Nanosurgical Manipulation of Titin and Its M-Complex
Abstract
1. Introduction
2. Materials and Methods
2.1. Titin Purification
2.2. Titin Sample Preparation for AFM
2.3. Nanosurgical Manipulation
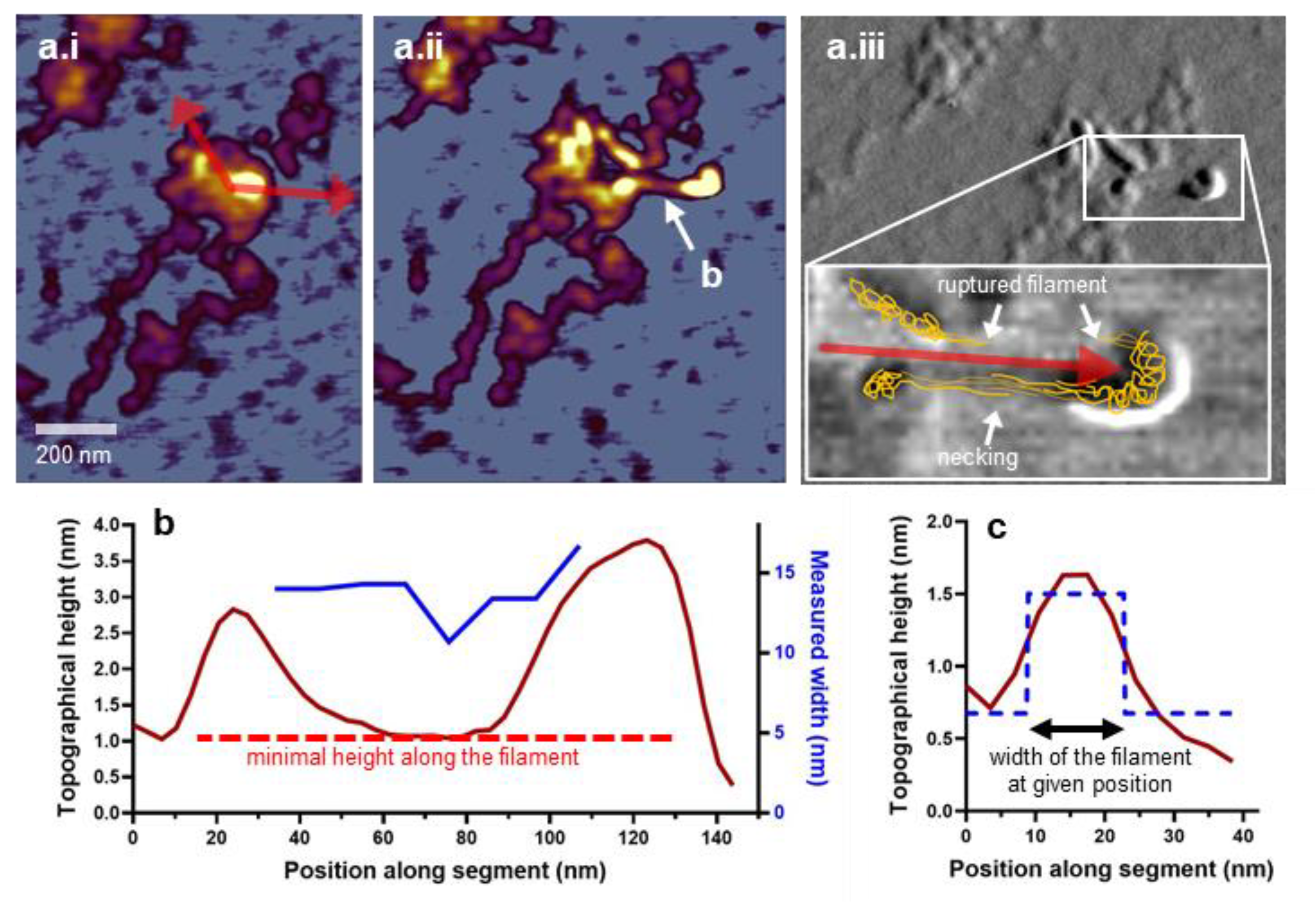
2.4. Image Analysis, Processing, and Statistics
3. Results and Discussion
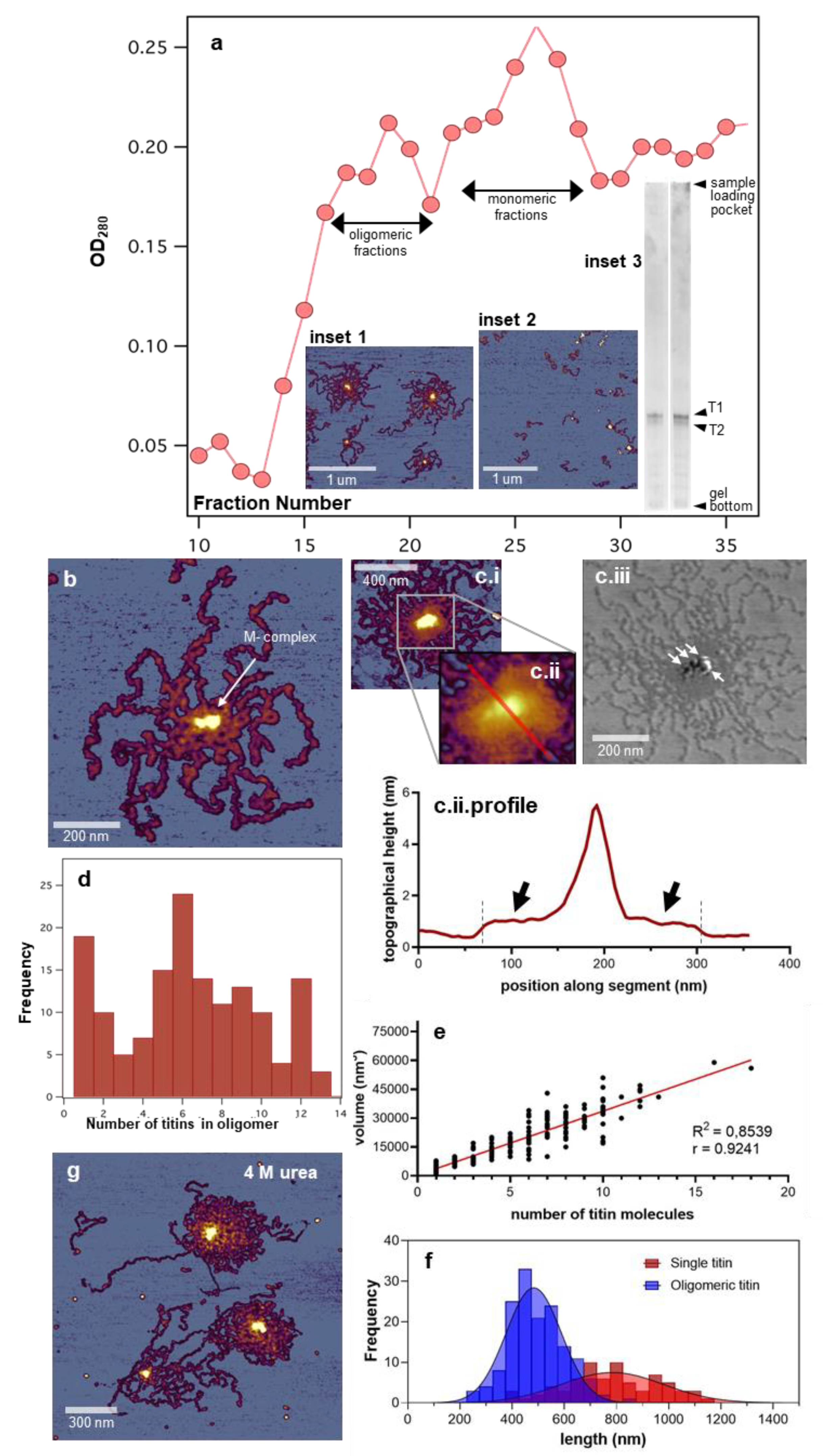
3.1. Topographical Structure, Stability, and Organization of the Titin M-Complex
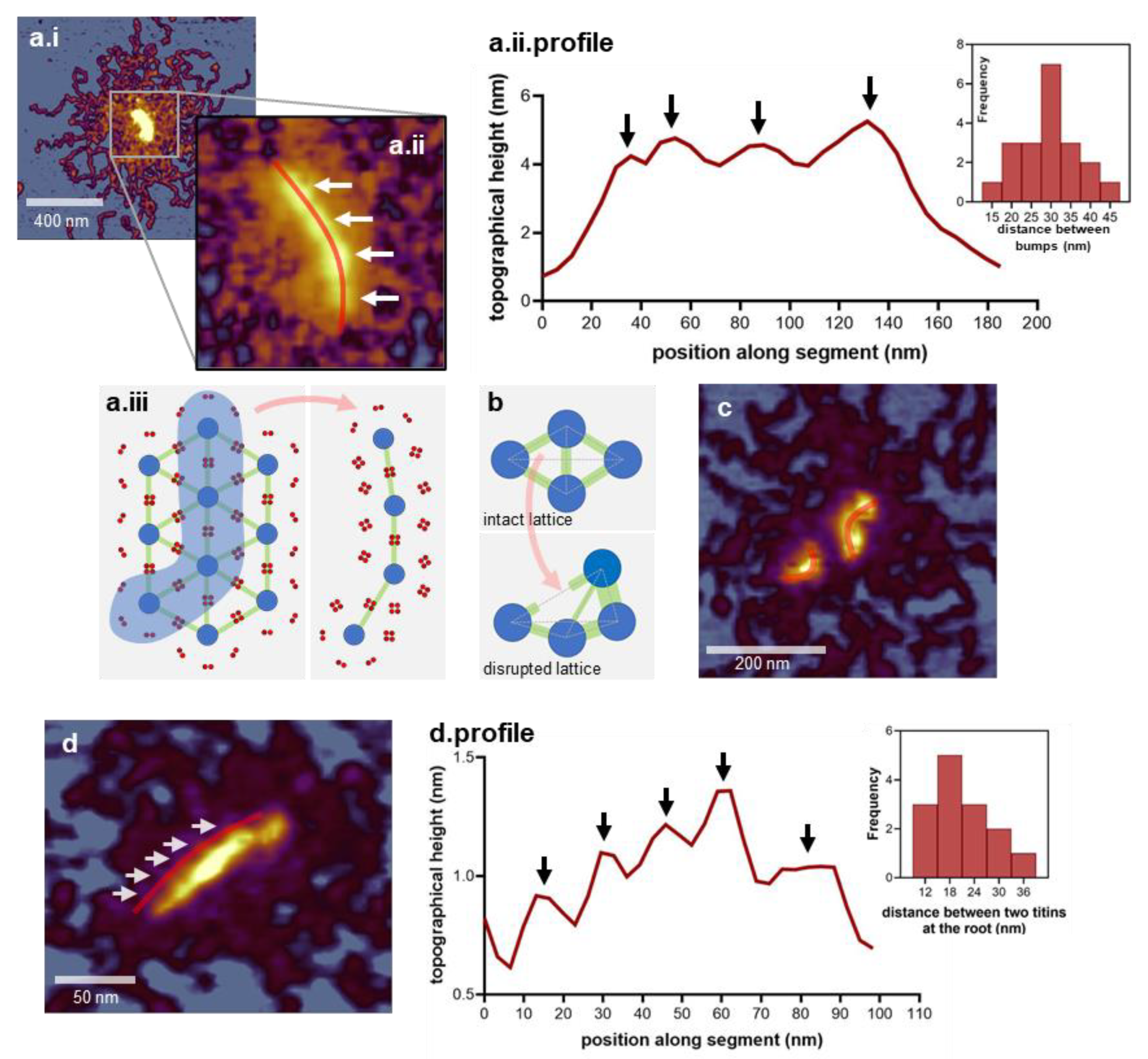
3.2. Architecture of the M-Complex
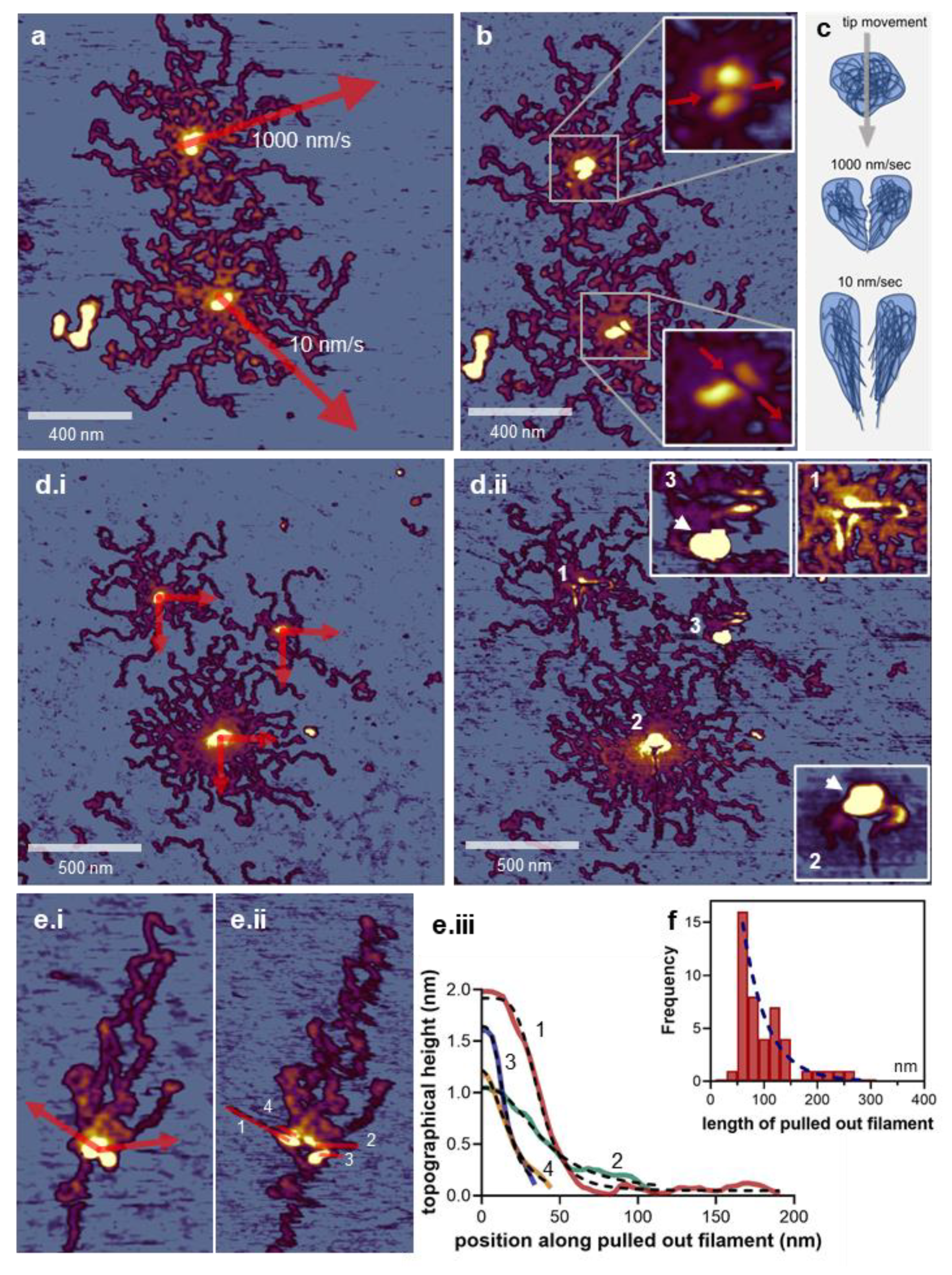
3.3. Nanosurgical Manipulation of Titin Molecules
3.4. Nanosurgical Manipulation of the M-Complex
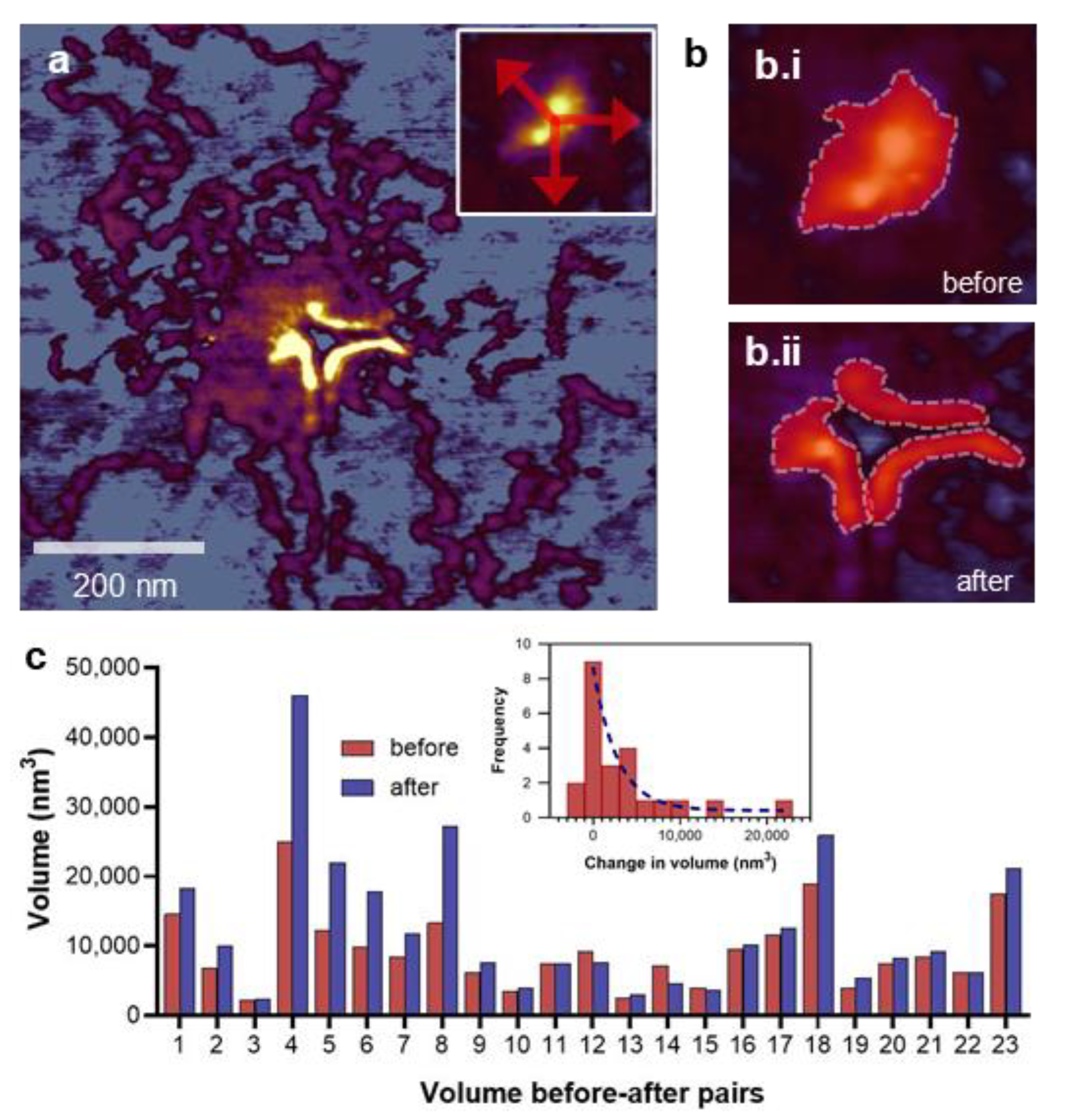
3.5. Manipulation-induced Volume Change of the M-Complex
3.6. Bulk Material Properties of the M-Complex
3.7. Nanosurgical Manipulation of the Titin Oligomer in Aqueous Buffer
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Labeit, S.; Kolmerer, B. Titins: Giant proteins in charge of muscle ultrastructure and elasticity. Science 1995, 270, 293–296. [Google Scholar] [CrossRef] [PubMed]
- Kellermayer, D.; Smith, J.E.; Granzier, H. Titin mutations and muscle disease. Pflügers Arch.-Eur. J. Physiol. 2019, 471, 673–682. [Google Scholar] [CrossRef] [PubMed]
- Linke, W.A. Titin Gene and Protein Functions in Passive and Active Muscle. Annu. Rev. Physiol. 2018, 80, 389–411. [Google Scholar] [CrossRef] [PubMed]
- Crocini, C.; Gotthardt, M. Cardiac sarcomere mechanics in health and disease. Biophys. Rev. 2021, 13, 637–652. [Google Scholar] [CrossRef] [PubMed]
- Tonino, P.; Kiss, B.; Strom, J.; Methawasin, M.; Smith, J.E.; Kolb, J.; Labeit, S.; Granzier, H. The giant protein titin regulates the length of the striated muscle thick filament. Nat. Commun. 2017, 8, 1041. [Google Scholar] [CrossRef]
- Tskhovrebova, L.; Trinick, J. Molecular rulers? Curr. Biol. 2012, 22, R317–R318. [Google Scholar] [CrossRef]
