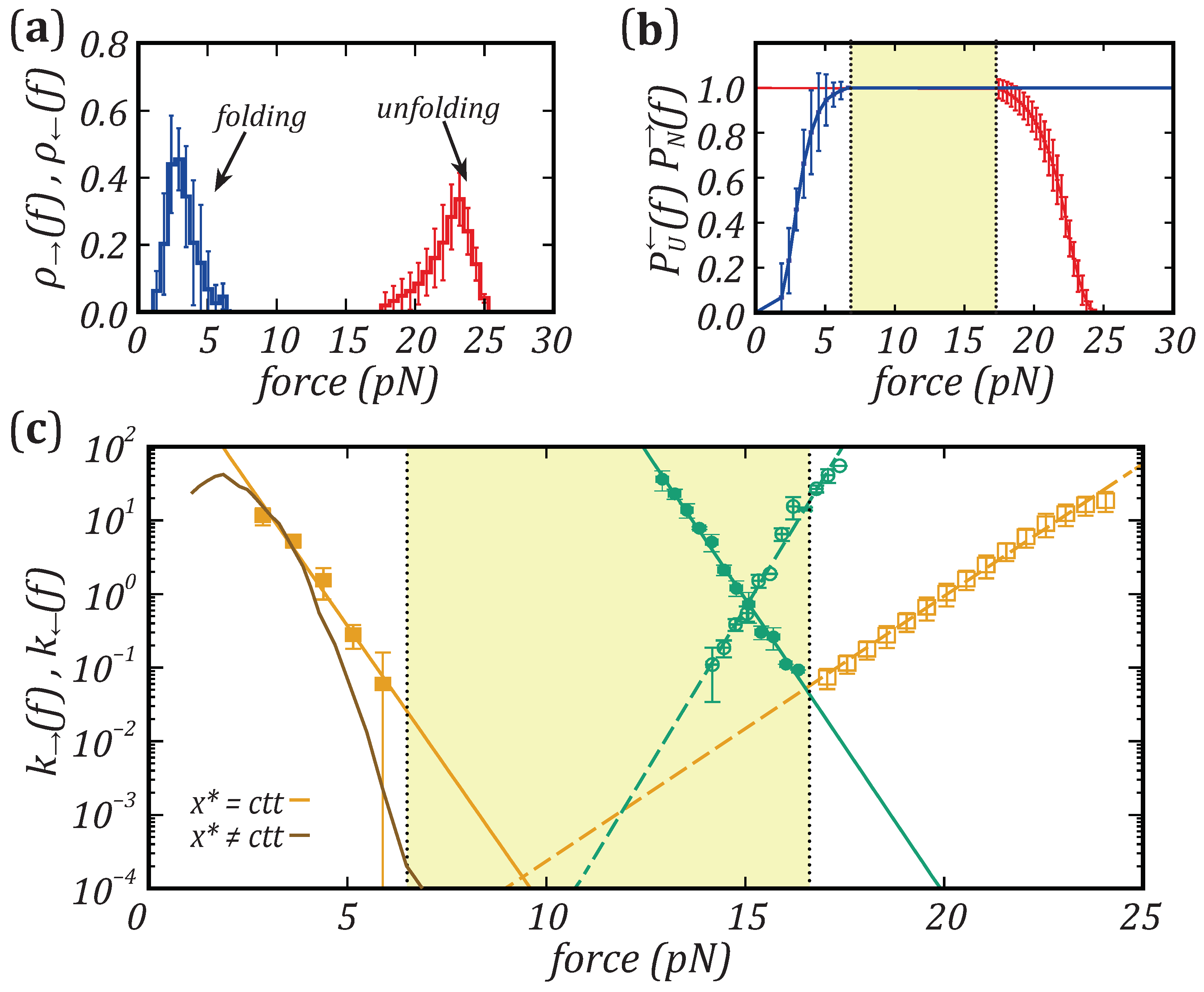
3.1. Unfolding and Folding Kinetic Rates
3.2. The Transition State Position and the Leffler–Hammond Postulate
4. Discussion
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Frauenfelder, H.; Sligar, S.; Wolynes, P. The energy landscapes and motions of proteins. Science 1991, 254, 1598–1603. [Google Scholar] [CrossRef]
- Bryngelson, J.D.; Onuchic, J.N.; Socci, N.D.; Wolynes, P.G. Funnels, pathways, and the energy landscape of protein folding: A synthesis. Proteins Struct. Funct. Bioinform. 1995, 21, 167–195. [Google Scholar] [CrossRef]
- Baldwin, R.L. The nature of protein folding pathways: The classical versus the new view. J. Biomol. NMR 1995, 5, 103–109. [Google Scholar] [CrossRef]
- Maity, H.; Maity, M.; Krishna, M.M.G.; Mayne, L.; Englander, S.W. Protein folding: The stepwise assembly of foldon units. Proc. Natl. Acad. Sci. USA 2005, 102, 4741–4746. [Google Scholar] [CrossRef]
- Camacho, C.J.; Thirumalai, D. Kinetics and thermodynamics of folding in model proteins. Proc. Natl. Acad. Sci. USA 1993, 90, 6369–6372. [Google Scholar] [CrossRef] [PubMed]
- Shakhnovich, E.I. Proteins with selected sequences fold into unique native conformation. Phys. Rev. Lett. 1994, 72, 3907–3910. [Google Scholar] [CrossRef] [PubMed]
- Chan, H.S.; Dill, K.A. Transition states and folding dynamics of proteins and heteropolymers. J. Chem. Phys. 1994, 100, 9238–9257. [Google Scholar] [CrossRef]
- Bai, Y.; Sosnick, T.; Mayne, L.; Englander, S. Protein folding intermediates: Native-state hydrogen exchange. Science 1995, 269, 192–197. [Google Scholar] [CrossRef]
- Rief, M.; Gautel, M.; Oesterhelt, F.; Fernandez, J.M.; Gaub, H.E. Reversible Unfolding of Individual Titin Immunoglobulin Domains by AFM. Science 1997, 276, 1109–1112. [Google Scholar] [CrossRef] [PubMed]
- Kellermayer, M.S.Z.; Smith, S.B.; Granzier, H.L.; Bustamante, C. Folding-Unfolding Transitions in Single Titin Molecules Characterized with Laser Tweezers. Science 1997, 276, 1112–1116. [Google Scholar] [CrossRef] [PubMed]
- Neumann, C.K.; Nagy, A. Single-molecule force spectroscopy: Optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods 2008, 5, 491–505. [Google Scholar] [CrossRef]
- Zaltron, A.; Merano, M.; Mistura, G.; Sada, C.; Seno, F. Optical tweezers in single-molecule experiments. Eur. Phys. J. Plus 2020, 135, 1–33. [Google Scholar] [CrossRef]
- Bustamante, C.; Alexander, L.; Maciuba, K.; Kaiser, C.M. Single-Molecule Studies of Protein Folding with Optical Tweezers. Annu. Rev. Biochem. 2020, 89, 443–470. [Google Scholar] [CrossRef] [PubMed]
- Gebhardt, J.C.M.; Bornschlögl, T.; Rief, M. Full distance-resolved folding energy landscape of one single protein molecule. Proc. Natl. Acad. Sci. USA 2010, 107, 2013–2018. [Google Scholar] [CrossRef] [PubMed]