- Tskhovrebova, L.; Trinick, J. Titin and Nebulin in Thick and Thin Filament Length Regulation. Fibrous Proteins Struct. Mech. 2017, 82, 285–318. [Google Scholar] [CrossRef]
- Puchner, E.M.; Alexandrovich, A.; Kho, A.L.; Hensen, U.; Schäfer, L.V.; Brandmeier, B.; Gräter, F.; Grubmüller, H.; Gaub, H.E.; Gautel, M. Mechanoenzymatics of titin kinase. Proc. Natl. Acad. Sci. USA 2008, 105, 13385–13390. [Google Scholar] [CrossRef]
- Tskhovrebova, L.; Trinick, J. Muscle disease: A giant feels the strain. Nat. Med. 2005, 11, 478–479. [Google Scholar] [CrossRef]
- Tskhovrebova, L.; Trinick, J. Giant proteins: Sensing tension with titin kinase. Curr. Biol. 2008, 18, R1141–R1142. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bang, M.-L.; Centner, T.; Fornoff, F.; Geach, A.J.; Gotthardt, M.; McNabb, M.; Witt, C.C.; Labeit, D.; Gregorio, C.C.; Granzier, H.; et al. The Complete Gene Sequence of Titin, Expression of an Unusual ≈700-kDa Titin Isoform, and Its Interaction With Obscurin Identify a Novel Z-Line to I-Band Linking System. Circ. Res. 2001, 89, 1065–1072. [Google Scholar] [CrossRef]
- Eckels, E.C.; Haldar, S.; Tapia-Rojo, R.; Rivas-Pardo, J.A.; Fernández, J.M. The Mechanical Power of Titin Folding. Cell Rep. 2019, 27, 1836–1847. [Google Scholar] [CrossRef]
- Freundt, J.K.; Linke, W.A. Titin as a force-generating muscle protein under regulatory control. J. Appl. Physiol. 2019, 126, 1474–1482. [Google Scholar] [CrossRef]
- Mártonfalvi, Z.; Bianco, P.; Naftz, K.; Ferenczy, G.; Kellermayer, M. Force generation by titin folding. Protein Sci. 2017, 26, 1380–1390. [Google Scholar] [CrossRef]
- Agarkova, I.; Perriard, J.C. The M-band: An elastic web that crosslinks thick filaments in the center of the sarcomere. Trends Cell Biol. 2005, 15, 477–485. [Google Scholar] [CrossRef] [PubMed]
- Vinkemeier, U.; Obermann, W.M.; Weber, K.; Fürst, D.O. The globular head domain of titin extends into the center of the sarcomeric M band. cDNA cloning, epitope mapping and immunoelectron microscopy of two titin-associated proteins. J. Cell Sci. 1993, 106, 319–330. [Google Scholar] [CrossRef] [PubMed]
- Nave, R.; Fürst, D.O.; Weber, K. Visualization of the polarity of isolated titin molecules: A single globular head on a long thin rod as the M band anchoring domain? J. Cell Biol. 1989, 109, 2177–2187. [Google Scholar] [CrossRef]
- Lange, S.; Pinotsis, N.; Agarkova, I.; Ehler, E. The M-band: The underestimated part of the sarcomere. Biochim. Biophys. Acta-Mol. Cell Res. 2020, 1867, 118440. [Google Scholar] [CrossRef] [PubMed]
- Knappeis, G.G.; Carlsen, F. The ultrastructure of the M line in skeletal muscle. J. Cell Biol. 1968, 38, 202–211. [Google Scholar] [CrossRef]
- Luther, P.; Squire, J. Three-dimensional structure of the vertebrate muscle M-region. J. Mol. Biol. 1978, 125, 313–324. [Google Scholar] [CrossRef]
- Obermann, W.M.; Gautel, M.; Steiner, F.; van der Ven, P.F.; Weber, K.; Fürst, D.O. The structure of the sarcomeric M band: Localization of defined domains of myomesin, M-protein, and the 250-kD carboxy-terminal region of titin by immunoelectron microscopy. J. Cell Biol. 1996, 134, 1441–1453. [Google Scholar] [CrossRef] [PubMed]
- Lange, S.; Himmel, M.; Auerbach, D.; Agarkova, I.; Hayess, K.; Fürst, D.O.; Perriard, J.-C.; Ehler, E. Dimerisation of Myomesin: Implications for the Structure of the Sarcomeric M-band. J. Mol. Biol. 2005, 345, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Pernigo, S.; Fukuzawa, A.; Bertz, M.; Holt, M.; Rief, M.; Steiner, R.A.; Gautel, M. Structural insight into M-band assembly and mechanics from the titin-obscurin-like-1 complex. Proc. Natl. Acad. Sci. USA 2010, 107, 2908–2913. [Google Scholar] [CrossRef] [PubMed]
- Szikora, S.; Gajdos, T.; Novák, T.; Farkas, D.; Földi, I.; Lenart, P.; Erdélyi, M.; Mihály, J. Nanoscopy reveals the layered organization of the sarcomeric H-zone and I-band complexes. J. Cell Biol. 2019, 219, e201907026. [Google Scholar] [CrossRef]