- Neupane, K.; Manuel, A.P.; Woodside, M.T. Protein folding trajectories can be described quantitatively by one-dimensional diffusion over measured energy landscapes. Nat. Phys. 2016, 12, 700–703. [Google Scholar] [CrossRef]
- Rebane, A.A.; Ma, L.; Zhang, Y. Structure-Based Derivation of Protein Folding Intermediates and Energies from Optical Tweezers. Biophys. J. 2016, 110, 441–454. [Google Scholar] [CrossRef][Green Version]
- Sharma, D.; Perisic, O.; Peng, Q.; Cao, Y.; Lam, C.; Lu, H.; Li, H. Single-molecule force spectroscopy reveals a mechanically stable protein fold and the rational tuning of its mechanical stability. Proc. Natl. Acad. Sci. USA 2007, 104, 9278–9283. [Google Scholar] [CrossRef] [PubMed]
- Zheng, P.; Wang, Y.; Li, H. Reversible Unfolding–Refolding of Rubredoxin: A Single-Molecule Force Spectroscopy Study. Angew. Chem. Int. Ed. 2014, 53, 14060–14063. [Google Scholar] [CrossRef]
- Goldman, D.H.; Kaiser, C.M.; Milin, A.; Righini, M.; Tinoco, I.; Bustamante, C. Mechanical force releases nascent chain–mediated ribosome arrest in vitro and in vivo. Science 2015, 348, 457–460. [Google Scholar] [CrossRef]
- Cecconi, C.; Shank, E.A.; Bustamante, C.; Marqusee, S. Direct Observation of the Three-State Folding of a Single Protein Molecule. Science 2005, 309, 2057–2060. [Google Scholar] [CrossRef] [PubMed]
- Stigler, J.; Ziegler, F.; Gieseke, A.; Gebhardt, J.C.M.; Rief, M. The Complex Folding Network of Single Calmodulin Molecules. Science 2011, 334, 512–516. [Google Scholar] [CrossRef]
- Stockmar, F.; Kobitski, A.Y.; Nienhaus, G.U. Fast Folding Dynamics of an Intermediate State in RNase H Measured by Single-Molecule FRET. J. Phys. Chem. B 2016, 120, 641–649. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Siewny, M.G.W.; Edwards, D.T.; Sanders, A.W.; Perkins, T.T. Hidden dynamics in the unfolding of individual bacteriorhodopsin proteins. Science 2017, 355, 945–950. [Google Scholar] [CrossRef]
- Evans, E.; Ritchie, K. Dynamic strength of molecular adhesion bonds. Biophys. J. 1997, 72, 1541–1555. [Google Scholar] [CrossRef]
- Bell, G.I. Models for the specific adhesion of cells to cells. Science 1978, 200, 618–627. [Google Scholar] [CrossRef] [PubMed]
- Merkel, R.; Nassoy, P.; Leung, A.; Ritchie, K.; Evans, E. Energy landscapes of receptor–ligand bonds explored with dynamic force spectroscopy. Nature 1999, 397, 50. [Google Scholar] [CrossRef]
- Evans, E. Probing the relation between force—Lifetime—And chemistry in single molecular bonds. Annu. Rev. Biophys. Biomol. Struct. 2001, 30, 105–128. [Google Scholar] [CrossRef]