- Pinotsis, N.; Chatziefthimiou, S.D.; Berkemeier, F.; Beuron, F.; Mavridis, I.M.; Konarev, P.V.; Svergun, D.I.; Morris, E.; Rief, M.; Wilmanns, M. Superhelical Architecture of the Myosin Filament-Linking Protein Myomesin with Unusual Elastic Properties. PLoS Biol. 2012, 10, e1001261. [Google Scholar] [CrossRef]
- Xiao, S.; Gräter, F. Molecular basis of the mechanical hierarchy in myomesin dimers for sarcomere integrity. Biophys. J. 2014, 107, 965–973. [Google Scholar] [CrossRef]
- Freitas, R.A., Jr. Nanotechnology, nanomedicine and nanosurgery. Int. J. Surg. 2005, 3, 243–246. [Google Scholar] [CrossRef]
- Uchugonova, A.; Lessel, M.; Nietzsche, S.; Zeitz, C.; Jacobs, K.; Lemke, C.; König, K. Nanosurgery of cells and chromosomes using near-infrared twelve-femtosecond laser pulses. J. BioMed. Opt. 2012, 17, 101502. [Google Scholar] [CrossRef]
- Müller, D.; Hagenah, D.; Biswanath, S.; Coffee, M.; Kampmann, A.; Zweigerdt, R.; Heisterkamp, A.; Kalies, S.M.K. Femtosecond laser-based nanosurgery reveals the endogenous regeneration of single Z-discs including physiological consequences for cardiomyocytes. Sci. Rep. 2019, 9, 3625. [Google Scholar] [CrossRef]
- Eversole, D.; Subramanian, K.; Harrison, R.K.; Bourgeois, F.; Yuksel, A.; Ben-Yakar, A. Femtosecond Plasmonic Laser Nanosurgery (fs-PLN) mediated by molecularly targeted gold nanospheres at ultra-low pulse fluences. Sci. Rep. 2020, 10, 12387. [Google Scholar] [CrossRef]
- Blázquez-Castro, A.; Fernández-Piqueras, J.; Santos, J. Genetic Material Manipulation and Modification by Optical Trapping and Nanosurgery-A Perspective. Front. Bioeng. Biotechnol. 2020, 8, 580937. [Google Scholar] [CrossRef]
- Fotiadis, D.; Scheuring, S.; Muller, S.A.; Engel, A.; Muller, D.J. Imaging and manipulation of biological structures with the AFM. Micron 2002, 33, 385–397. [Google Scholar] [CrossRef]
- Lü, J.H. Nanomanipulation of extended single-DNA molecules on modified mica surfaces using the atomic force microscopy. Colloids Surf. B Biointerfaces 2004, 39, 177–180. [Google Scholar] [CrossRef]
- Geisler, B.; Noll, F.; Hampp, N. Nanodissection and noncontact imaging of plasmid DNA with an atomic force microscope. Scanning 2000, 22, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Henderson, E. Imaging and nanodissection of individual supercoiled plasmids by atomic force microscopy. Nucleic Acids Res. 1992, 20, 445–447. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Di Bucchianico, S.; Poma, A.M.; Giardi, M.F.; Di Leandro, L.; Valle, F.; Biscarini, F.; Botti, D. Atomic Force Microscope nanolithography on chromosomes to generate single-cell genetic probes. J. Nanobiotechnol. 2011, 9, 27. [Google Scholar] [CrossRef] [PubMed]
- Hakari, T.; Sekiguchi, H.; Osada, T.; Kishimoto, K.; Afrin, R.; Ikai, A. Nonlinear displacement of ventral stress fibers under externally applied lateral force by an atomic force microscope. Cytoskeleton 2011, 68, 628–638. [Google Scholar] [CrossRef]
- Wen, C.K.; Goh, M.C. Fibrous long spacing type collagen fibrils have a hierarchical internal structure. Proteins Struct. Funct. Bioinform. 2006, 64, 227–233. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Song, B.; Sun, Z.; Lai, K.W.C.; Fung, C.K.M.; Patterson, K.C.; Seiffert-Sinha, K.; Sinha, A.A.; Xi, N. Cellular level robotic surgery: Nanodissection of intermediate filaments in live keratinocytes. Nanomed. Nanotechnol. Biol. Med. 2015, 11, 137–145. [Google Scholar] [CrossRef]
- Mahdjour Firouzi, M.A.; Nejat Pishkenari, H.; Mahboobi, S.H.; Meghdari, A. Manipulation of biomolecules: A molecular dynamics study. Curr. Appl. 2014, 14, 1216–1227. [Google Scholar] [CrossRef]
- Lea, A.S.; Pungor, A.; Hlady, V.; Andrade, J.D.; Herron, J.N.; Voss, E.W., Jr. Manipulation of Proteins on Mica by Atomic Force Microscopy. Langmuir 1992, 8, 68–73. [Google Scholar] [CrossRef]
- Kheirodin, M.; Pishkenari, H.N.; Mahboobi, S.H. Molecular dynamics study of bio-manipulation in aqueous media. Micro Nano Lett. 2016, 11, 9–14. [Google Scholar] [CrossRef]