- Heidarsson, P.O.; Naqvi, M.M.; Otazo, M.R.; Mossa, A.; Kragelund, B.B.; Cecconi, C. Direct single-molecule observation of calcium-dependent misfolding in human neuronal calcium sensor-1. Proc. Natl. Acad. Sci. USA 2014, 111, 13069–13074. [Google Scholar] [CrossRef]
- Jahn, M.; Tych, K.; Girstmair, H.; Steinmaßl, M.; Hugel, T.; Buchner, J.; Rief, M. Folding and Domain Interactions of Three Orthologs of Hsp90 Studied by Single-Molecule Force Spectroscopy. Structure 2018, 26, 96–105.e4. [Google Scholar] [CrossRef]
- Mehlich, A.; Fang, J.; Pelz, B.; Li, H.; Stigler, J. Slow Transition Path Times Reveal a Complex Folding Barrier in a Designed Protein. Front. Chem. 2020, 8, 587824. [Google Scholar] [CrossRef]
- Shank, E.A.; Cecconi, C.; Dill, J.W.; Marqusee, S.; Bustamante, C. The folding cooperativity of a protein is controlled by its chain topology. Nature 2010, 465, 637–640. [Google Scholar] [CrossRef]
- Motlagh, H.N.; Toptygin, D.; Kaiser, C.M.; Hilser, V.J. Single-Molecule Chemo-Mechanical Spectroscopy Provides Structural Identity of Folding Intermediates. Biophys. J. 2016, 110, 1280–1290. [Google Scholar] [CrossRef][Green Version]
- Alemany, A.; Rey-Serra, B.; Frutos, S.; Cecconi, C.; Ritort, F. Mechanical Folding and Unfolding of Protein Barnase at the Single-Molecule Level. Biophys. J. 2016, 110, 63–74. [Google Scholar] [CrossRef]
- Liu, K.; Chen, X.; Kaiser, C.M. Energetic dependencies dictate folding mechanism in a complex protein. Proc. Natl. Acad. Sci. USA 2019, 116, 25641–25648. [Google Scholar] [CrossRef] [PubMed]
- Forns, N.; de Lorenzo, S.; Manosas, M.; Hayashi, K.; Huguet, J.; Ritort, F. Improving Signal/Noise Resolution in Single-Molecule Experiments Using Molecular Constructs with Short Handles. Biophys. J. 2011, 100, 1765–1774. [Google Scholar] [CrossRef]
- Palassini, M.; Ritort, F. Improving free-energy estimates from unidirectional work measurements: Theory and experiment. Phys. Rev. Lett. 2011, 107, 060601. [Google Scholar] [CrossRef]
- Rico-Pasto, M.; Pastor, I.; Ritort, F. Force feedback effects on single molecule hopping and pulling experiments. J. Chem. Phys. 2018, 148, 123327. [Google Scholar] [CrossRef]
- Leffler, J.E. Parameters for the description of transition states. Science 1953, 117, 340–341. [Google Scholar] [CrossRef] [PubMed]
- Hammond, G.S. A correlation of reaction rates. J. Am. Chem. Soc. 1955, 77, 334–338. [Google Scholar] [CrossRef]
- Martin, C.; Richard, V.; Salem, M.; Hartley, R.; Mauguen, Y. Refinement and structural analysis of barnase at 1.5 Å resolution. Acta Crystallogr. Sect. D 1999, 55, 386–398. [Google Scholar] [CrossRef] [PubMed]
- van der Sleen, L.M.; Tych, K.M. Bioconjugation Strategies for Connecting Proteins to DNA-Linkers for Single-Molecule Force-Based Experiments. Nanomaterials 2021, 11, 2424. [Google Scholar] [CrossRef]