- Kreplak, L.; Bär, H.; Leterrier, J.F.; Herrmann, H.; Aebi, U. Exploring the Mechanical Behavior of Single Intermediate Filaments. J. Mol. Biol. 2005, 354, 569–577. [Google Scholar] [CrossRef] [PubMed]
- Kellermayer, M.S.; Smith, S.B.; Granzier, H.L.; Bustamante, C. Folding-unfolding transitions in single titin molecules characterized with laser tweezers. Science 1997, 276, 1112–1116. [Google Scholar] [CrossRef]
- Soteriou, A.; Gamage, M.; Trinick, J. A survey of interactions made by the giant protein titin. J. Cell Sci. 1993, 104 Pt 1, 119–123. [Google Scholar] [CrossRef]
- Warren, C.M.; Krzesinski, P.R.; Greaser, M.L. Vertical agarose gel electrophoresis and electroblotting of high-molecular-weight proteins. Electrophoresis 2003, 24, 1695–1702. [Google Scholar] [CrossRef]
- Kellermayer, M.; Sziklai, D.; Papp, Z.; Decker, B.; Lakatos, E.; Mártonfalvi, Z. Topology of interaction between titin and myosin thick filaments. J. Struct. Biol. 2018, 203, 46–53. [Google Scholar] [CrossRef]
- Mártonfalvi, Z.; Kellermayer, M. Individual globular domains and domain unfolding visualized in overstretched titin molecules with atomic force microscopy. PLoS ONE 2014, 9, e85847. [Google Scholar] [CrossRef]
- Schoenauer, R.; Lange, S.; Hirschy, A.; Ehler, E.; Perriard, J.C.; Agarkova, I. Myomesin 3, a Novel Structural Component of the M-band in Striated Muscle. J. Mol. Biol. 2008, 376, 338–351. [Google Scholar] [CrossRef] [PubMed]
- Fukuzawa, A.; Lange, S.; Holt, M.; Vihola, A.; Carmignac, V.; Ferreiro, A.; Udd, B.; Gautel, M. Interactions with titin and myomesin target obscurin and obscurin-like 1 to the M-band—Implications for hereditary myopathies. J. Cell Sci. 2008, 121, 1841–1851. [Google Scholar] [CrossRef] [PubMed]
- Kellermayer, M.S.Z.; Bustamante, C.; Granzier, H.L. Mechanics and structure of titin oligomers explored with atomic force microscopy. Biochim. Biophys. Acta-Bioenerg. 2003, 1604, 105–114. [Google Scholar] [CrossRef]