- Huguet, J.M.; Bizarro, C.V.; Forns, N.; Smith, S.B.; Bustamante, C.; Ritort, F. Single-molecule derivation of salt dependent base-pair free energies in DNA. Proc. Natl. Acad. Sci. USA 2010, 107, 15431–15436. [Google Scholar] [CrossRef]
- de Lorenzo, S.; Ribezzi-Crivellari, M.; Arias-Gonzalez, J.R.; Smith, S.B.; Ritort, F. A Temperature-Jump Optical Trap for Single-Molecule Manipulation. Biophys. J. 2015, 108, 2854–2864. [Google Scholar] [CrossRef]
- Li, P.T.X.; Collin, D.; Smith, S.B.; Bustamante, C.; Tinoco, I.J. Probing the Mechanical Folding Kinetics of TAR RNA by Hopping, Force-Jump, and Force-Ramp Methods. Biophys. J. 2006, 130, 250. [Google Scholar] [CrossRef][Green Version]
- Woodside, M.T.; Behnke-Parks, W.M.; Larizadeh, K.; Travers, K.; Herschlag, D.; Block, S.M. Nanomechanical measurements of the sequence-dependent folding landscapes of single nucleic acid hairpins. Proc. Natl. Acad. Sci. USA 2006, 103, 6190–6195. [Google Scholar] [CrossRef]
- Gao, Y.; Sirinakis, G.; Zhang, Y. Highly Anisotropic Stability and Folding Kinetics of a Single Coiled Coil Protein under Mechanical Tension. J. Am. Chem. Soc. 2011, 133, 12749. [Google Scholar] [CrossRef]
- Gao, Y.; Zorman, S.; Gundersen, G.; Xi, Z.; Ma, L.; Sirinakis, G.; Rothman, J.E.; Zhang, Y. Single Reconstituted Neuronal SNARE Complexes Zipper in Three Distinct Stages. Science 2012, 337, 1340. [Google Scholar] [CrossRef]
- Yu, H.; Guptaa, A.N.; Liua, X.; Neupanea, K.; Brigleyb, A.M.; Sosovab, I.; Woodside, M.T. Energy landscape analysis of native folding of the prion protein yields the diffusion constant, transition path time, and rates. Proc. Natl. Acad. Sci. USA 2012, 109, 14452. [Google Scholar] [CrossRef]
- Hummer, G.; Szabo, A. Kinetics from Nonequilibrium Single-Molecule Pulling Experiments. Biophys. J. 2003, 85, 5–15. [Google Scholar] [CrossRef]
- Dudko, O.K.; Hummer, G.; Szabo, A. Intrinsic Rates and Activation Free Energies from Single-Molecule Pulling Experiments. Phys. Rev. Lett. 2006, 96, 108101. [Google Scholar] [CrossRef]
- Bustamante, C.; Marko, J.; Siggia, E.; Smith, S. Entropic elasticity of lambda-phage DNA. Science 1994, 265, 1599–1600. [Google Scholar] [CrossRef]
- Viader-Godoy, X.; Manosas, M.; Ritort, F. Sugar-Pucker Force-Induced Transition in Single-Stranded DNA. Int. J. Mol. Sci. 2021, 22, 4745. [Google Scholar] [CrossRef] [PubMed]
- Alemany, A.; Ritort, F. Determination of the elastic properties of short ssDNA molecules by mechanically folding and unfolding DNA hairpins. Biopolymers 2014, 101, 1193–1199. [Google Scholar] [CrossRef]
- Makarov, A.A.; Protasevich, I.I.; Kuznetsova, N.V.; Fedorov, B.B.; Korolev, S.V.; Struminskaya, N.K.; Bazhulina, N.P.; Leshchinskaya, I.B.; Hartley, R.W.; Kirpichnikov, M.P.; et al. Comparative Study of Thermostability and Structure of Close Homologues—Barnase and Binase. J. Biomol. Struct. Dyn. 1993, 10, 1047–1065. [Google Scholar] [CrossRef] [PubMed]