- Tskhovrebova, L.; Trinick, J. Flexibility and Extensibility in the Titin Molecule: Analysis of Electron Microscope Data. J. Mol. Biol. 2001, 310, 755–771. [Google Scholar] [CrossRef]
- Hu, L.-Y.R.; Ackermann, M.A.; Kontrogianni-Konstantopoulos, A. The Sarcomeric M-Region: A Molecular Command Center for Diverse Cellular Processes. BioMed. Res. Int. 2015, 2015, 714197. [Google Scholar] [CrossRef] [PubMed]
- Al-Khayat, H.A.; Kensler, R.W.; Squire, J.; Marston, S.B.; Morris, E. Atomic model of the human cardiac muscle myosin filament. Proc. Natl. Acad. Sci. USA 2013, 110, 318–323. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, N.; Wu, Y.; Farman, G.; Irving, T.C.; Granzier, H.L. Titin-based modulation of active tension and interfilament lattice spacing in skinned rat cardiac muscle. Eur. J. Physiol. 2005, 449, 449–457. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lang, P.; Linke, W.A. Titin stiffness modifies the force-generating region of muscle sarcomeres. Sci. Rep. 2016, 6, 24492. [Google Scholar] [CrossRef] [PubMed]
- Millman, B.M.; Irving, T.C. Filament lattice of frog striated muscle. Biophys. J. 1988, 54, 437–447. [Google Scholar] [CrossRef]
- Berkemeier, F.; Bertz, M.; Xiao, S.; Pinotsis, N.; Willmans, M.; Grater, F.; Rief, M. Fast-folding α-helices as reversible strain absorbers in the muscle protein myomesin. Proc. Natl. Acad. Sci. USA 2011, 108, 14139–14144. [Google Scholar] [CrossRef]
- Tskhovrebova, L.; Trinick, J. Making muscle elastic: The structurl basis of myomesin stretching. PLoS Biol. 2012, 10, e1001264. [Google Scholar] [CrossRef]
- Horowits, R.; Podolsky, R.J. The positional stability of thick filaments in activated skeletal muscle depends on sarcomere length: Evidence for the role of titin filaments. J. Cell Biol. 1987, 105, 2217–2223. [Google Scholar] [CrossRef]
- Buehler, M.J.; Ackbarow, T. Fracture mechanics of protein materials. Mater. Today 2007, 10, 46–58. [Google Scholar] [CrossRef]
- Hsin, J.; Strümpfer, J.; Lee, E.H.; Schulten, K. Molecular origin of the hierarchical elasticity of titin: Simulation, experiment, and theory. Annu. Rev. Biophys. 2011, 40, 187–203. [Google Scholar] [CrossRef] [PubMed]
- Pfuhl, M.; Pastore, A. Tertiary structure of an immunoglobulin-like domain from the giant muscle protein titin: A new member of the I set. Structure 1995, 3, 391–401. [Google Scholar] [CrossRef]
- Rief, M.; Gautel, M.; Oesterhelt, F.; Fernandez, J.M.; Gaub, H.E. Reversible Unfolding of Individual Titin Immunoglobulin Domains by AFM. Science 1997, 276, 1109–1112. [Google Scholar] [CrossRef]
- Agarkova, I.; Ehler, E.; Lange, S.; Schoenauer, R.; Perriard, J.C. M-band: A safeguard for sarcomere stability? J. Muscle Res. Cell Motil. 2003, 24, 191–203. [Google Scholar] [CrossRef] [PubMed]
- Michler, G.H.; Baltá-Calleja, F.J. General Mechanisms. In Nano- and Micromechanics of Polymers; Michler, G.H., Baltá-Calleja, F.J., Eds.; Carl Hanser Verlag: Munich, Germany, 2012; pp. 95–117. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sziklai, D.; Sallai, J.; Papp, Z.; Kellermayer, D.; Mártonfalvi, Z.; Pires, R.H.; Kellermayer, M.S.Z. Nanosurgical Manipulation of Titin and Its M-Complex. Nanomaterials 2022, 12, 178. https://doi.org/10.3390/nano12020178
Sziklai D, Sallai J, Papp Z, Kellermayer D, Mártonfalvi Z, Pires RH, Kellermayer MSZ. Nanosurgical Manipulation of Titin and Its M-Complex. Nanomaterials. 2022; 12(2):178. https://doi.org/10.3390/nano12020178
Chicago/Turabian StyleSziklai, Dominik, Judit Sallai, Zsombor Papp, Dalma Kellermayer, Zsolt Mártonfalvi, Ricardo H. Pires, and Miklós S. Z. Kellermayer. 2022. "Nanosurgical Manipulation of Titin and Its M-Complex" Nanomaterials 12, no. 2: 178. https://doi.org/10.3390/nano12020178
APA StyleSziklai, D., Sallai, J., Papp, Z., Kellermayer, D., Mártonfalvi, Z., Pires, R. H., & Kellermayer, M. S. Z. (2022). Nanosurgical Manipulation of Titin and Its M-Complex. Nanomaterials, 12(2), 178. https://doi.org/10.3390/nano12020178