- Griko, Y.V.; Makhatadze, G.I.; Privalov, P.L.; Hartley, R.W. Thermodynamics of barnase unfolding. Protein Sci. 1994, 3, 669–676. [Google Scholar] [CrossRef] [PubMed]
- Galano-Frutos, J.J.; Sancho, J. Accurate Calculation of Barnase and SNase Folding Energetics Using Short Molecular Dynamics Simulations and an Atomistic Model of the Unfolded Ensemble: Evaluation of Force Fields and Water Models. J. Chem. Inf. Model. 2019, 59, 4350–4360. [Google Scholar] [CrossRef] [PubMed]
- Hyeon, C.; Thirumalai, D. Forced-Unfolding and Force-Quench Refolding of RNA Hairpins. Biophys. J. 2006, 90, 3410–3427. [Google Scholar] [CrossRef]
- Manosas, M.; Collin, D.; Ritort, F. Force-Dependent Fagility in RNA hairpins. Phys. Rev. Lett. 2006, 96, 218301. [Google Scholar] [CrossRef]
- Alemany, A.; Ritort, F. Force-Dependent Folding and Unfolding Kinetics in DNA Hairpins Reveals Transition-State Displacements along a Single Pathway. J. Phys. Chem. Lett. 2017, 8, 895–900. [Google Scholar] [CrossRef]
- Matouschek, A.; Fersht, A. Applications of physical organic chemistry to engineered mutants of barnase: Hammond postulate behaviour in the transition state of protein folding. Proc. Natl. Acad. Sci. USA 1993, 90, 7814. [Google Scholar] [CrossRef]
- Waite, J. 11-Quinone-Tanned Scleroproteins. In Metabolic Biochemistry and Molecular Biomechanics; Hochachka, P.W., Ed.; Academic Press: Cambridge, MA, USA, 1983; pp. 467–504. [Google Scholar] [CrossRef]
- Lu, Y.; Yeung, N.; Sieracki, N.; Marshall, N.M. Design of functional metalloproteins. Nature 2009, 460, 855–862. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, S.; Wu, H.C. Lipoproteins in bacteria. J. Bioenerg. Biomembr. 1990, 22, 451–471. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rico-Pasto, M.; Zaltron, A.; Ritort, F. Force Dependence of Proteins’ Transition State Position and the Bell–Evans Model. Nanomaterials 2021, 11, 3023. https://doi.org/10.3390/nano11113023
Rico-Pasto M, Zaltron A, Ritort F. Force Dependence of Proteins’ Transition State Position and the Bell–Evans Model. Nanomaterials. 2021; 11(11):3023. https://doi.org/10.3390/nano11113023
Chicago/Turabian StyleRico-Pasto, Marc, Annamaria Zaltron, and Felix Ritort. 2021. "Force Dependence of Proteins’ Transition State Position and the Bell–Evans Model" Nanomaterials 11, no. 11: 3023. https://doi.org/10.3390/nano11113023
APA StyleRico-Pasto, M., Zaltron, A., & Ritort, F. (2021). Force Dependence of Proteins’ Transition State Position and the Bell–Evans Model. Nanomaterials, 11(11), 3023. https://doi.org/10.3390/nano11113023







